Tuberous Sclerosis Complex Axis Controls Renal Extracellular Vesicle Production and Protein Content
Abstract
:1. Introduction
2. Results
2.1. Renal Collecting Duct Cells Produce EVs In Vitro
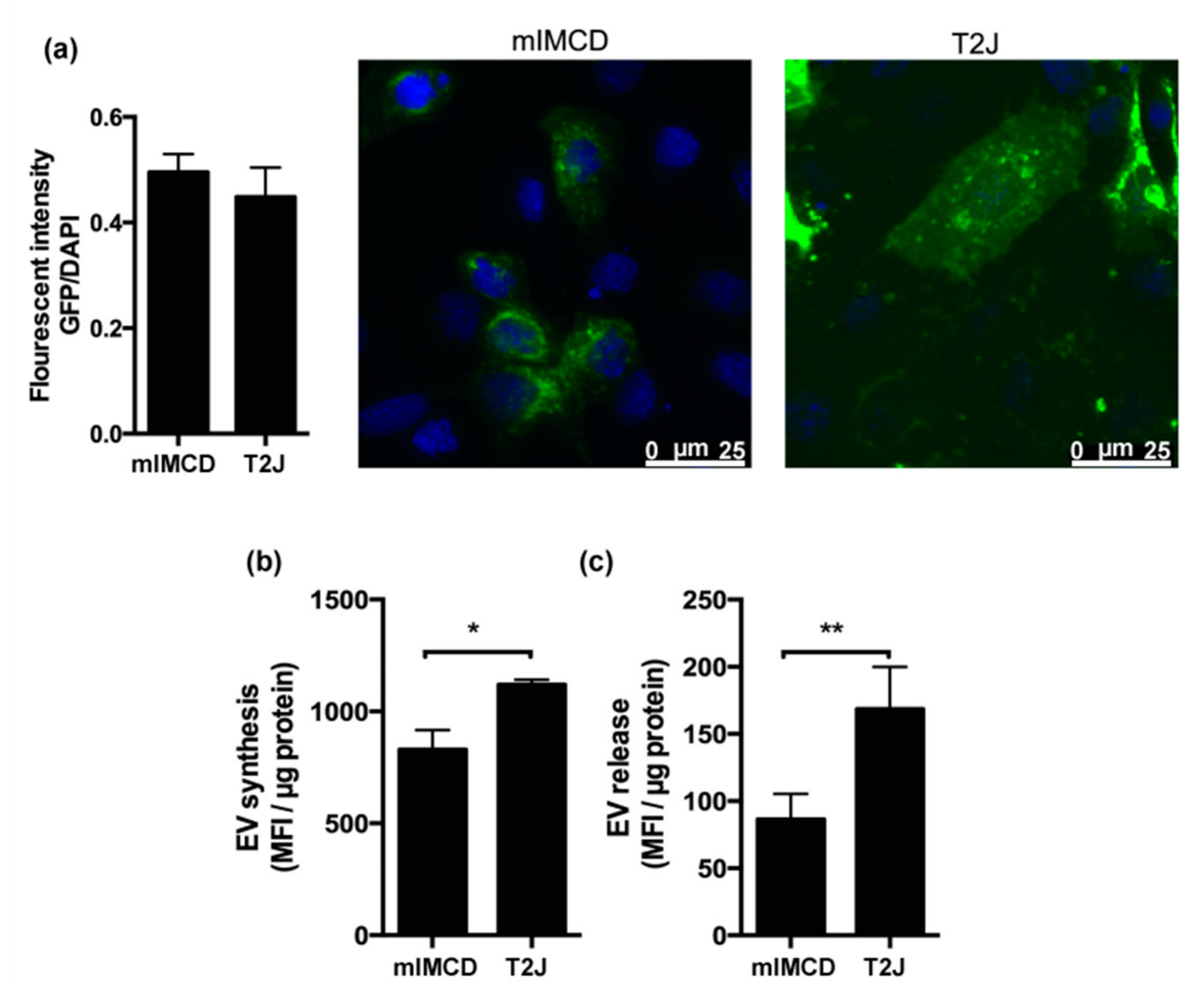
2.2. Loss of Tsc2 Increases the Production of EVs
2.3. Proteomic Analysis of EVs
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Isolation of EVs by Size Exclusion Chromatography (SEC)
4.3. Characterizations of EVs
4.4. Protein Isolation and Western Blot
4.5. EV Synthesis and Release
4.6. EV Mass Spectrometry (LC/MSMS) and Proteomics Analysis
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bissler, J.J.; Christopher Kingswood, J. Renal manifestation of tuberous sclerosis complex. Am. J. Med. Genet. Part C Semin. Med. Genet. 2018, 178, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Ito, N.; Rubin, G.M. Gigas, a Drosophila homolog of tuberous sclerosis gene product-2, regulates the cell cycle. Cell 1999, 96, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Land, S.C.; Scott, C.L.; Walker, D. MTOR signalling, embryogenesis and the control of lung development. Semin. Cell Dev. Biol. 2014, 36, 68–78. [Google Scholar]
- Tian, Y.; Morrisey, E.E. Kidney Development and Disease. In Results and Problems in Cell Differentiation; Miller, R.K., Ed.; Springer International Publishing: Cham, Germany, 2017; Volume 60, ISBN 978-3-319-51435-2. [Google Scholar]
- Stillwell, T.J.; Gomez, M.R.; Kelalis, P.P. Renal lesions in tuberous sclerosis. J. Urol. 1987, 138, 477–481. [Google Scholar] [CrossRef]
- Bissler, J.J.; Kingswood, J.C. Optimal treatment of tuberous sclerosis complex associated renal angiomyolipomata: A systematic review. Ther. Adv. Urol. 2016, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Lam, H.C.; Nijmeh, J.; Henske, E.P. New developments in the genetics and pathogenesis of tumours in tuberous sclerosis complex. J. Pathol. 2017, 241, 219–225. [Google Scholar] [CrossRef] [Green Version]
- Giannikou, K.; Malinowska, I.A.; Pugh, T.J.; Yan, R.; Tseng, Y.Y.; Oh, C.; Kim, J.; Tyburczy, M.E.; Chekaluk, Y.; Liu, Y.; et al. Whole Exome Sequencing Identifies TSC1/TSC2 Biallelic Loss as the Primary and Sufficient Driver Event for Renal Angiomyolipoma Development. PLoS Genet. 2016, 12, e1006242. [Google Scholar] [CrossRef] [Green Version]
- Onda, H.; Lueck, A.; Marks, P.W.; Warren, H.B.; Kwiatkowski, D.J. Tsc2(+/-) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J. Clin. Invest. 1999, 104, 687–695. [Google Scholar] [CrossRef] [Green Version]
- Wilson, C.; Bonnet, C.; Guy, C.; Idziaszczyk, S.; Colley, J.; Humphreys, V.; Maynard, J.; Sampson, J.R.; Cheadle, J.P. Tsc1 haploinsufficiency without mammalian target of rapamycin activation is sufficient for renal cyst formation in Tsc1+/- mice. Cancer Res. 2006, 66, 7934–7938. [Google Scholar] [CrossRef] [Green Version]
- Bonsib, S.M.; Boils, C.; Gokden, N.; Grignon, D.; Gu, X.; Higgins, J.P.T.; Leroy, X.; McKenney, J.K.; Nasr, S.H.; Phillips, C.; et al. Tuberous sclerosis complex: Hamartin and tuberin expression in renal cysts and its discordant expression in renal neoplasms. Pathol. Res. Pract. 2016, 212, 972–979. [Google Scholar] [CrossRef] [Green Version]
- Badenas, C.; Torra, R.; Pérez-Oller, L.; Mallolas, J.; Talbot-Wright, R.; Torregrosa, V.; Darnell, A. Loss of heterozygosity in renal and hepatic epithelial cystic cells from ADPKD1 patients. Eur. J. Hum. Genet. 2000, 8, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Brasier, J.L.; Henske, E.P. Loss of the polycystic kidney disease (PKD1) region of chromosome 16p13 in renal cyst cells supports a loss-of-function model for cyst pathogenesis. J. Clin. Invest. 1997, 99, 194–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bissler, J.J.; Zadjali, F.; Bridges, D.; Astrinidis, A.; Barone, S.; Yao, Y.; Redd, J.A.R.; Siroky, B.J.; Wang, Y.; Finley, J.T.; et al. Tuberous sclerosis complex exhibits a new renal cystogenic mechanism. Physiol. Rep. 2019, 7, e13983. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.; Patel, J.; Cho, J.-H.; Manne, S.; Bonala, S.; Henske, E.; Roegiers, F.; Markiewski, M.; Karbowniczek, M. Exosomes mediate the acquisition of the disease phenotypes by cells with normal genome in tuberous sclerosis complex. Oncogene 2015, 35, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rikkert, L.G.; Nieuwland, R.; Terstappen, L.W.M.M.; Coumans, F.A.W. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracell. Vesicles 2019, 8, 1–10. [Google Scholar] [CrossRef]
- Garnier, D.; Magnus, N.; Meehan, B.; Kislinger, T.; Rak, J. Qualitative changes in the proteome of extracellular vesicles accompanying cancer cell transition to mesenchymal state. Exp. Cell Res. 2013, 319, 2747–2757. [Google Scholar] [CrossRef] [Green Version]
- Hallal, S.; Russell, B.P.; Wei, H.; Lee, M.Y.T.; Toon, C.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Kaufman, K.L. Extracellular Vesicles from Neurosurgical Aspirates Identifies Chaperonin Containing TCP1 Subunit 6A as a Potential Glioblastoma Biomarker with Prognostic Significance. Proteomics 2019, 19, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.E.; Moon, P.G.; Lee, I.K.; Baek, M.C. Proteomic Analysis of Extracellular Vesicles Released by Adipocytes of Otsuka Long-Evans Tokushima Fatty (OLETF) Rats. Protein J. 2015, 34, 220–235. [Google Scholar] [CrossRef]
- Dixon, B.P.B.P.P.; Hulbert, J.C.J.C.; Bissler, J.J.J. Tuberous sclerosis complex renal disease. Nephron. Exp. Nephrol. 2011, 118, e15–e20. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen Wen, S.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Böing, A.N.; Van Der Pol, E.; Grootemaat, A.E.; Coumans, F.A.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles from plasma by size-exclusion chromatography. Int. Meet. ISEV Rotterdam 2014, 3, 118. [Google Scholar]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. 2016, 113, E968–E977. [Google Scholar] [CrossRef] [Green Version]
- McAndrews, K.M.; Kalluri, R. Mechanisms associated with biogenesis of exosomes in cancer. Mol. Cancer 2019, 18, 1–11. [Google Scholar] [CrossRef]
- Hor, C.H.; Goh, E.L. Small GTPases in hedgehog signalling: Emerging insights into the disease mechanisms of Rab23-mediated and Arl13b-mediated ciliopathies. Curr. Opin. Genet. Dev. 2019, 56, 61–68. [Google Scholar] [CrossRef]
- Higginbotham, H.; Eom, T.Y.; Mariani, L.E.; Bachleda, A.; Hirt, J.; Gukassyan, V.; Cusack, C.L.; Lai, C.; Caspary, T.; Anton, E.S. Arl13b in Primary Cilia Regulates the Migration and Placement of Interneurons in the Developing Cerebral Cortex. Dev. Cell 2012, 23, 925–938. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.M.; Jacobs, D.T.; Allard, B.A.; Fields, T.A.; Sharma, M.; Wallace, D.P.; Tran, P.V. Inhibition of Hedgehog signaling suppresses proliferation and microcyst formation of human Autosomal Dominant Polycystic Kidney Disease cells. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Bhuria, V.; Xing, J.; Scholta, T.; Bui, K.C.; Nguyen, M.L.T.; Malek, N.P.; Bozko, P.; Plentz, R.R. Hypoxia induced Sonic Hedgehog signaling regulates cancer stemness, epithelial-to-mesenchymal transition and invasion in cholangiocarcinoma. Exp. Cell Res. 2019, 385, 111671. [Google Scholar] [CrossRef]
- Zonneveld, M.I.; Keulers, T.G.H.; Rouschop, K.M.A. Extracellular vesicles as transmitters of hypoxia tolerance in solid cancers. Cancers (Basel). 2019, 11, 154. [Google Scholar] [CrossRef] [Green Version]
- Evans, R.G.; Smith, J.A.; Wright, C.; Gardiner, B.S.; Smith, D.W.; Cochrane, A.D. Urinary oxygen tension: A clinical window on the health of the renal medulla? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 306, 45–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witzgall, R.; Brown, D.; Schwarz, C.; Bonventre, J.V. Localization of proliferating cell nuclear antigen, vimentin, c-Fos, and clusterin in the postischemic kidney. Evidence for a heterogenous genetic response among nephron segments, and a large pool of mitotically active and dedifferentiated cells. J. Clin. Invest. 1994, 93, 2175–2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Yang, Y.; Huang, Y.; Zhang, L.; Ling, Z.; Zhu, Y.; Wang, F.; Zou, X.; Chen, M. Hypoxia-induced extracellular vesicles mediate protection of remote ischemic preconditioning for renal ischemia-reperfusion injury. Biomed. Pharmacother. 2017, 90, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, U.; Ozcan, L.; Yilmaz, E.; Düvel, K.; Sahin, M.; Manning, B.D.; Hotamisligil, G.S. Loss of the Tuberous Sclerosis Complex Tumor Suppressors Triggers the Unfolded Protein Response to Regulate Insulin Signaling and Apoptosis. Mol. Cell 2008, 29, 541–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siroky, B.J.; Yin, H.; Babcock, J.T.; Lu, L.; Hellmann, A.R.; Dixon, B.P.; Quilliam, L.A.; Bissler, J.J. Human TSC-associated renal angiomyolipoma cells are hypersensitive to ER stress. AJP Ren. Physiol. 2012, 303, F831–F844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanemoto, S.; Nitani, R.; Murakami, T.; Kaneko, M.; Asada, R.; Matsuhisa, K.; Saito, A.; Imaizumi, K. Multivesicular body formation enhancement and exosome release during endoplasmic reticulum stress. Biochem. Biophys. Res. Commun. 2016, 480, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Toschi, A.; Lee, E.; Gadi, N.; Ohh, M.; Foster, D.A. Differential dependence of hypoxia-inducible factors 1α and 2α on mTORC1 and mTORC2. J. Biol. Chem. 2008, 283, 34495–34499. [Google Scholar] [CrossRef] [Green Version]
- Dodd, K.M.; Yang, J.; Shen, M.H.; Sampson, J.R.; Tee, A.R. mTORC1 drives HIF-1α and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene 2015, 34, 2239–2250. [Google Scholar] [CrossRef] [Green Version]
- Kilic, T.; Valinhas, A.T.D.S.; Wall, I.; Renaud, P.; Carrara, S. Label-free detection of hypoxia-induced extracellular vesicle secretion from MCF-7 cells. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Shima, N.; Alcaraz, A.; Liachko, I.; Buske, T.R.; Andrews, C.A.; Munroe, R.J.; Hartford, S.A.; Tye, B.K.; Schimenti, J.C. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 2007, 39, 93–98. [Google Scholar] [CrossRef]
- Bochman, M.L.; Schwacha, A. The Mcm Complex: Unwinding the Mechanism of a Replicative Helicase. Microbiol. Mol. Biol. Rev. 2009, 73, 652–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bissler, J.J.; McCormack, F.X.; Young, L.R.; Elwing, J.M.; Chuck, G.; Leonard, J.M.; Schmithorst, V.J.; Laor, T.; Brody, A.S.; Bean, J.; et al. Sirolimus for Angiomyolipoma in Tuberous Sclerosis Complex or Lymphangioleiomyomatosis. N. Engl. J. Med. 2008, 358, 140–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bissler, J.J.; Kingswood, J.C.; Radzikowska, E.; Zonnenberg, B.A.B.A.; Frost, M.; Belousova, E.; Sauter, M.; Nonomura, N.; Brakemeier, S.; de Vries, P.J.P.J.; et al. Everolimus for Angiomyolipoma Associated With Tuberous Sclerosis Complex or Sporadic Lymphangioleiomyomatosis (EXIST-2): A Multicentre, Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 2013, 381, 817–824. [Google Scholar] [CrossRef]
- Govindarajan, B.; Willoughby, L.; Band, H.; Curatolo, A.S.; Veledar, E.; Chen, S.; Bonner, M.Y.; Abel, M.-G.; Moses, M.A.; Arbiser, J.L. Cooperative benefit for the combination of rapamycin and imatinib in tuberous sclerosis complex neoplasia. Vasc. Cell 2012, 4, 11. [Google Scholar] [CrossRef] [Green Version]
- Mineo, M.; Garfield, S.H.; Taverna, S.; Flugy, A.; De Leo, G.; Alessandro, R.; Kohn, E.C. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a src-dependent fashion. Angiogenesis 2012, 15, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Kaunitz, J.D.; Cummins, V.P.S.; Mishler, D.; Nagami, G.T. Inhibition of Gentamicin Uptake into Cultured Mouse Proximal Tubule Epithelial Cells by L-Lysine. J. Clin. Pharmacol. 1993, 33, 63–69. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen Wen, S.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16, 1–9. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| #Term ID | Term Description | Observed Gene Count | Background Gene Count | False Discovery Rate |
|---|---|---|---|---|
| Linked to Cell Proliferation | ||||
| MMU-68827 | CDT1 association with the CDC6:ORC:origin complex | 8 | 53 | 6.19 × 10–10 |
| MMU-69229 | Ubiquitin-dependent degradation of Cyclin D1 | 8 | 46 | 6.19 × 10–10 |
| MMU-69481 | G2/M Checkpoints | 10 | 128 | 6.19 × 10–10 |
| MMU-69601 | Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | 8 | 47 | 6.19 × 10–10 |
| MMU-174154 | APC/C:Cdc20 mediated degradation of Securin | 8 | 61 | 9.92 × 10–10 |
| MMU-69206 | G1/S Transition | 9 | 96 | 9.92 × 10–10 |
| MMU-8948751 | Regulation of PTEN stability and activity | 8 | 64 | 1.25 × 10–9 |
| MMU-174178 | APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | 8 | 66 | 1.43 × 10–9 |
| MMU-174184 | Cdc20:Phospho-APC/C mediated degradation of Cyclin A | 8 | 66 | 1.43 × 10–9 |
| MMU-69017 | CDK-mediated phosphorylation and removal of Cdc6 | 8 | 66 | 1.43 × 10–9 |
| MMU-5687128 | MAPK6/MAPK4 signaling | 8 | 67 | 1.52 × 10–9 |
| MMU-8852276 | The role of GTSE1 in G2/M progression after G2 checkpoint | 8 | 68 | 1.58 × 10–9 |
| MMU-69620 | Cell Cycle Checkpoints | 11 | 240 | 6.48 × 10–9 |
| MMU-69278 | Cell Cycle, Mitotic | 10 | 435 | 1.18 × 10–5 |
| MMU-5663213 | RHO GTPases Activate WASPs and WAVEs | 2 | 32 | 0.0154 |
| MMU-5674135 | MAP2K and MAPK activation | 2 | 36 | 0.0186 |
| MMU-6806834 | Signaling by MET | 2 | 63 | 0.0465 |
| Linked to Primary Cilia | ||||
| MMU-5358346 | Hedgehog ligand biogenesis | 9 | 58 | 2.87 × 10–10 |
| MMU-4086400 | PCP/CE pathway | 9 | 82 | 6.19 × 10–10 |
| MMU-4641258 | Degradation of DVL | 8 | 52 | 6.19 × 10–10 |
| MMU-5610780 | Degradation of GLI1 by the proteasome | 8 | 52 | 6.19 × 10–10 |
| MMU-5610785 | GLI3 is processed to GLI3R by the proteasome | 8 | 54 | 6.19 × 10–10 |
| MMU-4608870 | Asymmetric localization of PCP proteins | 8 | 57 | 7.09 × 10–10 |
| MMU-5632684 | Hedgehog ‘on’ state | 8 | 105 | 2.79 × 10–10 |
| Stress Response | ||||
| MMU-1234176 | Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | 9 | 60 | 2.87 × 10–10 |
| MMU-349425 | Autodegradation of the E3 ubiquitin ligase COP1 | 8 | 47 | 6.19 × 10–10 |
| MMU-2262752 | Cellular responses to stress | 13 | 327 | 1.43 × 10–9 |
| MMU-3299685 | Detoxification of Reactive Oxygen Species | 3 | 32 | 0.00072 |
| ID | mIMCD Mean | T2J Mean | Difference | Fold Change | p-Value |
|---|---|---|---|---|---|
| Myosin-9 | 12.54 ± 1.74 | 5.63 ± 3.26 | 0.45 | 2.23 | 0.02 |
| T-complex protein 1 subunit γ | 4.42 ± 0.59 | 2.49 ± 1.00 | 0.56 | 1.77 | 0.03 |
| Adseverin | 5.27 ± 1.41 | 2.06 ± 0.84 | 0.39 | 2.56 | 0.04 |
| Protein disulfide-isomerase A3 | 2.95 ± 0.28 | 1.81 ± 0.57 | 0.61 | 1.63 | 0.02 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zadjali, F.; Kumar, P.; Yao, Y.; Johnson, D.; Astrinidis, A.; Vogel, P.; Gross, K.W.; Bissler, J.J. Tuberous Sclerosis Complex Axis Controls Renal Extracellular Vesicle Production and Protein Content. Int. J. Mol. Sci. 2020, 21, 1729. https://doi.org/10.3390/ijms21051729
Zadjali F, Kumar P, Yao Y, Johnson D, Astrinidis A, Vogel P, Gross KW, Bissler JJ. Tuberous Sclerosis Complex Axis Controls Renal Extracellular Vesicle Production and Protein Content. International Journal of Molecular Sciences. 2020; 21(5):1729. https://doi.org/10.3390/ijms21051729
Chicago/Turabian StyleZadjali, Fahad, Prashant Kumar, Ying Yao, Daniel Johnson, Aristotelis Astrinidis, Peter Vogel, Kenneth W. Gross, and John J. Bissler. 2020. "Tuberous Sclerosis Complex Axis Controls Renal Extracellular Vesicle Production and Protein Content" International Journal of Molecular Sciences 21, no. 5: 1729. https://doi.org/10.3390/ijms21051729
APA StyleZadjali, F., Kumar, P., Yao, Y., Johnson, D., Astrinidis, A., Vogel, P., Gross, K. W., & Bissler, J. J. (2020). Tuberous Sclerosis Complex Axis Controls Renal Extracellular Vesicle Production and Protein Content. International Journal of Molecular Sciences, 21(5), 1729. https://doi.org/10.3390/ijms21051729






