The Complex Network between MYC Oncogene and microRNAs in Gastric Cancer: An Overview
Abstract
:1. Introduction
2. Biological Significance of MYC
3. The Complex Relationship between microRNAs and MYC Expression
3.1. MYC Is Regulated by Epigenetic Modifications
3.2. MicroRNAs Regulate MYC Oncogenic Pathways
3.3. MYC can Promote Angiogenesis through the Regulation of microRNAs
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GC | Gastric cancer |
| miRNA | microRNA |
| RISC | RNA-induced silencing complex |
| H. pylori | Helicobacter pylori |
| IARC | International Agency for Research on Cancer |
| EMT | Epithelial-mesenchymal matrix |
| GBM | Glioblastoma multiforme |
| CRISPRi | CRISPR interference |
References
- Ferro, A.; Peleteiro, B.; Malvezzi, M.; Bosetti, C.; Bertuccio, P.; Levi, F.; Negri, E.; La Vecchia, C.; Lunet, N. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur. J. Cancer 2014, 50, 1330–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA A Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carcas, L.P. Gastric cancer review. J. Carcinog. 2014, 13, 14. [Google Scholar] [CrossRef]
- Group, H.A.C.C. Gastric cancer and Helicobacter pylori: A combined analysis of 12 case control studies nested within prospective cohorts. Gut 2001, 49, 347–353. [Google Scholar] [CrossRef] [Green Version]
- Hino, R.; Uozaki, H.; Murakami, N.; Ushiku, T.; Shinozaki, A.; Ishikawa, S.; Morikawa, T.; Nakaya, T.; Sakatani, T.; Takada, K.; et al. Activation of DNA Methyltransferase 1 by EBV Latent Membrane Protein 2A Leads to Promoter Hypermethylation of PTEN Gene in Gastric Carcinoma. Cancer Res. 2009, 69, 2766–2774. [Google Scholar] [CrossRef] [Green Version]
- D’Elia, L.; Galletti, F.; Strazzullo, P. Dietary salt intake and risk of gastric cancer. Cancer Treat. Res. 2014, 159, 83–95. [Google Scholar] [CrossRef]
- Woo, H.D.; Park, S.; Oh, K.; Kim, H.J.; Shin, H.R.; Moon, H.K.; Kim, J. Diet and cancer risk in the Korean population: A meta- analysis. Asian Pac. J. Cancer Prev. 2014, 15, 8509–8519. [Google Scholar] [CrossRef] [Green Version]
- Duell, E.J.; Travier, N.; Lujan-Barroso, L.; Clavel-Chapelon, F.; Boutron-Ruault, M.C.; Morois, S.; Palli, D.; Krogh, V.; Panico, S.; Tumino, R.; et al. Alcohol consumption and gastric cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort. Am. J. Clin. Nutr. 2011, 94, 1266–1275. [Google Scholar] [CrossRef]
- Ladeiras-Lopes, R.; Pereira, A.K.; Nogueira, A.; Pinheiro-Torres, T.; Pinto, I.; Santos-Pereira, R.; Lunet, N. Smoking and gastric cancer: Systematic review and meta-analysis of cohort studies. Cancer Causes Control 2008, 19, 689–701. [Google Scholar] [CrossRef]
- Takeno, S.S.; Leal, M.F.; Lisboa, L.C.; Lipay, M.V.; Khayat, A.S.; Assumpção, P.P.; Burbano, R.R.; Smith Mde, A. Genomic alterations in diffuse-type gastric cancer as shown by high-resolution comparative genomic hybridization. Cancer Genet. Cytogenet. 2009, 190. [Google Scholar] [CrossRef]
- Fu, D.G. Epigenetic alterations in gastric cancer (Review). Mol. Med. Rep. 2015, 12, 3223–3230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Y.; Dang, S.; Hou, P. Gene methylation in gastric cancer. Clin. Chim. Acta 2013, 424, 53–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burbano, R.R.; Assumpcao, P.P.; Leal, M.F.; Calcagno, D.Q.; Guimaraes, A.C.; Khayat, A.S.; Takeno, S.S.; Chen, E.S.; De Arruda Cardoso Smith, M. C-MYC locus amplification as metastasis predictor in intestinal-type gastric adenocarcinomas: CGH study in Brazil. Anticancer Res. 2006, 26, 2909–2914. [Google Scholar]
- Calcagno, D.Q.; Guimarães, A.C.; Leal, M.F.; Seabra, A.D.; Khayat, A.S.; Pontes, T.B.; Assumpção, P.P.; Smith, M.D.A.C.; Burbano, R.R. MYC Insertions in Diffuse-type Gastric Adenocarcinoma. Anticancer Res. 2009, 29, 2479–2483. [Google Scholar]
- Calcagno, D.Q.; Leal, M.F.; Assumpção, P.P.; Smith, M.A.C.; Burbano, R. MYC and gastric adenocarcinoma carcinogenesis. World J. Gastroenterol. 2008, 14, 5962–5968. [Google Scholar] [CrossRef] [Green Version]
- Calcagno, D.Q.; Leal, M.F.; Seabra, A.D.; Khayat, A.S.; Chen, E.S.; Demachki, S.; Assumpcao, P.P.; Faria, M.H.; Rabenhorst, S.H.; Ferreira, M.V.; et al. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World J. Gastroenterol. 2006, 12, 6207–6211. [Google Scholar] [CrossRef] [Green Version]
- Venkateswaran, N.; Conacci-Sorrell, M. MYC leads the way. Small Gtpases 2017, 11, 1–9. [Google Scholar] [CrossRef]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, Metabolism, and Cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef] [Green Version]
- Lancho, O.; Herranz, D. The MYC Enhancer-ome: Long-Range Transcriptional Regulation of MYC in Cancer. Trends Cancer 2018, 4, 810–822. [Google Scholar] [CrossRef]
- Calcagno, D.Q.; Freitas, V.M.; Leal, M.F.; de Souza, C.R.; Demachki, S.; Montenegro, R.; Assumpcao, P.P.; Khayat, A.S.; Smith, M.D.; Dos Santos, A.K.; et al. MYC, FBXW7 and TP53 copy number variation and expression in gastric cancer. BMC Gastroenterol. 2013, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, D.Q.; Leal, M.F.; Demachki, S.; Araujo, M.T.; Freitas, F.W.; Oliveira e Souza, D.; Assumpcao, P.P.; Ishak, G.; de Smith, M.; Burbano, R.R. MYC in gastric carcinoma and intestinal metaplasia of young adults. Cancer Genet. Cytogenet. 2010, 202, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Costa Raiol, L.C.; Figueira Silva, E.C.; Mendes da Fonseca, D.; Leal, M.F.; Guimarães, A.C.; Calcagno, D.Q.; Burbano, R.R. Interrelationship between MYC gene numerical aberrations and protein expression in individuals from northern Brazil with early gastric adenocarcinoma. Cancer Genet. Cytogenet. 2008, 181, 31–35. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.R.; Leal, M.F.; Calcagno, D.Q.; Costa Sozinho, E.K.; Borges Bdo, N.; Montenegro, R.C.; Dos Santos, A.K.; Dos Santos, S.E.; Ribeiro, H.F.; Assumpcao, P.P.; et al. MYC deregulation in gastric cancer and its clinicopathological implications. PLoS ONE 2013, 8, e64420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leal, M.F.; Calcagno, D.Q.; Borges da Costa Jde, F.; Silva, T.C.; Khayat, A.S.; Chen, E.S.; Burbano, R.R. MYC, TP53, and chromosome 17 copy-number alterations in multiple gastric cancer cell lines and in their parental primary tumors. J. Biomed. Biotechnol. 2011, 2011, 631268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges da Costa, J.; Leal, M.F.; Silva, T.C.R.; Andrade Junior, E.F.; Rezende, A.P.; Carneiro Muniz Jé, A.P.; Burbano, R.R. Experimental Gastric Carcinogenesis in Cebus apella Nonhuman Primates. PLoS ONE 2011, 6, e21988. [Google Scholar] [CrossRef] [Green Version]
- Silva, T.C.R.; Leal, M.F.; Calcagno, D.Q.; de Souza, C.R.T.; Khayat, A.S.; dos Santos, N.P.C.; Montenegro, R.C.; Rabenhorst, S.H.B.; Nascimento, M.Q.; Assumpção, P.P.; et al. hTERT, MYC and TP53 deregulation in gastric preneoplastic lesions. BMC Gastroenterol. 2012, 12, 85. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, H.F.; Alcântara, D.F.A.; Matos, L.A.; Sousa, J.M.C.; Leal, M.F.; Smith, M.A.C.; Burbano, R.R.; Bahia, M.O. Cytogenetic characterization and evaluation of c-MYC gene amplification in PG100, a new Brazilian gastric cancer cell line. Braz. J. Med. Biol. Res. 2010, 43, 717–721. [Google Scholar] [CrossRef] [Green Version]
- Calcagno, D.; Leal, M.; Takeno, S.; Assumpção, P.; Demachki, S.; Smith, M.; Burbano, R. Aneuploidy of Chromosome 8 and C-MYC Amplification in Individuals from Northern Brazil with Gastric Adenocarcinoma. Anticancer Res. 2005, 25, 4069–4074. [Google Scholar]
- Colombo, T.; Farina, L.; Macino, G.; Paci, P. PVT1: A Rising Star among Oncogenic Long Noncoding RNAs. Biomed. Res. Int. 2015. [Google Scholar] [CrossRef]
- Cui, M.; You, L.; Ren, X.; Zhao, W.; Liao, Q.; Zhao, Y. Long non-coding RNA PVT1 and cancer. Biochem. Biophys. Res. Commun. 2016, 471, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.P.; Cho, G.A.; Han, S.W.; Shin, J.Y.; Jeong, E.G.; Song, S.H.; Lee, W.C.; Lee, K.H.; Bang, D.; Seo, J.S.; et al. Novel fusion transcripts in human gastric cancer revealed by transcriptome analysis. Oncogene 2013, 33, 5434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derderian, C.; Orunmuyi, A.T.; Olapade-Olaopa, E.O.; Ogunwobi, O.O. PVT1 Signaling Is a Mediator of Cancer Progression. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, R.; Zhang, E.; Yin, D.; You, L.; Xu, T.; Chen, W.; Xia, R.; Wan, L.; Sun, M.; Wang, Z.; et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol. Cancer 2015, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.; Liu, H.W.; Chen, J.Q.; Wang, S.H.; Hao, L.Q.; Liu, M.; Wang, B. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed. Pharmacother. 2017, 88, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Dan-Dan, Z.; Xiu-fen, L.; Cheng-wei, L.; Prakash, P.O.; Xiao-dong, L. Long non-coding RNA PVT1: Emerging biomarker in digestive system cancer. Cell Prolif. 2017, 50, e12398. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Kang, W.; Lu, X.; Ma, S.; Dong, L.; Zou, B. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle 2018, 17, 1886–1900. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Liu, H.-M.; Wu, W.-H.; Liu, H.; Pan, Y.; Li, W.-J. Upregulation of long noncoding RNA CCAT1-L promotes epithelial-mesenchymal transition in gastric adenocarcinoma. Onco Targets 2018, 11, 5647–5655. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Jiang, K.; Fang, L.P.; Yao, L.L.; Yu, Z. Knockdown of long noncoding RNA CCAT1 inhibits cell growth, invasion and peritoneal metastasis via downregulation of Bmi-1 in gastric cancer. Neoplasma 2018, 65, 736–744. [Google Scholar] [CrossRef]
- Wu, S.W.; Hao, Y.P.; Qiu, J.H.; Zhang, D.B.; Yu, C.G.; Li, W.H. High expression of long non-coding RNA CCAT2 indicates poor prognosis of gastric cancer and promotes cell proliferation and invasion. Minerva Med. 2017, 108, 317–323. [Google Scholar] [CrossRef]
- Hayashi, H.; Arao, T.; Togashi, Y.; Kato, H.; Fujita, Y.; De Velasco, M.A.; Kimura, H.; Matsumoto, K.; Tanaka, K.; Okamoto, I.; et al. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene 2013, 34, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.Y.; Wang, Z.; Lee, K.Y.; Yuan, P.; Ding, J. Effect of silencing colon cancer-associated transcript 2 on the proliferation, apoptosis and autophagy of gastric cancer BGC-823 cells. Oncol. Lett. 2018, 15, 3127–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.U.; Lee, H.E.; Park do, J.; Jung, E.J.; Song, J.; Kim, H.H.; Choe, G.; Kim, W.H.; Lee, H.S. MYC quantitation in cell-free plasma DNA by real-time PCR for gastric cancer diagnosis. Clin. Chem. Lab. Med. 2009, 47, 530–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Shintaku, K.; Ishii, Y.; Asai, S.; Ishikawa, K.; Fujii, M. Analysis of MYC and chromosome 8 copy number changes in gastrointestinal cancers by dual-color fluorescence in situ hybridization. Cancer Genet. Cytogenet. 1998, 107, 61–64. [Google Scholar] [CrossRef]
- Ishak, G.; Leal, M.F.; dos Santos, N.P.C.; Demachki, S.; Nunes, C.A.M.; do Nascimento Borges, B.; Calcagno, D.Q.; Smith, M.C.; Assumpção, P.P.; Burbano, R.R. Deregulation of MYC and TP53 through genetic and epigenetic alterations in gallbladder carcinomas. Clin. Exp. Med. 2015, 15, 421–426. [Google Scholar] [CrossRef] [PubMed]
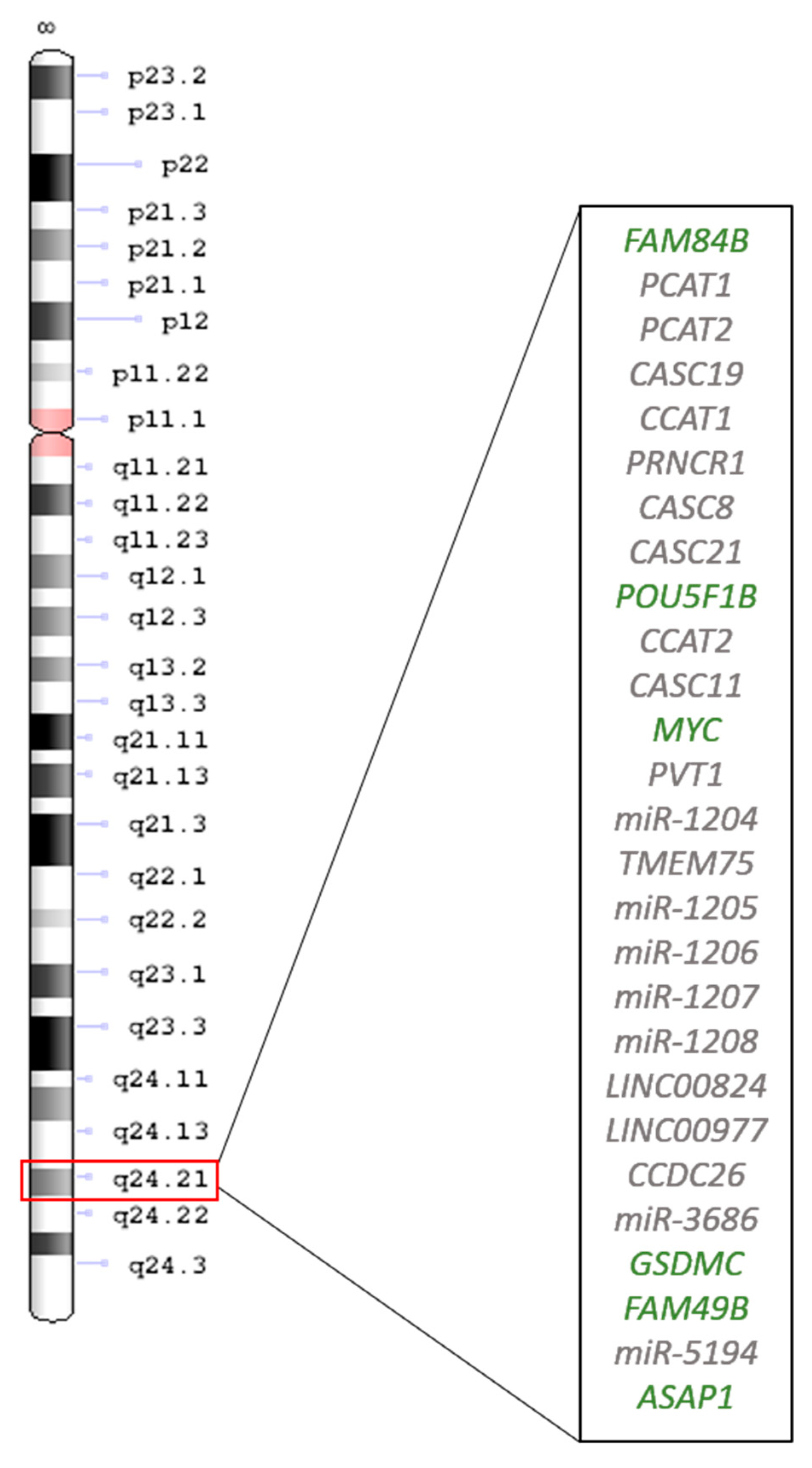
- Ma, G.; Gu, D.; Lv, C.; Chu, H.; Xu, Z.; Tong, N.; Wang, M.; Tang, C.; Xu, Y.; Zhang, Z.; et al. Genetic variant in 8q24 is associated with prognosis for gastric cancer in a Chinese population. J. Gastroenterol. Hepatol. 2015, 30, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Anauate, A.C.; Leal, M.F.; Wisnieski, F.; Santos, L.C.; Gigek, C.O.; Chen, E.S.; Calcagno, D.Q.; Assumpção, P.P.; Demachki, S.; Arasaki, C.H.; et al. Analysis of 8q24.21 miRNA cluster expression and copy number variation in gastric cancer. Future Med. Chem. 2019, 11, 947–958. [Google Scholar] [CrossRef]
- Li, L.; Jia, F.; Bai, P.; Liang, Y.; Sun, R.; Yuan, F.; Zhang, L.; Gao, L. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of gastric cancer. Tumor Biol. 2016, 37, 299–303. [Google Scholar] [CrossRef]
- Li, T.; Meng, X.L.; Yang, W.Q. Long Noncoding RNA PVT1 Acts as a “Sponge” to Inhibit microRNA-152 in Gastric Cancer Cells. Dig. Dis. Sci. 2017, 62, 3021–3028. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Piletič, K.; Kunej, T. MicroRNA epigenetic signatures in human disease. Arch. Toxicol. 2016, 90, 2405–2419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Du, X. Noncoding RNAs in gastric cancer: Research progress and prospects. World J. Gastroenterol. 2016, 22, 6610–6618. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.H.; Chen, Z.N.; Li, L.; Chen, J.; Wei, W.E.; Mo, X.W.; Qin, Y.Z.; Lin, Y.; Chen, J.S. miR-135a promotes gastric cancer progression and resistance to oxaliplatin. Oncotarget 2016, 7, 70699–70714. [Google Scholar] [CrossRef] [Green Version]
- Venturutti, L.; Cordo Russo, R.I.; Rivas, M.A.; Mercogliano, M.F.; Izzo, F.; Oakley, R.H.; Pereyra, M.G.; De Martino, M.; Proietti, C.J.; Yankilevich, P.; et al. MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene 2016, 35, 6189–6202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, Y.; Huang, T.; Wu, F.; Pan, Y.; Dong, Y.; Wang, Y.; Chan, A.K.Y.; Liu, L.; Kwan, J.S.H.; et al. FGF18, a prominent player in FGF signaling, promotes gastric tumorigenesis through autocrine manner and is negatively regulated by miR-590-5p. Oncogene 2019, 38, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Wu, J.; Jiao, K.; Wu, Q.; Ma, J.; Chen, D.; Kang, J.; Zhao, G.; Shi, Y.; Fan, D.; et al. MicroRNA-495-3p inhibits multidrug resistance by modulating autophagy through GRP78/mTOR axis in gastric cancer. Cell Death Dis. 2018, 9, 1070. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Swier, L.; Dzikiewicz-Krawczyk, A.; Winkle, M.; van den Berg, A.; Kluiver, J. Intricate crosstalk between MYC and non-coding RNAs regulates hallmarks of cancer. Mol. Oncol. 2019, 13, 26–45. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, B.; Morgenbesser, S.D.; DePinho, R.A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: Cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992, 6, 1480–1492. [Google Scholar] [CrossRef] [Green Version]
- Tao, J.; Zhao, X.; Tao, J. c-MYC–miRNA circuitry. Cell Cycle 2014, 13, 191–198. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Soutto, M.; Rahman, B.; Fazili, M.W.; Peng, D.; Blanca Piazuelo, M.; Chen, H.; Kay Washington, M.; Shyr, Y.; El-Rifai, W. Integrated expression analysis identifies transcription networks in mouse and human gastric neoplasia. Genes Chromosomes Cancer 2017, 56, 535–547. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, I.J.; Kim, C.G.; Kim, H.S.; Oshima, A.; Yamada, Y.; Arao, T.; Nishio, K.; Michalowski, A.; Green, J.E. Three-gene predictor of clinical outcome for gastric cancer patients treated with chemotherapy. Pharm. J. 2012, 12, 119–127. [Google Scholar] [CrossRef]
- Wisnieski, F.; Calcagno, D.Q.; Leal, M.F.; Chen, E.S.; Gigek, C.O.; Santos, L.C.; Pontes, T.B.; Rasmussen, L.T.; Payão, S.L.M.; Assumpção, P.P.; et al. Differential expression of histone deacetylase and acetyltransferase genes in gastric cancer and their modulation by trichostatin A. Tumor Biol. 2014, 35, 6373–6381. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.Y.; Loven, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012, 151, 56–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999, 19, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.Y.; Chun, Y.S.; Shin, H.W.; Park, J.W. Potential role of the N-MYC downstream-regulated gene family in reprogramming cancer metabolism under hypoxia. Oncotarget 2016, 7, 57442–57451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, V.P.; de Lima, M.A.; Andre, A.R.; Ferreira, M.V.; Barros, M.A.; Rabenhorst, S.H. H pylori (CagA) and Epstein-Barr virus infection in gastric carcinomas: Correlation with p53 mutation and c-Myc, Bcl-2 and Bax expression. World J. Gastroenterol. 2008, 14, 884–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bueno, M.J.; Gómez de Cedrón, M.; Gómez-López, G.; Pérez de Castro, I.; Di Lisio, L.; Montes-Moreno, S.; Martínez, N.; Guerrero, M.; Sánchez-Martínez, R.; Santos, J.; et al. Combinatorial effects of microRNAs to suppress the Myc oncogenic pathway. Blood 2011, 117, 6255–6266. [Google Scholar] [CrossRef]
- Alzahrani, S.; Lina, T.T.; Gonzalez, J.; Pinchuk, I.V.; Beswick, E.J.; Reyes, V.E. Effect of Helicobacter pylori on gastric epithelial cells. World J. Gastroenterol. 2014, 20, 12767–12780. [Google Scholar] [CrossRef] [PubMed]
- Ott, G.; Rosenwald, A.; Campo, E. Understanding MYC-driven aggressive B-cell lymphomas: Pathogenesis and classification. Blood 2013, 122, 3884–3891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- IARC. Helicobacter pylori Eradication as a Strategy for Preventing Gastric Cancer. Helicobacter pylori Working Group. International Agency for Research on Cancer. IARC Work. Group Rep. 2014, 8. [Google Scholar]
- Correa, P. New strategies for the prevention of gastric cancer: Helicobacter pylori and genetic susceptibility. J. Surg. Oncol. 2005, 90, 134–138. [Google Scholar] [CrossRef]
- Keates, S.; Keates, A.C.; Warny, M.; Peek, R.M.; Murray, P.G.; Kelly, C.P. Differential Activation of Mitogen-Activated Protein Kinases in AGS Gastric Epithelial Cells by cag+ and cag− Helicobacter pylori. J. Immunol. 1999, 163, 5552–5559. [Google Scholar]
- Hayashi, Y.; Tsujii, M.; Wang, J.; Kondo, J.; Akasaka, T.; Jin, Y.; Li, W.; Nakamura, T.; Nishida, T.; Iijima, H.; et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut 2013, 62, 1536–1546. [Google Scholar] [CrossRef]
- Silva-Fernandes, I.d.J.L.; Alves, M.K.S.; Lima, V.P.; de Lima, M.A.P.; Barros, M.A.P.; Ferreira, M.V.P.; Rabenhorst, S.H.B. Differential expression of MYC in H. pylori-related intestinal and diffuse gastric tumors. Virchows Arch. 2011, 458, 725–731. [Google Scholar] [CrossRef]
- Nardone, G.; Staibano, S.; Rocco, A.; Mezza, E.; D’Armiento, F.; Insabato, L.; Coppola, A.; Salvatore, G.; Lucariello, A.; Figura, N.; et al. Effect of Helicobacter pylori infection and its eradication on cell proliferation, DNA status, and oncogene expression in patients with chronic gastritis. Gut 1999, 44, 789–799. [Google Scholar] [CrossRef] [Green Version]
- Byun, E.; Park, B.; Lim, J.W.; Kim, H. Activation of NF-κB and AP-1 Mediates Hyperproliferation by Inducing β-Catenin and c-Myc in Helicobacter pylori-Infected Gastric Epithelial Cells. Yonsei Med. J. 2016, 57, 647–651. [Google Scholar] [CrossRef]
- Niwa, T.; Tsukamoto, T.; Toyoda, T.; Mori, A.; Tanaka, H.; Maekita, T.; Ichinose, M.; Tatematsu, M.; Ushijima, T. Inflammatory Processes Triggered by Helicobacter pylori Infection Cause Aberrant DNA Methylation in Gastric Epithelial Cells. Cancer Res. 2010, 70, 1430–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Qian, Y.; Li, F.; Bei, S.; Li, M.; Feng, L. microRNA-9 selectively targets LMX1A to promote gastric cancer cell progression. Biochem. Biophys. Res. Commun. 2018, 505, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, K.C.D.S.; Bona, A.B.; Da Silva, F.J.; Pinheiro, T.M.; Alcantara, D.D.F.A.; Lamarao, L.M.; Moreira-Nunes, C.A.; Assumpção, P.P.; Burbano, R.R.; Calcagno, D.Q. Expression of hsa-miR-9 and MYC Copy Number Variation in Hereditary Diffuse Gastric Cancer. Anticancer Res. 2017, 37, 2401–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Chen, X.; Lin, J.; Lwin, T.; Wright, G.; Moscinski, L.C.; Dalton, W.S.; Seto, E.; Wright, K.; Sotomayor, E.; et al. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene 2012, 31, 3002–3008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, W.; Tong, J.H.M.; Lung, R.W.M.; Dong, Y.; Zhao, J.; Liang, Q.; Zhang, L.; Pan, Y.; Yang, W.; Pang, J.C.S.; et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol. Cancer 2015, 14, 52. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Peng, Z.; Zhao, Y.; Chen, L. microRNA-25 Inhibits Cell Apoptosis of Human Gastric Adenocarcinoma Cell Line AGS via Regulating CCNE1 and MYC. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 1415–1420. [Google Scholar] [CrossRef]
- Kim, Y.K.; Yu, J.; Han, T.S.; Park, S.Y.; Namkoong, B.; Kim, D.H.; Hur, K.; Yoo, M.W.; Lee, H.J.; Yang, H.K.; et al. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009, 37, 1672–1681. [Google Scholar] [CrossRef]
- Park, D.; Lee, S.C.; Park, J.W.; Cho, S.Y.; Kim, H.K. Overexpression of miR-17 in gastric cancer is correlated with proliferation-associated oncogene amplification. Pathol. Int. 2014, 64, 309–314. [Google Scholar] [CrossRef]
- Wu, W.; Takanashi, M.; Borjigin, N.; Ohno, S.; Fujita, K.; Hoshino, S.; Osaka, Y.; Tsuchida, A.; Kuroda, M. MicroRNA-18a modulates STAT3 activity through negative regulation of PIAS3 during gastric adenocarcinogenesis. Br. J. Cancer 2013, 108, 653–661. [Google Scholar] [CrossRef]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Li, S.; Liang, X.; Ma, L.; Shen, L.; Li, T.; Zheng, L.; Sun, A.; Shang, W.; Chen, C.; Zhao, W.; et al. MiR-22 sustains NLRP3 expression and attenuates H. pylori-induced gastric carcinogenesis. Oncogene 2017, 37, 884. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, R.; He, Z.; Qiu, Q.; Lu, X.; Yin, J.; Liu, H.; Jia, X.; He, Z. TET-Mediated Sequestration of miR-26 Drives EZH2 Expression and Gastric Carcinogenesis. Cancer Res. 2017, 77, 6069–6082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Fan, Z.; Liu, F.; Zuo, J. Hsa-miR-21 and Hsa-miR-29 in Tissue as Potential Diagnostic and Prognostic Biomarkers for Gastric Cancer. Cell. Physiol. Biochem. 2015, 37, 1454–1462. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839. [Google Scholar] [CrossRef] [PubMed]
- Jafari, N.; Abediankenari, S. MicroRNA-34 dysregulation in gastric cancer and gastric cancer stem cell. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christoffersen, N.R.; Shalgi, R.; Frankel, L.B.; Leucci, E.; Lees, M.; Klausen, M.; Pilpel, Y.; Nielsen, F.C.; Oren, M.; Lund, A.H. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2009, 17, 236. [Google Scholar] [CrossRef] [PubMed]
- Shin, V.Y.; Siu, M.T.; Liu, X.; Ng, E.K.O.; Kwong, A.; Chu, K.M. MiR-92 suppresses proliferation and induces apoptosis by targeting EP4/Notch1 axis in gastric cancer. Oncotarget 2018, 9, 24209–24220. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Xia, Y.; Li, L.; Zhang, G. MiR-101 inhibits cell growth and tumorigenesis of Helicobacter pylori related gastric cancer by repression of SOCS2. Cancer Biol. 2015, 16, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.; Zhang, M.; Qiao, H. Diagnostic significance of miR-106a in gastric cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 13096–13101. [Google Scholar]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef]
- You, W.; Zhang, X.; Ji, M.; Yu, Y.; Chen, C.; Xiong, Y.; Liu, Y.; Sun, Y.; Tan, C.; Zhang, H.; et al. MiR-152-5p as a microRNA passenger strand special functions in human gastric cancer cells. Int. J. Biol. Sci. 2018, 14, 644–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.; Li, Y.; Zhao, Q.; Fan, L.; Wang, D. ZNF139 increases multidrug resistance in gastric cancer cells by inhibiting miR-185. Biosci. Rep. 2018, 38, BSR20181023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, C.; Sun, D.; Zhang, L.; Song, L. miR-186 affects the proliferation, invasion and migration of human gastric cancer by inhibition of Twist1. Oncotarget 2016, 7, 79956–79963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Nie, Y.; Tu, S.; Wang, H.; Zhou, Y.; Du, Y.; Cao, J.; Ye, M. Epigenetically deregulated miR-200c is involved in a negative feedback loop with DNMT3a in gastric cancer cells. Oncol. Rep. 2016, 36, 2108–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, K.W.; Wang, A.M.; Ping, Y.H.; Huang, K.H.; Huang, T.T.; Lee, H.C.; Lo, S.S.; Chi, C.W.; Yeh, T.S. Downregulation of tumor suppressor MBP-1 by microRNA-363 in gastric carcinogenesis. Carcinogenesis 2014, 35, 208–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, R.; Zhang, X.; Qian, H.; Gu, H.; Sun, Z.; Mao, F.; Yan, Y.; Chen, J.; Liang, Z.; Xu, W. miR-374 mediates the malignant transformation of gastric cancer-associated mesenchymal stem cells in an experimental rat model. Oncol. Rep. 2017, 38, 1473–1481. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Wang, C.; Xing, J.; Wu, D. miR-429 Modulates the expression of c-myc in human gastric carcinoma cells. Eur. J. Cancer 2011, 47, 2552–2559. [Google Scholar] [CrossRef]
- He, W.; Li, Y.; Chen, X.; Lu, L.; Tang, B.; Wang, Z.; Pan, Y.; Cai, S.; He, Y.; Ke, Z. miR-494 acts as an anti-oncogene in gastric carcinoma by targeting c-myc. J. Gastroenterol. Hepatol. 2014, 29, 1427–1434. [Google Scholar] [CrossRef]
- Li, R.; Yuan, W.; Mei, W.; Yang, K.; Chen, Z. MicroRNA 520d-3p inhibits gastric cancer cell proliferation, migration, and invasion by downregulating EphA2 expression. Mol. Cell. Biochem. 2014, 396, 295–305. [Google Scholar] [CrossRef]
- Qian, K.; Mao, B.; Zhang, W.; Chen, H. MicroRNA-561 inhibits gastric cancercell proliferation and invasion by downregulating c-Myc expression. Am. J. Transl. Res. 2016, 8, 3802–3811. [Google Scholar] [PubMed]
- Yang, M.; Cui, G.; Ding, M.; Yang, W.; Liu, Y.; Dai, D.; Chen, L. miR-935 promotes gastric cancer cell proliferation by targeting SOX7. Biomed. Pharmacother. 2016, 79, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Wei, W.; Zhan, Z.; Xie, Y.; Xiao, Q. MiR-1284 modulates multidrug resistance of gastric cancer cells by targeting EIF4A1. Oncol. Rep. 2016, 35, 2583–2591. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.X.; Yu, Y.Y.; Zhou, A.Q.; Zhu, J.L.; Luo, L.N.; Chen, W.Q.; Hu, L.; Chen, G.X. Yangzheng Sanjie decoction regulates proliferation and apoptosis of gastric cancer cells by enhancing let-7a expression. World J. Gastroenterol. 2017, 23, 5538–5548. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, F.; Xu, X.F.; Mo, W.H.; Xia, Y.J.; Wan, R.; Wang, X.P.; Guo, C.Y. Down-regulation of miR-212 expression by DNA hypermethylation in human gastric cancer cells. Med. Oncol. 2011, 28, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Song, P.; Su, R.; Yang, G.; Dong, L.; Luo, M.; Wang, B.; Gong, B.; Liu, C.; Song, W.; et al. DNA Methylation mediated down-regulating of MicroRNA-33b and its role in gastric cancer. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Hong, X.; Xu, Y.; Qiu, X.; Zhu, Y.; Feng, X.; Ding, Z.; Zhang, S.; Zhong, L.; Zhuang, Y.; Su, C.; et al. MiR-448 promotes glycolytic metabolism of gastric cancer by downregulating KDM2B. Oncotarget 2016, 7, 22092–22102. [Google Scholar] [CrossRef]
- Yoon, J.H.; Choi, Y.J.; Choi, W.S.; Ashktorab, H.; Smoot, D.T.; Nam, S.W.; Lee, J.Y.; Park, W.S. GKN1–miR-185–DNMT1 Axis Suppresses Gastric Carcinogenesis through Regulation of Epigenetic Alteration and Cell Cycle. Clin. Cancer Res. 2013, 19, 4599–4610. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Xiao, D.; Li, G.; Ma, J.; Chen, P.; Yuan, W.; Hou, F.; Ge, J.; Zhong, M.; Tang, Y.; et al. EphA2 promotes epithelial-mesenchymal transition through the Wnt/[beta]-catenin pathway in gastric cancer cells. Oncogene 2014, 33, 2737–2747. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Wang, L.; Song, W.; Cui, H.; Chen, G.; Qiao, F.; Hu, J.; Zhou, R.; Fan, H. Reduced miR-29a-3p expression is linked to the cell proliferation and cell migration in gastric cancer. World J. Surg. Oncol. 2015, 13, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Wang, F.; Zhao, H.; Gong, J.; Zhang, J.; Dong, L.; Luo, M.; Song, W.; Yu, J.; Li, J.; et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis 2013, 35, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Kawano, M.; Tanaka, K.; Itonaga, I.; Iwasaki, T.; Tsumura, H. c-Myc Represses Tumor-Suppressive microRNAs, let-7a, miR-16 and miR-29b, and Induces Cyclin D2-Mediated Cell Proliferation in Ewing’s Sarcoma Cell Line. PLoS ONE 2015, 10, e0138560. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Guo, Q.; Fu, F.J.; Wang, Z.; Yin, Z.; Wei, Y.B.; Yang, J.R. The role of miR-29b in cancer: Regulation, function, and signaling. Onco Targets 2015, 8, 539–548. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.; Suzuki, H.; Imaeda, H.; Matsuzaki, J.; Hirata, K.; Tsugawa, H.; Hibino, S.; Kanai, Y.; Saito, H.; Hibi, T. The tumor suppressor microRNA-29c is downregulated and restored by celecoxib in human gastric cancer cells. Int. J. Cancer 2013, 132, 1751–1760. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Luo, M.; Zhang, Z.; Gong, J.; Li, J.; You, L.; Dong, L.; Su, R.; Lin, H.; et al. Chemotherapy-Induced miRNA-29c/Catenin-δ Signaling Suppresses Metastasis in Gastric Cancer. Cancer Res. 2015, 75, 1332–1344. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhu, H.; Yang, S.; Wang, Z.; Bai, J.; Xu, N. c-Myc suppressed E-cadherin through miR-9 at the post-transcriptional level. Cell Biol. Int. 2013, 37, 197–202. [Google Scholar] [CrossRef]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Wu, J.; Wu, S.H.; Thakur, A.; Bollig, A.; Huang, Y.; Liao, D.J. Expression profile of microRNAs in c-Myc induced mouse mammary tumors. Breast Cancer Res. Treat. 2009, 118, 185–196. [Google Scholar] [CrossRef] [Green Version]
- Peter, M.E. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle 2009, 8, 843–852. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Mo, X.; Fu, L.; Xiao, B.; Guo, J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016, 7, 8601–8612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, E.M.; Leal, M.F.; Burbano, R.R.; Khayat, A.S.; Assumpção, P.P.; Bello, M.J.; Rey, J.A.; Smith, M.A.C.; Casartelli, C. Methylation status of ANAPC1, CDKN2A and TP53 promoter genes in individuals with gastric cancer. Braz. J. Med. Biol. Res. 2008, 41, 539–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisnieski, F.; Calcagno, D.Q.; Leal, M.F.; Santos, L.C.; Gigek, C.O.; Chen, E.S.; Demachki, S.; Artigiani, R.; Assumpção, P.P.; Lourenço, L.G.; et al. CDKN1A histone acetylation and gene expression relationship in gastric adenocarcinomas. Clin. Exp. Med. 2017, 17, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.O.; Lam, S.; Wong, B.C.; Wong, W.; Yuen, M.; Yeung, Y.; Hui, W.; Rashid, A.; Kwong, Y. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut 2003, 52, 502–506. [Google Scholar] [CrossRef] [Green Version]
- Choi, W.S.; Seo, H.S.; Song, K.Y.; Yoon, J.H.; Kim, O.; Nam, S.W.; Lee, J.Y.; Park, W.S. Gastrokine 1 Expression in the Human Gastric Mucosa Is Closely Associated with the Degree of Gastritis and DNA Methylation. J. Gastric Cancer 2013, 13, 232–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.J.; Ito, K.; Ala, U.; Kats, L.; Webster, K.; Sun, S.M.; Jongen-Lavrencic, M.; Manova-Todorova, K.; Teruya-Feldstein, J.; Avigan, D.E.; et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 2013, 13, 87–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Chadalapaka, G.; Lee, S.O.; Yamada, D.; Sastre-Garau, X.; Defossez, P.A.; Park, Y.Y.; Lee, J.S.; Safe, S. Identification of oncogenic microRNA-17–92/ZBTB4/specificity protein axis in breast cancer. Oncogene 2012, 31, 1034–1044. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Li, S.; Ding, Y.; Wang, Q.; Luo, H.; Shi, Q.; Hao, Z.; Xiao, G.; Tong, S. The role of miR-18a in gastric cancer angiogenesis. Hepatogastroenterology 2013, 60, 1809–1813. [Google Scholar]
- Chen, Y.J.; Wu, H.; Zhu, J.M.; Li, X.D.; Luo, S.W.; Dong, L.; Liu, T.T.; Shen, X.Z. MicroRNA-18a modulates P53 expression by targeting IRF2 in gastric cancer patients. J. Gastroenterol. Hepatol. 2016, 31, 155–163. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, Z.; An, Y.; Hu, H.; Yin, J.; Zhang, P.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014, 5, e1144. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, Q. Poor expression of microRNA-135b results in the inhibition of cisplatin resistance and proliferation and induces the apoptosis of gastric cancer cells through MST1-mediated MAPK signaling pathway. FASEB J. 2019, 33, 3420–3436. [Google Scholar] [CrossRef] [PubMed]
- Knies-Bamforth, U.E.; Fox, S.B.; Poulsom, R.; Evan, G.I.; Harris, A.L. c-Myc Interacts with Hypoxia to Induce Angiogenesis In vivo by a Vascular Endothelial Growth Factor-Dependent Mechanism. Cancer Res. 2004, 64, 6563–6570. [Google Scholar] [CrossRef] [Green Version]
- El Baroudi, M.; Corà, D.; Bosia, C.; Osella, M.; Caselle, M. A curated database of miRNA mediated feed-forward loops involving MYC as master regulator. PLoS ONE 2011, 6, e14742. [Google Scholar] [CrossRef] [PubMed]
- Thakore, P.I.; D’Ippolito, A.M.; Song, L.; Safi, A.; Shivakumar, N.K.; Kabadi, A.M.; Reddy, T.E.; Crawford, G.E.; Gersbach, C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 2015, 12, 1143–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355, aah7111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowdy, S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017, 35, 222–229. [Google Scholar] [CrossRef]
- Mercatelli, N.; Coppola, V.; Bonci, D.; Miele, F.; Costantini, A.; Guadagnoli, M.; Bonanno, E.; Muto, G.; Frajese, G.V.; De Maria, R.; et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS ONE 2008, 3, e4029. [Google Scholar] [CrossRef]
- Sicard, F.; Gayral, M.; Lulka, H.; Buscail, L.; Cordelier, P. Targeting miR-21 for the therapy of pancreatic cancer. Mol. Ther. 2013, 21, 986–994. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, A.L.C.; Simões, S.; de Almeida, L.P.; Plesnila, N.; Pedroso de Lima, M.C.; Wagner, E.; Culmsee, C. Tf-lipoplexes for neuronal siRNA delivery: A promising system to mediate gene silencing in the CNS. J. Control. Release 2008, 132, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Winters, M.; Holodniy, M.; Dai, H. siRNA Delivery into Human T Cells and Primary Cells with Carbon-Nanotube Transporters. Angew. Chem. Int. Ed. 2007, 46, 2023–2027. [Google Scholar] [CrossRef]


| Gene | Gene Name | Expression in GC | Reference |
|---|---|---|---|
| Coding genes | |||
| FAM84B | Family with sequence similarity 84, member B | ? | |
| POU5F1B | POU domain, class 5, transcription Factor 1B | Down | [42] |
| Up | [41] | ||
| MYC | MYC proto-oncogene | Up | [14,16,21,22,24,25,26,27,43,44,45] |
| GSDMC | Gasdermin C | ? | |
| FAM49B | Family with sequence similarity 49 member B | ? | |
| ASAP1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 | ? | |
| Noncoding genes | |||
| CASC8 | Cancer susceptibility candidate 8 | ? | [46] |
| CASC11 | Cancer susceptibility candidate 11 | Up | [37] |
| CASC21 | Cancer susceptibility candidate 21 | ? | |
| CASC19 | Cancer susceptibility candidate 19 | ? | |
| CCAT1 | Colon cancer-associated transcript 1 | Up | [38,39] |
| CCAT2 | Colon cancer-associated transcript 2 | Up | [40] |
| LINC00824 | Long intergenic non-protein coding RNA 824 | ? | |
| LINC00977 | Long intergenic non-protein coding RNA 977 | ? | |
| miR-1204 | MicroRNA 1204 | ? | |
| miR-1205 | MicroRNA 1205 | Did not differ | [47] |
| miR-1206 | MicroRNA 1206 | ? | |
| miR-1207 | MicroRNA 1207 | Did not differ | [47] |
| miR-1208 | MicroRNA 1208 | Did not differ | [47] |
| miR-5194 | MicroRNA 5794 | ? | |
| miR-3686 | MicroRNA 3686 | ? | |
| CCDC26 | Coiled-coil domain-containing protein 26 | ? | |
| TMEM75 | Transmembrane protein 75 | ? | |
| PCAT1 | Prostate cancer-associated transcript 1 | ? | |
| PCAT2 | Prostate cancer-associated transcript 2 | ? | |
| PRNCR1 | Prostate cancer associated noncoding RNA 1 | ? | [48] |
| PVT1 | Plasmacytoma variant translocation 1, MYC activator | Up | [49] |
| miRNA | miRNA Expression | MYC Expression | Reference | |
|---|---|---|---|---|
| miR-9 | Up | Up | [82,83] | |
| miR-15a/16-1 | Down | Up | [84,85] | |
| miR-25 | Up | Up | [86,87] | |
| Cluster miR-17-92 | miR-17 | Up | Up | [87,88] |
| miR-18a | Up | Up | [87,89] | |
| miR-19a/miR-19b | Up | Up | [87,90] | |
| miR-20 | Up | ? | [87] | |
| miR-22 | Down | ? | [91] | |
| miR-26 | Down | Up | [92] | |
| miR-29 | Down | ? | [93] | |
| miR-33b | Down | Down | [94] | |
| miR-34a | Down | Up | [95,96] | |
| miR-92 | Down | ? | [97] | |
| miR-93 | Up | Down | [98] | |
| miR-101 | Down | Up | [99] | |
| miR-106a/miR-106b | Up | ? | [87,100] | |
| miR-150 | Down | Up | [101] | |
| miR-152 | Down | [102] | ||
| miR-185 | Down | Up | [103] | |
| miR-186 | Down | ? | [104] | |
| miR-200c | Down | ? | [105] | |
| miR-212 | Up | Up | [106] | |
| miR-363 | Down | Up | [106] | |
| miR-374 | Up | ? | [107] | |
| miR-429 | Down | Up | [108] | |
| miR-494 | Down | Up | [109] | |
| miR-520d-3p | Down | Up | [110] | |
| miR-561 | Down | Up | [111] | |
| miR-935 | Up | Up | [112] | |
| miR-1284 | Down | ? | [113] | |
| let-7a | Up | Up | [114] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anauate, A.C.; Leal, M.F.; Calcagno, D.Q.; Gigek, C.O.; Karia, B.T.R.; Wisnieski, F.; dos Santos, L.C.; Chen, E.S.; Burbano, R.R.; Smith, M.A.C. The Complex Network between MYC Oncogene and microRNAs in Gastric Cancer: An Overview. Int. J. Mol. Sci. 2020, 21, 1782. https://doi.org/10.3390/ijms21051782
Anauate AC, Leal MF, Calcagno DQ, Gigek CO, Karia BTR, Wisnieski F, dos Santos LC, Chen ES, Burbano RR, Smith MAC. The Complex Network between MYC Oncogene and microRNAs in Gastric Cancer: An Overview. International Journal of Molecular Sciences. 2020; 21(5):1782. https://doi.org/10.3390/ijms21051782
Chicago/Turabian StyleAnauate, Ana Carolina, Mariana Ferreira Leal, Danielle Queiroz Calcagno, Carolina Oliveira Gigek, Bruno Takao Real Karia, Fernanda Wisnieski, Leonardo Caires dos Santos, Elizabeth Suchi Chen, Rommel Rodríguez Burbano, and Marília Arruda Cardoso Smith. 2020. "The Complex Network between MYC Oncogene and microRNAs in Gastric Cancer: An Overview" International Journal of Molecular Sciences 21, no. 5: 1782. https://doi.org/10.3390/ijms21051782
APA StyleAnauate, A. C., Leal, M. F., Calcagno, D. Q., Gigek, C. O., Karia, B. T. R., Wisnieski, F., dos Santos, L. C., Chen, E. S., Burbano, R. R., & Smith, M. A. C. (2020). The Complex Network between MYC Oncogene and microRNAs in Gastric Cancer: An Overview. International Journal of Molecular Sciences, 21(5), 1782. https://doi.org/10.3390/ijms21051782






