Abstract
Abiotic stresses, such as drought, salinity, and extreme temperatures, are major limiting factors in global crop productivity and are predicted to be exacerbated by climate change. The overproduction of reactive oxygen species (ROS) is a common consequence of many abiotic stresses. Ascorbate, also known as vitamin C, is the most abundant water-soluble antioxidant in plant cells and can combat oxidative stress directly as a ROS scavenger, or through the ascorbate–glutathione cycle—a major antioxidant system in plant cells. Engineering crops with enhanced ascorbate concentrations therefore has the potential to promote broad abiotic stress tolerance. Three distinct strategies have been utilized to increase ascorbate concentrations in plants: (i) increased biosynthesis, (ii) enhanced recycling, or (iii) modulating regulatory factors. Here, we review the genetic pathways underlying ascorbate biosynthesis, recycling, and regulation in plants, including a summary of all metabolic engineering strategies utilized to date to increase ascorbate concentrations in model and crop species. We then highlight transgene-free strategies utilizing genome editing tools to increase ascorbate concentrations in crops, such as editing the highly conserved upstream open reading frame that controls translation of the GDP-L-galactose phosphorylase gene.
1. Introduction
Providing adequate, safe, and nutritious food for the growing human population under changing climatic conditions represents one of the greatest challenges of the 21st century. Adverse environmental conditions such as extreme temperatures, drought, and soil salinity are the primary cause of crop losses worldwide and are predicted to be exacerbated by climate change [1]. With the human population expected to reach 9.7 billion by the year 2050, the need for more abiotic stress tolerant crops is critical for ensuring global food security [2].
A common consequence of many abiotic stresses is the overproduction of reactive oxygen species (ROS) such as singlet oxygen (1O2), superoxide (O2−), hydrogen peroxide (H2O2), and the hydroxyl radical (HO−) [3]. For example, stomatal closure during drought and salt stress limits carbon dioxide uptake, which has both direct and indirect effects on ROS production. For instance, reduced NADP+ regeneration through the Calvin cycle or enhanced photorespiration results in a higher leakage of photosynthetic or respiratory electrons, that in turn stimulate ROS production [4,5]. Similarly, Na+ and Cl- toxicity during salt stress can disrupt the photosynthetic electron transport chain, further promoting electron leakage and ROS accumulation [6]. Likewise, temperature stresses are capable of uncoupling temperature-sensitive pathways, such as photosynthesis and respiration, consequently increasing ROS formation [7]. Identifying mechanisms to enhance the capacity of plants to mitigate the harmful effects of the overproduction of ROS will greatly facilitate the production of crops with broad abiotic stress tolerance.
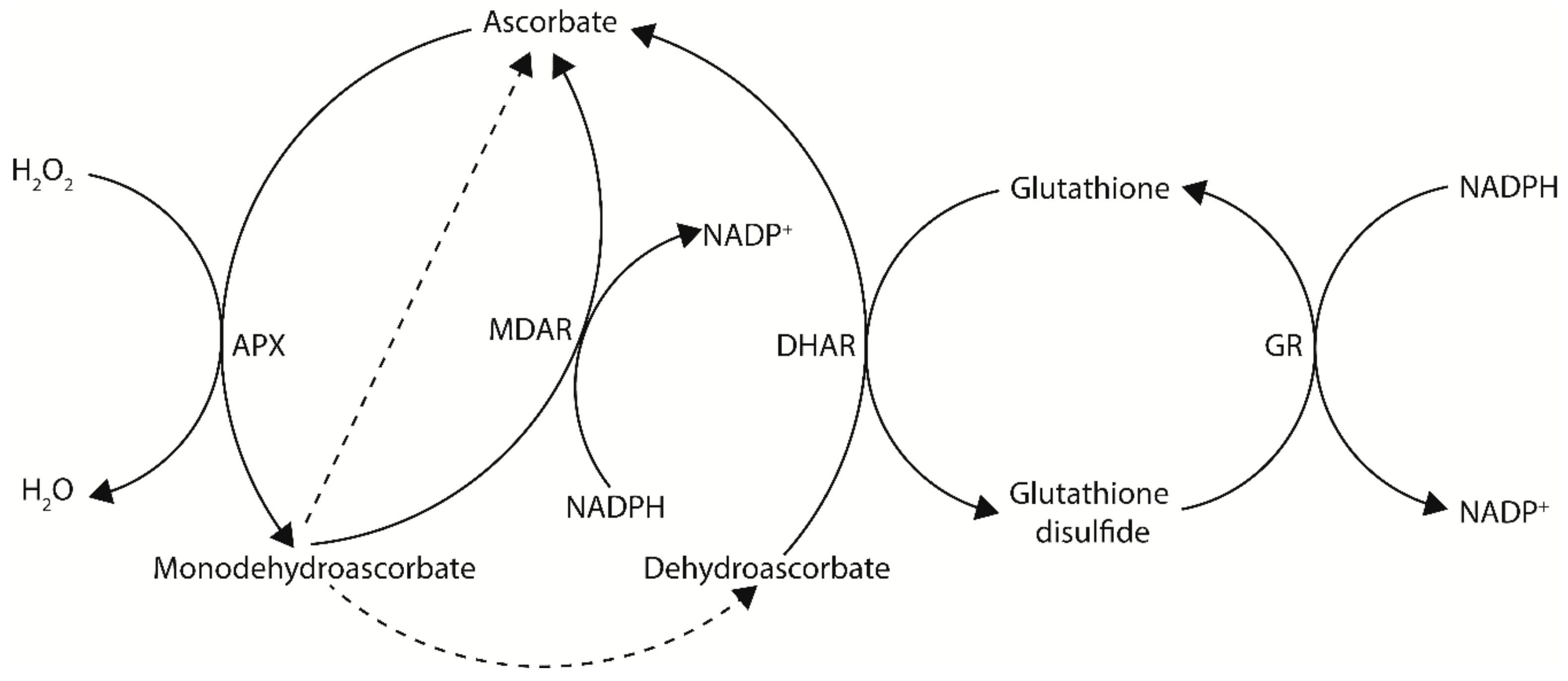
Ascorbate, also known as vitamin C, is an important multifunctional molecule for both plants and animals. Ascorbate is a reducing agent capable of donating electrons and primarily functions as a cellular antioxidant and enzymatic co-factor. In plants, ascorbate is the most abundant water-soluble antioxidant and is found in high concentrations in peroxisomes (22.8 mM), the cytosol (21.7 mM), and nuclei (16.3 mM), intermediate concentrations in mitochondria (10.4 mM) and chloroplasts (10.8 mM), low concentrations in the vacuole (2.3 mM), and very low concentrations in the apoplast [8,9]. Ascorbate is particularly well-known for its roles in photosynthetic functions and stress tolerance. This is largely due to its ability to counteract oxidative stress produced by normal or stressed cellular metabolism, either directly as a ROS scavenger or through the ascorbate-glutathione cycle—a major antioxidant system of plants cells (Figure 1) [10,11,12]. In this cycle, H2O2 is detoxified to H2O by ascorbate peroxidase (APX), using ascorbate as an electron donor. The oxidized ascorbate can then be regenerated by the ascorbate recycling enzymes monodehydroascorbate reductase (MDAR) or dehydroascorbate reductase (DHAR) using NADPH and glutathione, respectively, as electron donors (see Section 4) [3,11].
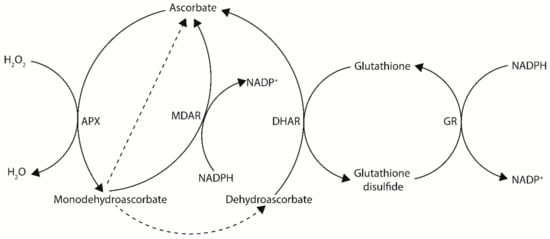
Figure 1.
The ascorbate–glutathione cycle—a major antioxidant system of plants cells. In this cycle, electrons flow from NADPH to H2O2. Dashed arrows indicate non-enzymatic disproportionation. APX, ascorbate peroxidase; MDAR, monodehydroascorbate reductase; DHAR, dehydroascorbate reductase; GR, glutathione reductase.
To date, four ascorbate biosynthetic pathways have been proposed in plants: the L-galactose, L-gulose, myo-inositol, and D-galacturonate pathways [13,14]. The L-galactose pathway, however, is the only pathway to have all the enzymatic steps characterized. Substantial genetic evidence now also supports the L-galactose pathway as the dominant pathway towards ascorbate biosynthesis in plants. Increased expression of ascorbate biosynthetic genes—particularly L-galactose pathway genes—has proven to be an effective strategy to significantly increase ascorbate concentrations in plants. In addition to de novo biosynthesis, cellular ascorbate concentrations are also maintained via the ascorbate recycling enzymes MDAR and DHAR. Increased expression of these ascorbate recycling genes has also proven to be an effective strategy for increasing the concentrations of reduced, active ascorbate in plants. While the genetic pathways underlying the biosynthesis and recycling of ascorbate have largely been understood for over a decade, the genetic pathways underlying the regulation of ascorbate biosynthesis are only now starting to be unraveled. Manipulating ascorbate regulatory factors is emerging as a promising new strategy to increase ascorbate concentrations in plants.
Here, we review the genetic pathways underlying ascorbate biosynthesis, recycling, and regulation in plants, including a summary of all metabolic engineering strategies utilized to date to increase ascorbate concentrations in model and crop species.
2. Biosynthesis of Ascorbate in Plants
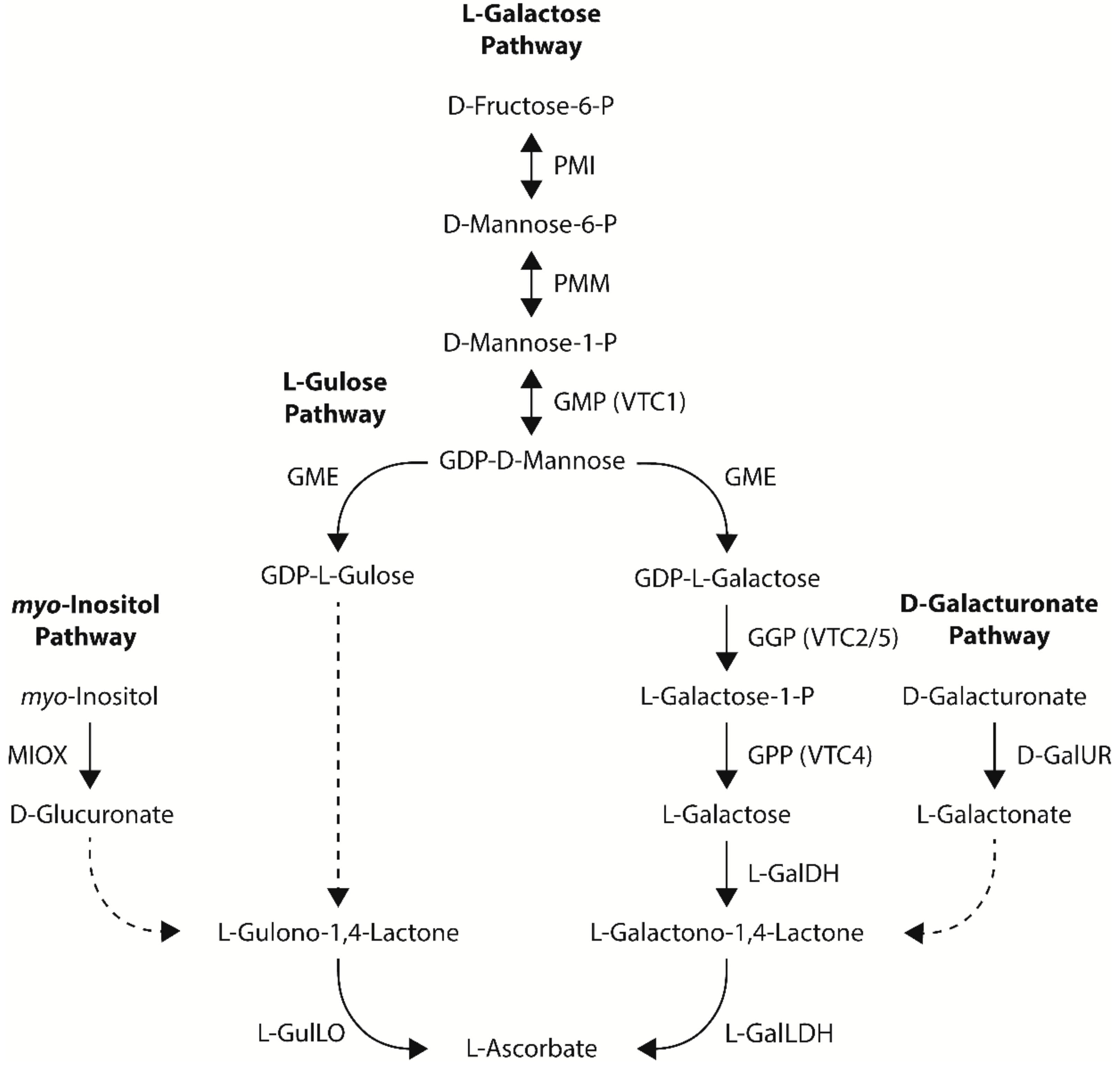
Four pathways towards ascorbate biosynthesis have been proposed in plants: the L-galactose, L-gulose, myo-inositol, and D-galacturonate pathways [13,14] (Figure 2). All four pathways share an aldonolactone as the direct precursor to ascorbate (L-galactono-1,4-lactone for the L-galactose and D-galacturonate pathways and L-gulono-1,4-lactone for the L-gulose and myo-inositol pathways). While there is strong evidence for the L-galactose pathway as the dominant ascorbate biosynthetic pathway in plants, the contribution of the L-gulose, myo-inositol, and D-galacturonate pathways towards ascorbate biosynthesis in plants is not as well documented and often controversial. An overview of metabolic engineering strategies utilizing ascorbate biosynthetic genes to increase ascorbate concentrations in model and crop species, including the tissue examined and any stress tolerances observed, is summarized in Table 1.
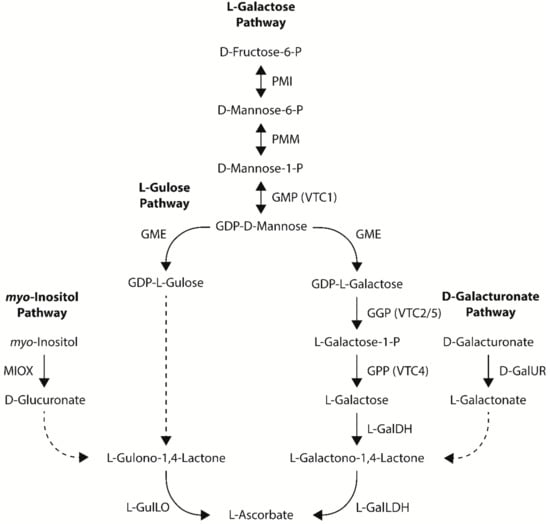
Figure 2.
The four proposed ascorbate biosynthetic pathways in higher plants—the L-galactose, L-gulose, myo-inositol, and D-galacturonate pathways. Dashed arrows indicate multiple and/or unknown biosynthetic steps in higher plants. PMI, phosphomannose isomerase; PMM, phosphomannose mutase; GMP, GDP-D-mannose pyrophosphorylase; GME, GDP-D-mannose-3′,5′-epimerase; GGP, GDP-L-galactose phosphorylase; GPP, L-galactose-1-P phosphatase; L-GalDH, L-galactose dehydrogenase; L-GalLDH, L-GalLDH L-galactono-1,4-lactone dehydrogenase; L-GulLO, L-gulono-1,4-lactone oxidase; MIOX, myo-inositol oxygenase; D-GalUR, D-galacturonate reductase.

Table 1.
Overview of metabolic engineering strategies utilizing ascorbate biosynthetic genes to increase ascorbate concentrations in model and crop species.
2.1. The L-Galactose Pathway
The L-galactose pathway (also known as the D-mannose/L-galactose pathway or Smirnoff-Wheeler pathway) is the only ascorbate biosynthetic pathway in plants to have all its enzymatic steps characterized. The L-galactose pathway is responsible for converting D-fructose-6-P to ascorbate via eight enzymatic steps by phosphomannose isomerase (PMI), phosphomannose mutase (PMM), GDP-D-mannose pyrophosphorylase (GMP), GDP-D-mannose-3′,5′-epimerase (GME), GDP-L-galactose phosphorylase (GGP), L-galactose-1-phosphate phosphatase (GPP), L-galactose dehydrogenase (L-GalDH), and L-galactono-1,4-lactone dehydrogenase (L-GalLDH) [13,14] (Figure 2). All enzymatic steps of the L-galactose pathway take place in the cytosol, except for the conversion of L-galactono-1,4-lactone to ascorbate by L-GalLDH, which occurs in mitochondria [15].
2.1.1. Phosphomannose Isomerase (PMI)
The PMI enzyme is responsible for the first enzymatic step of the L-galactose pathway, catalyzing a reversible conversion of D-fructose-6-P to D-mannose-6-P (Figure 2). Knock-down of the PMI1 gene in Arabidopsis thaliana by RNAi reduced ascorbate to 47% of wild-type levels, supporting its role in L-galactose ascorbate biosynthesis [16]. To date, no studies have investigated the stable increased expression of the PMI gene in plants. However, transient expression of the PMI1 gene in Arabidopsis did not significantly change ascorbate concentrations, indicating that PMI is not a rate-limiting step in the L-galactose pathway in Arabidopsis [17].
2.1.2. Phosphomannose Mutase (PMM)
The PMM enzyme is responsible for the second enzymatic step of the L-galactose pathway, catalyzing a reversible conversion of D-mannose-6-P to D-mannose-1-P (Figure 2). Temperature-sensitive mutations in the Arabidopsis PMM gene reduced ascorbate to 20% of wild-type levels, supporting its important role in L-galactose ascorbate biosynthesis [18]. Increased expression of the PMM gene has increased ascorbate concentrations 1.3-fold in Arabidopsis [19] and 2.5-fold in tobacco (Nicotiana tabacum L.) [20] (Table 1). The small increase in ascorbate concentrations observed in the Arabidopsis plants with increased expression of the PMM gene was not proportionate to the highly upregulated PMM enzymatic activity, indicating that PMM is not a rate-limiting step of the L-galactose pathway in Arabidopsis [19]. Increased expression of the PMM gene in Arabidopsis was associated with enhanced tolerance to methyl viologen-induced stress [19] (Table 1).
2.1.3. GDP-D-Mannose Pyrophosphorylase (GMP)
The GMP (also known as VITAMIN C-1 [VTC1]) enzyme is responsible for the third enzymatic step of the L-galactose pathway, catalyzing a reversible conversion of D-mannose-1-P to GDP-D-mannose (Figure 2). Missense mutations in the Arabidopsis GMP gene reduced ascorbate to 25% of wild-type levels, supporting its important role in L-galactose ascorbate biosynthesis [21,22,23]. Increased expression of the GMP gene has increased ascorbate concentrations in a range of species, including 1.3-fold in Arabidopsis [24], 2.5-fold in tobacco [25], 1.4- to 1.7-fold in tomato (Solanum lycopersicum L.) [26,27], and 1.5-fold in rice [28] (Table 1). Increased co-expression of the GMP + GME genes in tomato increased ascorbate concentrations 2.0-fold, a greater increase than the individual genes alone, suggesting that GMP may act synergistically with GME to increase ascorbate concentrations in tomato [26]. Increased co-expression of the GMP + GME + GGP + GPP genes in tomato also increased ascorbate concentrations 2.0-fold; however, this was the same fold-change for increased expression of the GGP gene alone, suggesting that GMP is not a major rate-limiting step in the L-galactose pathway in tomato [26]. This conclusion is also supported by other studies increasing the expression of the GMP gene in Arabidopsis and tobacco and increasing the co-expression of the GMP + GME genes in tobacco that did not report significant changes in ascorbate concentrations [29,30] (Table 1).
2.1.4. GDP-D-Mannose-3′,5′-Epimerase (GME)
The GME enzyme is responsible for the fourth enzymatic step of the L-galactose pathway, catalyzing a reversible conversion of GDP-D-mannose to produce GDP-L-galactose (Figure 2). The GME enzyme also catalyzes a reversible conversion of GDP-D-mannose to produce GDP-L-gulose. Consequently, an alternative L-gulose pathway has been proposed to contribute towards ascorbate biosynthesis in plants (see Section 2.2) [31]. Knock-down of the GME gene in tomato by RNAi reduced ascorbate to 44% of wild-type levels, supporting its important role in L-galactose ascorbate biosynthesis [32]. Increased expression of the GME gene has increased ascorbate concentrations 1.4- to 1.9-fold in Arabidopsis [24,33,34], 1.4- to 1.8-fold in tomato [26,35], and 1.9-fold in rice [28]. Increased expression of the GMP + GME + GGP + GPP genes in tomato only increased ascorbate concentrations 2.0-fold, the same fold-change for increased expression of the GGP gene alone, suggesting that GME is not a major rate-limiting step in the L-galactose pathway in tomato [26]. Moreover, increased expression of the GME gene in tobacco did not significantly change ascorbate concentrations, suggesting the GME is not a rate-limiting step of the L-galactose pathway in tobacco [29]. Increased expression of the GME gene in plants has been associated with enhanced tolerance to salt, low pH, drought, cold, and methyl viologen-induced stress [28,33,35] (Table 1).
2.1.5. GDP-L-Galactose Phosphorylase (GGP)
The GGP (also known as VTC2/5) enzyme is responsible for the fifth enzymatic step of the L-galactose pathway, catalyzing the conversion of GDP-L-galactose to L-galactose1-P and represents the first committed step towards ascorbate biosynthesis, since GDP-D-mannose and GDP-L-galactose are also used as precursors for cell wall polysaccharides and glycoproteins (Figure 2). Splice junction and missense mutations in the Arabidopsis GGP gene reduced ascorbate to 10% of wild-type levels, supporting its critical role in L-galactose ascorbate biosynthesis [36,37,38,39]. The ggp1(vtc2)/ggp2(vtc5) double mutants in Arabidopsis also exhibit a seedling-lethal phenotype without the supplementation of L-galactose or ascorbate [38,39]. These results support the pivotal role of GGP in L-galactose ascorbate biosynthesis, as well as highlight the importance of L-galactose ascorbate biosynthesis for plant growth and development. Increased expression of the GGP gene has increased ascorbate concentrations in a wide range of species, including 2.9- to 4.1-fold in Arabidopsis [24,40], 3.1-fold in potato (Solanum tuberosum L.) [41], 2.1-fold in strawberry (Fragaria × ananassa) [41], 2.0- to 6.2-fold in tomato [26,41], 1.4-fold in tobacco [42], and 2.5- to 2.6-fold in rice [28,43] (Table 1). Increased co-expression of the GGP + L-GalLDH genes or the GGP + GPP genes increased ascorbate concentrations 3.6- and 4.1-fold, respectively, in Arabidopsis, a greater fold-change than increased expression of the GGP gene alone, suggesting that GGP may act synergistically with L-GalLDH and GPP in the L-galactose pathway [24]. However, increased co-expression of the GGP + GPP genes in tomato only increased ascorbate concentrations 1.8-fold, less than the fold-change for increased expression of the GGP gene alone, suggesting that GGP and GPP do not act synergistically in the L-galactose pathway in tomato [26]. Relative to other genes from the L-galactose pathway, increased expression of the GGP gene consistently leads to the greatest increase in ascorbate concentrations, providing strong genetic evidence that GGP represents the rate-limiting enzymatic step of the L-galactose pathway in many species [17,24,26,28,40]. Consistent with being the first committed step towards ascorbate biosynthesis and rate-limiting step of the L-galactose pathway, the strongest regulation of ascorbate concentrations in plants has been proposed to occur through the GGP gene [44]. Increased expression of the GGP gene in plants has been associated with enhanced tolerance to salt, ozone, and cold stress [28,42,43] (Table 1).
2.1.6. L-Galactose-1-Phosphate Phosphatase (GPP)
The GPP (also known as VTC4) enzyme is responsible for the sixth enzymatic step of the L-galactose pathway, catalyzing the conversion of L-galactose1-P to L-galactose (Figure 2). A missense mutation in the Arabidopsis GPP gene reduced ascorbate to 50% of wild-type levels, supporting its important role in L-galactose ascorbate biosynthesis [36,45]. Increased expression of the GPP gene has increased ascorbate concentrations 1.5-fold in Arabidopsis [24], 1.7-fold in tomato [26], and 1.4-fold in rice [28] (Table 1). Increased co-expression of the GGP + GPP genes or the GMP + GME + GGP + GPP genes in tomato did not increase ascorbate concentrations higher than increased expression of the GGP gene alone, indicating that GPP is not a major rate-limiting step in the L-galactose pathway in tomato [26].
2.1.7. L-Galactose Dehydrogenase (L-GalDH)
The L-GalDH enzyme is responsible for the penultimate enzymatic step of the L-galactose pathway, catalyzing the conversion of L-galactose to L-galactono-1,4-lactone (Figure 2). Knock-down of the L-GalDH gene in Arabidopsis with RNAi had no effect on ascorbate concentrations under low light conditions, but reduced ascorbate to 45% of wild-type levels under high light conditions when a larger pool of ascorbate is required, supporting its role in L-galactose ascorbate biosynthesis [46]. Increased expression of the L-GalDH gene increased ascorbate concentrations 1.2-fold in Arabidopsis [24] and 1.7-fold in rice [28] (Table 1). However, increased expression of the L-GalDH gene in tobacco did not significantly change ascorbate concentrations, despite having a 3.5-fold increase in L-GalDH enzymatic activity, indicating that L-GalDH is not a rate-limiting step in the L-galactose pathway in tobacco [46].
2.1.8. L-Galactono-1,4-Lactone Dehydrogenase (L-GalLDH)
The L-GalLDH enzyme is responsible for the final enzymatic step of the L-galactose pathway, catalyzing the conversion of L-galactono-1,4-lactone to ascorbate (Figure 2). Similar to the ggp1(vtc2)/ggp2(vtc5) double mutants in Arabidopsis, loss-of-function Arabidopsis l-galldh mutants exhibited a seedling lethal phenotype in the absence of ascorbate supplementation, supporting the critical role of L-GalLDH in L-galactose ascorbate biosynthesis [47]. Increased expression of the L-GalLDH gene has increased ascorbate concentrations in a range of species, including 1.8-fold in Arabidopsis [24], 1.3- to 3.2-fold in lettuce (Lactuca sativa L.) [48,49], 7.0-fold in lily (Lilium davidii) [50], 1.3- to 1.5-fold in rice [28,51], and 2.1-fold in tobacco [52] (Table 1). One study, however, found that increased expression of the L-GalLDH gene in tobacco did not significantly change ascorbate concentrations, despite having 10-fold higher L-GalLDH enzymatic activity, suggesting that L-GalLDH is not a major rate-limiting step of the L-galactose pathway in tobacco [53]. Increased co-expression of the GGP + L-GalLDH genes increased ascorbate concentrations 3.6-fold in Arabidopsis, a greater fold-change than increased expression of each gene alone, suggesting that L-GalLDH may act synergistically with GGP in the L-galactose pathway in Arabidopsis [24]. Increased expression of the L-GalLDH gene in tobacco was associated with enhanced tolerance to salt and methyl viologen-induced stress [52] (Table 1).
2.2. The L-Gulose Pathway
The L-gulose pathway (also known as the L-gulose shunt) is a result of the epimerization of GDP-D-mannose to GDP-L-gulose by GME, in addition to the epimerization of GDP-D-mannose to GDP-L-galactose [31] (Figure 2). In this pathway, GDP-L-gulose is proposed to be converted to the ascorbate precursor L-gulono-1,4-lactone via L-gulose-1-P and L-gulose intermediates; however, the genes and enzymes responsible for catalyzing these steps in plants have not yet been identified. The GGP and L-GalDH enzymes from the L-galactose pathway have been proposed to catalyze these reactions; however, GGP does not appear to have any enzymatic activity on GDP-L-gulose and L-GalDH has a much lower affinity for L-gulose than L-galactose [46,54,55]. L-gulono-1,4-lactone is then proposed to be converted to ascorbate by L-gulono-1,4-lactone oxidase (L-GulLO) (Figure 2). Increased expression of the rat (Rattus rattus L.) L-GulLO gene has increased ascorbate concentrations in a range of species, including 1.8- to 2.0-fold in Arabidopsis [56,57], 7.0-fold in lettuce [58], 2.4-fold in potato [59], 7.0-fold in tobacco [58], and 1.5-fold in tomato [60] (Table 1). However, increased expression of the Arabidopsis L-GulLOs genes in Arabidopsis did not change ascorbate concentrations [61] (Table 1) and increased expression of the Arabidopsis L-GulLOs genes in tobacco cells only increased ascorbate concentrations when supplied with L-gulono-1,4-lactone, together suggesting that the L-gulose pathway is not a major contributor to ascorbate biosynthesis in plants [62]. Increased expression of the rat L-GulLO gene in plants has been associated with enhanced tolerance to salt, cold, heat, drought, and pyrene- and methyl viologen-induced stress [56,59,60] (Table 1).
2.3. The Myo-Inositol Pathway
The myo-inositol pathway (also known as the D-gluconorate pathway), which operates in animals, has also been proposed to be present in plants (Figure 2). In this pathway, myo-inositol is proposed to be converted to the ascorbate precursor L-gulono-1,4-lactone via D-gluconorate and D-gulonate intermediates; however, the gene and enzyme responsible for catalyzing the conversion of D-gluconorate to D-gulonate have not yet been identified (Figure 2). L-gulono-1,4-lactone is then proposed to be converted to ascorbate by L-GulLO (Figure 2). The contribution of the myo-inositol pathway towards ascorbate biosynthesis in plants has been controversial. Supporting its contribution are studies that increased the expression of the myo-inositol oxygenase (MIOX) gene, which encodes the enzyme responsible for catalyzing the conversion of myo-inositol to D-glucuronate (Figure 2). For example, increased expression of the MIOX gene in Arabidopsis increased ascorbate concentrations 1.6- to 3.0-fold, and was associated with enhanced tolerance to salt, cold, heat, and pyrene-induced stress [56,63,64] (Table 1). Varying results were obtained when the expression of the MIOX gene was increased in tomato, with ascorbate reduced to 50% of wild-type levels in leaves, but increased 1.4- and 1.3-fold in green and red fruit, respectively, indicating that the myo-inositol pathway may be tissue-specific in tomato [27]. Further support for the myo-inositol pathway comes from increased expression of phytases that are proposed to increase the pool of myo-inositol, which can then serve as an entry point for ascorbate biosynthesis via the myo-inositol pathway. For example, increased expression of the Arabidopsis purple acid phosphatase 15 (PAP15) gene [65] and the bacterial beta-propeller phytase PHY-US417 gene [66] in Arabidopsis has increased ascorbate concentrations 2.0- to 2.3-fold and were associated with enhanced tolerance to drought and salt stress. Loss-of-function mutations in the Arabidopsis PAP15 gene reduced ascorbate to 70% of wild-type levels, supporting the contribution of PAP15 towards myo-inositol ascorbate biosynthesis [65]. The PAP15 gene, however, has also been proposed to encode a GPP in Arabidopsis, which may provide an alternative explanation for these results [13]. While there are several studies supporting the contribution of the myo-inositol pathway towards ascorbate biosynthesis in plants, there are equally as many studies opposing its contribution. For example, increased expression of the MIOX gene in Arabidopsis [67] and rice [68] did not change ascorbate concentrations, despite the Arabidopsis lines with increased MIOX expression having higher MIOX enzymatic activity (Table 1). Further, loss-of-function mutations in the Arabidopsis [69] and rice [70] MIOX genes did not change ascorbate concentrations relative to wild-type, challenging the view that the myo-inositol pathway contributes towards ascorbate biosynthesis. Further opposition comes from a recent study utilizing CRISPR/Cas9-induced knockout mutations in the Arabidopsis glucuronokinase1 (GlcAK1) gene and radiotracer experiments with 3H-myo-inositol. Knockout of GlcAK1 ought to increase the flux from myo-inositol to ascorbate if the myo-inositol pathway is operational; however, no changes in ascorbate concentrations were detected in the Arabidopsis glcak1 mutants. Moreover, radiolabeled 3H-myo-inositol in the Arabidopsis glcak1 mutants accumulated as D-gluconorate and ascorbate remained unlabeled, leading the authors to conclude that the myo-inositol pathway does not contribute towards ascorbate biosynthesis in plants [71].
2.4. The D-Galacturonate Pathway
Evidence for the D-galacturonate pathway was discovered in strawberry fruits [72]. In this pathway, D-galacturonate derived from pectin in the cell wall is proposed to be converted to the ascorbate precursor L-galactono-1,4-lactone via a L-galactonate intermediate; however, the gene and enzyme responsible for converting L-galactonate to L-galactono-1,4-lactone have not yet been identified in plants (Figure 2). L-galactono-1,4-lactone is then proposed to be converted to ascorbate by L-GalLDH, the final enzymatic step of the L-galactose pathway (Figure 2). To date, the D-galacturonate reductase (D-GalUR) gene, which encodes the enzyme responsible for catalyzing the conversion of D-galacturonate to L-galactonate, has only been identified in strawberry (Figure 2). Knock-out or knock-down studies of the D-GalUR gene in strawberry or other plant species have not yet been reported, making it difficult to determine the relative contribution of the D-galacturonate pathway towards ascorbate biosynthesis in plants. Nevertheless, increased expression of the D-GalUR gene has increased ascorbate concentrations 3.0-fold in Arabidopsis [72], 1.7- to 2.1-fold in potato [73,74,75], and 1.0- to 2.5-fold in tomato [76,77,78]. Increased expression of the D-GalUR gene in plants has been associated with enhanced tolerance to salt, drought, cold, zinc chloride, and methyl viologen-induced stress [73,74,75,77,78] (Table 1).
3. Recycling of Ascorbate in Plants
In addition to de novo biosynthesis of ascorbate, oxidized ascorbate can be regenerated via the recycling enzymes MDAR and DHAR to maintain cellular ascorbate levels (Figure 1). Following ascorbate oxidation, a short-lived monodehydroascorbate radical is produced. Monodehydroascorbate can then be recycled back to ascorbate by MDAR using NADPH as a reductant, however if not recycled monodehydroascorbate will rapidly undergo non-enzymatic disproportionation with another monodehydroascorbate molecule to produce an ascorbate and dehydroascorbate molecule [81]. Dehydroascorbate can then be recycled back to ascorbate by DHAR using glutathione as a reductant, however if not recycled dehydroascorbate will undergo an irreversible hydrolysis to generate 2,3-diketogulonic acid [81]. An overview of metabolic engineering strategies utilizing ascorbate recycling genes to increase ascorbate concentrations in model and crop species, including the tissue examined and any stress tolerances observed, is summarized in Table 2.

Table 2.
Overview of metabolic engineering strategies utilizing ascorbate recycling genes to increase ascorbate concentrations in model and crop species.
Loss-of-function mutations in the Arabidopsis MDAR4 gene reduced the ratio of ascorbate to dehydroascorbate but did not affect the pool size of total ascorbate (ascorbate + dehydroascorbate), supporting the role of MDAR in ascorbate recycling [82]. Increased expression of the MDAR gene has increased the concentrations of reduced ascorbate 1.2- to 2.2-fold in tobacco [83,84,85] and 1.2-fold in tomato [86] (Table 2). However, other studies increasing the expression of the MDAR gene in tobacco [87] and tomato [88] did not report any significant changes to reduced ascorbate concentrations, indicating that MDAR may not be a rate-limiting step in the recycling of ascorbate in these species (Table 2). Increased expression of the MDAR gene in plants has been associated with enhanced tolerance to salt, ozone, drought, cold, heat, and methyl viologen-induced stress [83,84,86,87] (Table 2).
Similar to the mdar4 mutant in Arabidopsis, loss-of-function mutations in the Arabidopsis DHAR3 gene reduced the ratio of ascorbate to dehydroascorbate but did not affect the pool size of total ascorbate, supporting the role of DHAR in ascorbate recycling [89]. Increased expression of the DHAR gene has increased the concentrations of reduced ascorbate in a wide range of species, including 1.1- to 3.3-fold in Arabidopsis [34,90,91,92,93,94], 1.9- to 6.1-fold in maize (Zea mays L.) [95,96], 1.3- to 2.8-fold in potato [97,98,99], 1.7-fold in rice [100], 1.3- to 3.9-fold in tobacco [85,95,101,102,103,104,105,106], and 1.5-fold to 1.9-fold in tomato [88,107,108] (Table 2). Increased expression of the DHAR gene in plants has been associated with enhanced tolerance to salt, drought, high light, heat, ozone, cold, aluminum, hydrogen peroxide, and methyl viologen-induced stress [85,90,91,92,93,94,98,101,102,103,104,105,106,107,108] (Table 2).
4. Regulation of Ascorbate Biosynthesis in Plants
The genes and enzymes responsible for the major biosynthetic route towards ascorbate biosynthesis—the L-galactose pathway—and the recycling of ascorbate in plants have been well characterized for over a decade now [37,55]. The regulation of ascorbate biosynthesis and recycling, on the other hand, has remained relatively unexplored until more recently. Several key regulators have now been identified, and it is becoming clear that ascorbate biosynthesis and recycling is tightly regulated at the transcriptional, translational, and post-translational level [44]. An overview of metabolic engineering strategies utilizing ascorbate regulatory factors to increase ascorbate concentrations in model and crop species, including the tissue examined and any stress tolerances observed, is summarized in Table 3.

Table 3.
Overview of metabolic engineering strategies utilizing ascorbate regulatory factors to increase ascorbate concentrations in model and crop species.
4.1. Ascorbic Acid Mannose Pathway Regulator 1 (AMR1)
The ascorbic acid mannose pathway regulator 1 (AMR1) gene was identified in an activation-tagged Arabidopsis mutant that contained 40% of wild-type ascorbate levels [109]. The AMR1 gene encodes an F-box protein that negatively regulates the transcription of genes of the L-galactose pathway, including GMP, GME, GGP1, GPP, L-GalDH, and L-GalLDH [109]. The expression of the AMR1 gene was developmentally and environmentally controlled with increased expression as the plants aged and decreased expression under high light [109]. Loss-of-function mutations in the AMR1 gene in Arabidopsis increased ascorbate concentrations 2.0-fold and were associated with enhanced ozone stress [109] (Table 3). Disrupting the function of the AMR1 gene with genome editing tools presents one possible transgene-free strategy to increase ascorbate concentrations in crops.
4.2. Basic Helix-Loop-Helix 59 Transcription Factor (bHLH59)
The tomato basic helix-loop-helix 59 (bHLH59) gene was recently identified as the likely candidate underlying the ascorbate quantitative trait locus TFA9 in tomato [110]. The bHLH59 transcription factor positively regulates the transcription of genes of the L-galactose pathway, including PMI, PMM, GMP1, GMP2, GMP3, GMP4, and GME1 [110]. Knock-down of the bHLH59 gene with RNAi in tomato reduced ascorbate to 65% of wild-type levels [110]. Increased expression of the bHLH59 gene in tomato increased ascorbate concentrations 1.5-fold and was associated with enhanced tolerance to methyl viologen-induced stress [110] (Table 3).
4.3. Calmodulin-Like 10 (CML10)
A calcium sensor, calmodulin-like 10 (CML10), was recently identified to interact with Arabidopsis PMM in a yeast two-hybrid screen [111]. The expression of the CML10 gene was upregulated under oxidative stress and the CML10 protein increased PMM enzymatic activity in a calcium-dependent manner [111]. Knock-down of the CML10 gene in Arabidopsis by RNAi reduced ascorbate to 70% of wild-type levels and was associated with decreased drought and H2O2-induced stress [111]. To date, no studies have investigated increased expression of the CML10 gene in plants.
4.4. COP9 Signalosome Subunit 5B and 8 (CSN5B and CSN8)
The Arabidopsis COP9 signalosome subunit 5B (CSN5B), a component of the photomorphogenic COP9 signalosome, was identified to interact with GMP in a yeast two-hybrid screen [112]. Under darkness, CSN5B promotes the ubiquitination and degradation of GMP through the 26S proteasome pathway [112]. Loss-of-function mutations in the CSN5B gene in Arabidopsis increased ascorbate concentrations 1.4-fold and were associated with enhanced salt stress [112] (Table 3). Loss-of-function mutations in the CSN8 gene in Arabidopsis also increased ascorbate concentrations 1.8-fold (Table 3); however, CSN8 did not interact with GMP in the yeast-two hybrid screen and its mechanism of action has yet to be determined [112]. Disrupting the function of the CSN5B or CSN8 genes with genome editing tools presents another transgene-free strategy to increase ascorbate concentrations in crops.
4.5. DNA-Binding with One Finger 22 (Dof22)
The DNA-binding with one finger 22 (Dof22), a member of the Dof family of plant specific transcription factors, has recently been proposed as a negative transcriptional regulator of L-galactose pathway, ascorbate recycling, and ascorbate-glutathione cycle genes in tomato, such as GGP1, GGP2, L-GalDH, L-GalLDH, MDAR, cytosolic APX, and glutathione reductase (GR) [113]. Knock-down of the Dof22 gene with RNAi in tomato increased ascorbate concentrations up to 1.6-fold (Table 3), but was associated with reduced tolerance to salt stress due to the transcriptional downregulation of the salt overly sensitive 1 (SOS1) gene, which encodes a plasma membrane Na+/H+ antiporter [113].
4.6. Ethylene Response Factor 98 (ERF98)
In a reverse genetic screen of a pool of Arabidopsis ethylene response factor (ERF) T-DNA insertion mutants, loss-of-function mutations in the ERF98 gene reduced ascorbate to 65% of wild-type levels [114]. The ERF98 gene encodes a transcription factor belonging to the AP2/ERF superfamily that positively regulates the transcription of ascorbate biosynthetic, ascorbate recycling, and ascorbate-glutathione cycle genes, including GMP, GGP1, L-GalDH, L-GalLDH, MIOX4, MDAR3, chloroplastic DHAR, cytosolic DHAR, and GR1 [114]. Increased expression of the ERF98 gene in Arabidopsis increased ascorbate concentrations 1.7-fold and was associated with enhanced tolerance to salt stress [114] (Table 3).
4.7. GGP Upstream Open Reading Frame (GGP uORF)
The translation of GGP has been discovered to be post-transcriptionally regulated through a highly conserved, cis-acting upstream open reading frame (uORF) in the long 5’ leader sequence of the GGP mRNA [115,116]. The GGP uORF is proposed to initiate from a non-canonical AUC or ACG start-codon and encode a 60- to 65-residue long peptide [115]. Disruption of the GGP uORF increased ascorbate concentrations when a GGP promoter-uORF-GGP construct was transiently transformed in Nicotiana benthamiana [115]. It is proposed that under high ascorbate concentrations, the GGP uORF is translated and causes ribosomal stalling, thereby preventing translation of the GGP major ORF (mORF), while under low ascorbate concentrations, the uORF is skipped and the GGP mORF is translated [115]. The precise mechanism of how ascorbate influences the translation of the GGP uORF or mORF, however, is yet to be determined. Recently, disruption of the GGP uORF with the CRISPR/Cas9 genome editing system increased ascorbate concentrations 1.7-fold in Arabidopsis [117], 1.4- to 2.6-fold in lettuce [117], and 1.4-fold in tomato [118] (Table 3). Moreover, disruption of the lettuce GGP uORFs was associated with enhanced tolerance to methyl viologen-induced stress [117] (Table 3). Disrupting the GGP uORF with the CRISPR/Cas9 genome editing system has established itself as a very promising transgene-free strategy to engineer plants with enhanced production of GGP for increased ascorbate biosynthesis.
4.8. HD-Zip I Family Transcription Factor 24 (HZ24)
The HD-Zip I family transcription factor 24 (HZ24) was recently identified based on a yeast-one hybrid assay to the promoter region of tomato GMP3 [119]. Knock-down mutants of the HZ24 gene with RNAi in tomato reduced ascorbate to 60% of wild-type levels [119]. The HZ24 transcription factor positively regulates the transcription of a wide range of genes from the L-galactose pathway in tomato, including PMM, GMP4, GME1, GME2, GGP, GPP1, GPP2, and L-GalDH [119]. Increased expression of the HZ24 gene in tomato increased ascorbate concentrations 1.5-fold and was associated with enhanced tolerance to methyl viologen-induced stress [119] (Table 3).
4.9. High-Pigment-1 (HP1)
The High-Pigment-1 (HP1) tomato mutants—previously characterized as having increased carotenoid and flavonoid concentrations—were found to have reduced ascorbate concentrations at various stages of fruit development and ripening relative to wild-type [120]. The HP1 gene is orthologous to the Arabidopsis UV-DAMAGED DNA-BINDING PROTEIN 1 (DDB1) gene which encodes a component of the CUL4-based E3 ligase complex [121]. Fourteen genes involved in ascorbate biosynthesis and recycling were differentially expressed in the developing fruit of the hp1 mutant and it was proposed that HP1 positively regulates the transcription of the GMP and GME1 genes, but negatively regulates the transcription of the L-GalLDH gene in tomato, ultimately resulting in the reduced ascorbate concentrations observed in the developing and ripening fruit [120]. To date, no studies have investigated the effect of increasing the expression of the HP1 gene has on ascorbate concentrations in plants.
4.10. KONJAC 1 and 2 (KJC1 and KJC2)
Two nucleotide sugar phosphorylase-like proteins, KONJAC 1 and 2 (KJC1 and KJC2), were identified based on their phylogenetic relationship to GMP [30]. Loss-of-function mutations in the KJC1 and KJC2 genes in Arabidopsis reduced ascorbate to 40% and 70% of wild-type levels, respectively, due to reducing GMP enzymatic activity to 10% and 70% of wild-type levels, respectively [30]. The KJC1 and KJC2 proteins are proposed to post-translationally regulate GMP enzymatic activity and may function opposite that of the COP9 signalosome in the regulation of GDP-D-mannose biosynthesis [30]. Increased expression of the KJC1 gene increased ascorbate concentrations 1.4-fold in Arabidopsis but increased co-expression of the KJC1 and GMP genes or increased expression of the KJC2 gene did not change ascorbate concentrations [30] (Table 3).
4.11. Myeloblastosis Transcription Factor 5 (MYB5)
Pear (Pyrus betulaefolia) myeloblastosis transcription factor 5 (MYB5) was recently identified in a yeast one-hybrid screen to interact with the promoter region of the DHAR2 gene [122]. The expression of the MYB5 gene was induced by a wide range of stresses, including cold, drought, and salt stress [122]. The MYB5 protein positively regulates the transcription of ascorbate recycling, ascorbate-glutathione cycle, and stress-associated genes, including MDAR, DHAR2, APX, cystathionine-β-synthase 1 (CBF1), CBF2, and CBF3 [122]. Increased expression of the MYB5 gene in tobacco increased ascorbate concentrations 1.3-fold and was associated with enhanced tolerance to cold stress [122] (Table 3).
4.12. NBS-LRR 33 (NL33)
The tomato NBS-LRR 33 (NL33) resistance protein was recently identified based on a candidate gene-based association study with ascorbate concentrations in tomato [123]. The NL33 protein negatively regulates the transcription of ascorbate biosynthetic, ascorbate recycling, and ascorbate-glutathione cycle genes, including PMM, GMP1, GMP4, GME1, GGP1, GPP1, GPP2, L-GalDH, L-GalLDH, MIOX, MDAR, DHAR, and cytosolic APX [123]. Knock-down of the NL33 gene with RNAi in tomato increased ascorbate concentrations 2.7-fold and was associated with enhanced tolerance to methyl viologen-induced stress and gray mold infection (Botrytis cinerea) [123] (Table 3). Disrupting the function of the NL33 gene with genome editing tools represents another transgene-free strategy to increase ascorbate concentrations in crops.
4.13. Non-specific Lipid Transfer Protein-1 (nsLTP1)
The potato thermo-tolerance gene non-specific lipid transfer protein-1 (nsLTP1) was first identified in a functional screen of potato high-temperature stress-responsive genes to impart heat tolerance in yeast [124]. The nsLTP1 protein in potato has now recently been identified to positively regulate the transcription of ascorbate-glutathione cycle and stress-related genes, including APX, catalase (CAT), superoxide dismutase (SOD), heat shock protein 20 (HSP20), HSP70, and HSP90 [125]. Increased expression of the nsLTP1 gene increased ascorbate concentrations 2.3-fold in potato and was associated with enhanced tolerance to heat, drought, and salt stress [125] (Table 3).
4.14. VITAMIN C-3 (VTC3)
The Arabidopsis vtc3 mutants were first identified in a forward genetic screen for ozone-sensitive mutants and contained 40% of wild-type ascorbate levels [36]. Over a decade later, the VTC3 gene was mapped to a unique dual-function polypeptide containing an N-terminal protein kinase domain and a C-terminal protein phosphatase domain [126]. The Arabidopsis vtc3 mutants were reported to be defective in their ability to increase ascorbate concentrations in response to light and heat [126]. Little is known about the mechanism of action for VTC3, but it is proposed to act as a signal transduction protein that can perceive a change in the environment to influence cellular ascorbate levels [126]. It has been suggested that VTC3 may play a role in the GGP uORF sensing mechanism of ascorbate levels [44]. Increased expression of the VTC3 gene in the Arabidopsis vtc3 mutants recovered ascorbate concentrations back to wild-type levels; however, increased expression of the VTC3 gene in wild-type Arabidopsis did not significantly change ascorbate concentrations [126] (Table 3).
4.15. WAX1
The saltwater cress (Eutrema salsugineum) WAX1 gene was identified based on being rapidly induced in abiotic stress-treated saltwater cress plants [127]. The WAX1 gene encodes a MYB transcription factor that positively regulates wax related genes, as well as ascorbate biosynthetic genes, including GGP1, L-GalDH, and MIOX4 [127]. Increased expression of the WAX1 gene with the constitutive super-promoter in Arabidopsis increased ascorbate concentrations 1.3-fold but severely disrupted plant growth and development [127] (Table 3). Increased expression of the WAX1 gene with the Arabidopsis RD29A stress-inducible promoter, instead of the constitutive super-promoter, increased ascorbate concentrations 1.4-fold under drought stress and was associated with enhanced tolerance to drought stress [127] (Table 3).
4.16. Zinc-Finger 3 (ZF3)
The tomato C2H2-type zinc finger 3 (ZF3) gene was first identified based on being induced in salt-stressed tomato plants [128]. The ZF3 protein in tomato has now recently been identified to interact with CSN5B and inhibit the interaction between CSN5B and GMP [129]. By doing so, ZF3 prevents the ubiquitination and degradation of GMP through the 26S proteasome pathway, promoting the accumulation of GMP [129]. Knock-down of the ZF3 gene with RNAi in tomato reduced ascorbate to 80% of wild-type levels [129]. Increased expression of the tomato ZF3 gene increased ascorbate concentrations 1.8-fold and 2.1-fold in Arabidopsis and tomato, respectively, and was associated with enhanced tolerance to salt stress [129] (Table 3).
5. Conclusions and Future Perspectives
Increased expression of single ascorbate biosynthetic genes has been the most utilized and effective strategy to increase ascorbate concentrations in plants and has been associated with enhanced tolerance to multiple abiotic stresses. Increased expression of the GGP gene from the L-galactose pathway has consistently generated the largest increases in ascorbate concentrations in a range of species relative to other ascorbate biosynthetic genes. Increased co-expression of the GGP gene with other biosynthetic genes from the L-galactose pathway should provide further opportunities to increase ascorbate concentrations in crops to greater than that of the GGP gene alone. For example, synergistic effects on ascorbate concentrations were observed when the co-expression of the GGP + GPP genes or the GGP + L-GalLDH genes were increased in Arabidopsis [24] and the transient co-expression of the GGP + GME genes were increased in N. benthamiana [40]. A recent study that pyramided GMP1 + GME2 + GGP1 + GPP1 in tomato, however, observed the same fold increase in ascorbate concentrations for that of the GGP gene alone. Further research is therefore warranted into increased co-expression of ascorbate biosynthetic genes in plants to better understand precisely which combination of genes leads to the greatest fold increases in ascorbate concentrations.
Increased expression of single ascorbate recycling genes has been the second most common strategy to increase ascorbate concentrations in plants and has also been associated with enhanced tolerance to a broad range of abiotic stress. Increased expression of the DHAR gene has consistently generated increased concentrations of reduced ascorbate in a range of species, whereas increased expression of the MDAR gene has not always increased reduced ascorbate concentrations. Increased co-expression of the DHAR gene with ascorbate biosynthetic genes, such as the GGP gene, could present one avenue to increase not only the concentration of total ascorbate, but also to maintain the ascorbate in the reduced, active form for improved stress tolerance.
More recently, the manipulation of ascorbate regulatory factors has emerged as a third strategy to increase ascorbate concentrations in crops. Typically, this strategy has focused on increased expression of a single transcriptional activator of ascorbate biosynthetic genes. However, we propose that more focus should be paid to the discovery and manipulation of transcriptional and translational repressors of ascorbate biosynthetic genes or regulatory factors that target ascorbate biosynthetic proteins for degradation, since they can be targeted for disruption with genome editing tools as an efficient approach to produce transgene-free crops. Genome edited plants offer many advantages over transgenic plants, for example, by avoiding the occurrence of transgene silencing and the random insertion of T-DNAs [130]. Moreover, since plants with genome edited-induced indels (small insertions and deletions) due to errors in non-homologous end joining do not rely on the presence of foreign DNA, they are less likely to be subjected to regulatory oversight. Disrupting the cis-acting GGP uORF in Arabidopsis, lettuce, and tomato with the CRISPR/Cas9 genome editing system was recently reported to significantly increase ascorbate concentrations, as well as improve methyl viologen-induced oxidative stress tolerance in the case of lettuce [117,118]. As the GGP uORF is highly conserved from mosses to angiosperms, editing the GGP uORF represents a promising transgene-free strategy to increase ascorbate concentrations for enhanced stress tolerance in most crops [115,116]. Other ascorbate regulatory factors that could be targeted with genome editing include AMR1, CSN5B, CSN8, and NL33, which have all been reported to increase ascorbate concentrations and enhance stress tolerance when disrupted with T-DNA insertions or downregulated with RNAi [109,112,123].
Finally, it is worth noting that increasing ascorbate concentrations in crops does not come without its challenges [131]. For example, increased expression of ascorbate biosynthetic and recycling genes has been reported to reduce guard cell responsiveness [132], disturb embryo development [133], and induce parthenocarpic (seedless) fruit in tomato [41]. A fine balance must therefore be struck when increasing ascorbate concentrations in crops in order to minimize any deleterious effects on plant growth and development that may arise due to perturbed ascorbate concentrations. Generating mutant alleles of varying strength in the GGP uORF with the CRISPR/Cas9 genome editing system represents an attractive strategy to fine-tune ascorbate concentrations in crops for enhanced stress tolerance, whilst minimizing any pleotropic effects on plant growth and development [117,118,130].
Author Contributions
R.C.B. drafted the manuscript. J.P.B., R.P.H., and A.A.T.J. reviewed the manuscript and supervised the research. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 1O2 | singlet oxygen |
| AMR | ascorbic acid mannose pathway regulator |
| APX | ascorbate peroxidase |
| bHLH | basic helix-loop-helix |
| CAT | catalase |
| CBF | cystathionine-β-synthase |
| CML | calmodulin-like |
| CSN | COP9 signalosome |
| DDB | UV-DAMAGED DNA-BINDING PROTEIN |
| D-GalUR | D-galacturonate reductase |
| DHAR | dehydroascorbate reductase |
| Dof | DNA-binding with one finger |
| ERF | ethylene response factor |
| GGP | GDP-L-galactose phosphorylase |
| GlcAK | glucuronokinase |
| GME | GDP-D-mannose-3′,5′-epimerase |
| GMP | GDP-D-mannose pyrophosphorylase |
| GPP | L-galactose-1-phosphate phosphatase |
| GR | glutathione reductase |
| H2O2 | hydrogen peroxide |
| HO− | hydroxyl radical |
| HP | High-Pigment |
| HSP | heat shock protein |
| HZ | HD-Zip I family transcription factor |
| KJC | KONJAC |
| L-GalDH | L-galactose dehydrogenase |
| L-GalLDH | L-galactono-1,4-lactone dehydrogenase |
| L-GulLO | L-gulono-1,4-lactone oxidase |
| MDAR | monodehydroascorbate reductase |
| MIOX | myo-inositol oxygenase |
| mORF | major ORF |
| MYB | myeloblastosis transcription factor |
| NL | NBS-LRR |
| nsLTP | non-specific lipid transfer protein |
| O2− | superoxide |
| PAP | purple acid phosphatase |
| PMI | phosphomannose isomerase |
| ROS | reactive oxygen species |
| SOD | superoxide dismutase |
| SOS | salt overly sensitive |
| uORF | upstream open reading frame |
| VTC | VITAMIN C |
| ZF | Zinc-finger |
References
- Ashraf, M.; Wu, L. Breeding for salinity tolerance in plants. Crit. Rev. Plant Sci. 1994, 13, 17–42. [Google Scholar] [CrossRef]
- United Nations Department of Economic and Social Affairs Population Division. World Population Prospects 2019: Highlights; ST/ESA/SER.A/423; United Nations: New York, NY, USA, 2019; pp. 1–39. [Google Scholar]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Carvalho, M.d. Drought stress and reactive oxygen species. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. The roles of reactive oxygen metabolism in drought: Not so cut and dried. Plant Physiol. 2014, 164, 1636–1648. [Google Scholar] [CrossRef]
- Abogadallah, G.M. Insights into the significance of antioxidative defense under salt stress. Plant Signal. Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef]
- Suzuki, N.; Mittler, R. Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction. Physiol. Plant 2006, 126, 45–51. [Google Scholar] [CrossRef]
- Zechmann, B. Subcellular distribution of ascorbate in plants. Plant Signal. Behav. 2011, 6, 360–363. [Google Scholar] [CrossRef]
- Zechmann, B.; Stumpe, M.; Mauch, F. Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta 2011, 233, 1–12. [Google Scholar] [CrossRef]
- Bartoli, C.G.; Buet, A.; Grozeff, G.G.; Galatro, A.; Simontacchi, M. Ascorbate-glutathione cycle and abiotic stress tolerance in plants. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M.A., Munné-Bosch, S., Eds.; Springer: New York, NY, USA, 2017; pp. 177–200. [Google Scholar]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Smirnoff, N.; Wheeler, G.L. Ascorbic acid in plants: Biosynthesis and function. Crit. Rev. Plant Sci. 2000, 19, 267–290. [Google Scholar] [CrossRef]
- Ishikawa, T.; Maruta, T.; Yoshimura, K.; Smirnoff, N. Biosynthesis and regulation of ascorbic acid in plants. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D.K., Palma, J.M., Eds.; Springer: New York, NY, USA, 2018; pp. 163–179. [Google Scholar]
- Yoshimura, K.; Ishikawa, T. Chemistry and metabolism of ascorbic acid in plants. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M.A., Munné-Bosch, S., Eds.; Springer: New York, NY, USA, 2017; pp. 1–23. [Google Scholar]
- Siendones, E.; González-Reyes, J.A.; Santos-Ocana, C.; Navas, P.; Córdoba, F. Biosynthesis of ascorbic acid in kidney bean. L-Galactono-γ-lactone dehydrogenase is an intrinsic protein located at the mitochondrial inner membrane. Plant Physiol. 1999, 120, 907–912. [Google Scholar] [CrossRef]
- Maruta, T.; Yonemitsu, M.; Yabuta, Y.; Tamoi, M.; Ishikawa, T.; Shigeoka, S. Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid biosynthesis. J. Biol. Chem. 2008, 283, 28842–28851. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, K.; Nakane, T.; Kume, S.; Shiomi, Y.; Maruta, T.; Ishikawa, T.; Shigeoka, S. Transient expression analysis revealed the importance of VTC2 expression level in light/dark regulation of ascorbate biosynthesis in Arabidopsis. Biosci. Biotechnol. Biochem. 2014, 78, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Hoeberichts, F.A.; Vaeck, E.; Kiddle, G.; Coppens, E.; Van De Cotte, B.; Adamantidis, A.; Ormenese, S.; Foyer, C.H.; Zabeau, M.; Inzé, D. A temperature-sensitive mutation in the Arabidopsis thaliana phosphomannomutase gene disrupts protein glycosylation and triggers cell death. J. Biol. Chem. 2008, 283, 5708–5718. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Yu, C.; Qin, H.; Liu, X.; Zhang, A.; Johansen, I.E.; Wang, D. Molecular and functional analysis of phosphomannomutase (PMM) from higher plants and genetic evidence for the involvement of PMM in ascorbic acid biosynthesis in Arabidopsis and Nicotiana benthamiana. Plant J. 2007, 49, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Badejo, A.A.; Eltelib, H.A.; Fukunaga, K.; Fujikawa, Y.; Esaka, M. Increase in ascorbate content of transgenic tobacco plants overexpressing the acerola (Malpighia glabra) phosphomannomutase gene. Plant Cell Physiol. 2009, 50, 423–428. [Google Scholar] [CrossRef]
- Conklin, P.L.; Norris, S.R.; Wheeler, G.L.; Williams, E.H.; Smirnoff, N.; Last, R.L. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 4198–4203. [Google Scholar] [CrossRef]
- Conklin, P.L.; Pallanca, J.E.; Last, R.L.; Smirnoff, N. L-ascorbic acid metabolism in the ascorbate-deficient Arabidopsis mutant vtc1. Plant Physiol. 1997, 115, 1277–1285. [Google Scholar] [CrossRef]
- Conklin, P.L.; Williams, E.H.; Last, R.L. Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proc. Natl. Acad. Sci. USA 1996, 93, 9970–9974. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, Q.; Wang, Z.; Fan, R.; Li, Y.; Sun, X.; Tang, K. Engineering ascorbic acid biosynthetic pathway in Arabidopsis leaves by single and double gene transformation. Biol. Plantarum. 2012, 56, 451–457. [Google Scholar] [CrossRef]
- Badejo, A.A.; Tanaka, N.; Esaka, M. Analysis of GDP-D-mannose pyrophosphorylase gene promoter from acerola (Malpighia glabra) and increase in ascorbate content of transgenic tobacco expressing the acerola gene. Plant Cell Physiol. 2008, 49, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, J.; Munir, S.; Yang, T.; Chen, W.; Liu, G.; Zheng, W.; Zhang, Y. Biosynthetic gene pyramiding leads to ascorbate accumulation with enhanced oxidative stress tolerance in tomato. Int. J. Mol. Sci. 2019, 20, 1558. [Google Scholar] [CrossRef] [PubMed]
- Cronje, C.; George, G.M.; Fernie, A.R.; Bekker, J.; Kossmann, J.; Bauer, R. Manipulation of L-ascorbic acid biosynthesis pathways in Solanum lycopersicum: Elevated GDP-mannose pyrophosphorylase activity enhances L-ascorbate levels in red fruit. Planta 2012, 235, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-Y.; Liu, R.-R.; Zhang, C.-Q.; Tang, K.-X.; Sun, M.-F.; Yan, G.-H.; Liu, Q.-Q. Manipulation of the rice L-galactose pathway: Evaluation of the effects of transgene overexpression on ascorbate accumulation and abiotic stress tolerance. PLoS ONE 2015, 10, e0125870. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Ban, Y.; Yamamoto, T.; Moriguchi, T. Ectopic overexpression of peach GDP-D-mannose pyrophosphorylase and GDP-D-mannose-3′, 5′-epimerase in transgenic tobacco. Plant Cell Tissue Organ Cult. 2012, 111, 1–13. [Google Scholar] [CrossRef]
- Sawake, S.; Tajima, N.; Mortimer, J.C.; Lao, J.; Ishikawa, T.; Yu, X.; Yamanashi, Y.; Yoshimi, Y.; Kawai-Yamada, M.; Dupree, P. KONJAC1 and 2 are key factors for GDP-mannose generation and affect L-ascorbic f and glucomannan biosynthesis in Arabidopsis. Plant Cell 2015, 27, 3397–3409. [Google Scholar] [CrossRef]
- Wolucka, B.A.; Van Montagu, M. GDP-mannose 3′, 5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J. Biol. Chem. 2003, 278, 47483–47490. [Google Scholar] [CrossRef]
- Gilbert, L.; Alhagdow, M.; Nunes-Nesi, A.; Quemener, B.; Guillon, F.; Bouchet, B.; Faurobert, M.; Gouble, B.; Page, D.; Garcia, V. GDP-D-mannose 3, 5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J. 2009, 60, 499–508. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Y.; Liu, W.; Liu, Z. Overexpression of an alfalfa GDP-mannose 3, 5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol. Lett. 2014, 36, 2331–2341. [Google Scholar] [CrossRef]
- Huang, M.; Xu, Q.; Deng, X.-X. L-Ascorbic acid metabolism during fruit development in an ascorbate-rich fruit crop chestnut rose (Rosa roxburghii Tratt). J. Plant Physiol. 2014, 171, 1205–1216. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhang, Y.; Cai, X.; Gong, P.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep. 2011, 30, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.L.; Saracco, S.A.; Norris, S.R.; Last, R.L. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 2000, 154, 847–856. [Google Scholar] [PubMed]
- Laing, W.A.; Wright, M.A.; Cooney, J.; Bulley, S.M. The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an L-galactose guanyltransferase, increases leaf ascorbate content. Proc. Natl. Acad. Sci. USA 2007, 104, 9534–9539. [Google Scholar] [CrossRef] [PubMed]
- Dowdle, J.; Ishikawa, T.; Gatzek, S.; Rolinski, S.; Smirnoff, N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 2007, 52, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Smirnoff, N.; Cobbett, C.S.; Golz, J.F. Ascorbate-deficient vtc2 mutants in Arabidopsis do not exhibit decreased growth. Front. Plant Sci. 2016, 7, 1025. [Google Scholar] [CrossRef] [PubMed]
- Bulley, S.M.; Rassam, M.; Hoser, D.; Otto, W.; Schünemann, N.; Wright, M.; MacRae, E.; Gleave, A.; Laing, W. Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J. Exp. Bot. 2009, 60, 765–778. [Google Scholar] [CrossRef]
- Bulley, S.; Wright, M.; Rommens, C.; Yan, H.; Rassam, M.; Lin-Wang, K.; Andre, C.; Brewster, D.; Karunairetnam, S.; Allan, A.C. Enhancing ascorbate in fruits and tubers through over-expression of the L-galactose pathway gene GDP-L-galactose phosphorylase. Plant Biotechnol. J. 2012, 10, 390–397. [Google Scholar] [CrossRef]
- Wang, L.; Meng, X.; Yang, D.; Ma, N.; Wang, G.; Meng, Q. Overexpression of tomato GDP-L-galactose phosphorylase gene in tobacco improves tolerance to chilling stress. Plant Cell Rep. 2014, 33, 1441–1451. [Google Scholar] [CrossRef]
- Ali, B.; Pantha, S.; Acharya, R.; Ueda, Y.; Wu, L.-B.; Ashrafuzzaman, M.; Ishizaki, T.; Wissuwa, M.; Bulley, S.; Frei, M. Enhanced ascorbate level improves multi-stress tolerance in a widely grown indica rice variety without compromising its agronomic characteristics. J. Plant Physiol. 2019, 240, 152998. [Google Scholar] [CrossRef]
- Bulley, S.; Laing, W. The regulation of ascorbate biosynthesis. Curr. Opin. Plant Biol. 2016, 33, 15–22. [Google Scholar] [CrossRef]
- Conklin, P.L.; Gatzek, S.; Wheeler, G.L.; Dowdle, J.; Raymond, M.J.; Rolinski, S.; Isupov, M.; Littlechild, J.A.; Smirnoff, N. Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J. Biol. Chem. 2006, 281, 15662–15670. [Google Scholar] [CrossRef] [PubMed]
- Gatzek, S.; Wheeler, G.L.; Smirnoff, N. Antisense suppression of L-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated L-galactose synthesis. Plant J. 2002, 30, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Pineau, B.; Layoune, O.; Danon, A.; De Paepe, R. L-galactono-1, 4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J. Biol. Chem. 2008, 283, 32500–32505. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.; Fambrini, M.; Basile, A.; Salvini, M.; Guidi, L.; Pugliesi, C. Overexpression of L-galactono-1, 4-lactone dehydrogenase (L-GalLDH) gene correlates with increased ascorbate concentration and reduced browning in leaves of Lactuca sativa L. after cutting. Plant Cell Tissue Organ Cult. 2015, 123, 109–120. [Google Scholar] [CrossRef]
- Guo, X.; Liu, R.H.; Fu, X.; Sun, X.; Tang, K. Over-expression of l-galactono-γ-lactone dehydrogenase increases vitamin C, total phenolics and antioxidant activity in lettuce through bio-fortification. Plant Cell Tissue Organ Cult. 2013, 114, 225–236. [Google Scholar] [CrossRef]
- Shi, S.; Ma, F.; Li, Y.; Feng, F.; Shang, Z. Overexpression of L-galactono-1, 4-lactone dehydrogenase (GLDH) in Lanzhou lily (Lilium davidii var. unicolor) via particle bombardment-mediated transformation. Vitr. Cell. Dev. Biol. Plant 2012, 48, 1–6. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, L.; Wang, R. Level of ascorbic acid in transgenic rice for L-galactono-1, 4-lactone dehydrogenase overexpressing or suppressed is associated with plant growth and seed set. Acta Physiol. Plant 2011, 33, 1353–1363. [Google Scholar] [CrossRef]
- Liu, W.; An, H.-M.; Yang, M. Overexpression of Rosa roxburghii L-galactono-1, 4-lactone dehydrogenase in tobacco plant enhances ascorbate accumulation and abiotic stress tolerance. Acta Physiol. Plant 2013, 35, 1617–1624. [Google Scholar] [CrossRef]
- Imai, T.; Niwa, M.; Ban, Y.; Hirai, M.; Ôba, K.; Moriguchi, T. Importance of the L-galactonolactone pool for enhancing the ascorbate content revealed by L-galactonolactone dehydrogenase-overexpressing tobacco plants. Plant Cell Tissue Organ Cult. 2009, 96, 105–112. [Google Scholar] [CrossRef]
- Linster, C.L.; Adler, L.N.; Webb, K.; Christensen, K.C.; Brenner, C.; Clarke, S.G. A second GDP-L-galactose phosphorylase in Arabidopsis en route to vitamin C covalent intermediate and substrate requirements for the conserved reaction. J. Biol. Chem. 2008, 283, 18483–18492. [Google Scholar] [CrossRef]
- Linster, C.L.; Gomez, T.A.; Christensen, K.C.; Adler, L.N.; Young, B.D.; Brenner, C.; Clarke, S.G. Arabidopsis VTC2 encodes a GDP-L-galactose phosphorylase, the last unknown enzyme in the Smirnoff-Wheeler pathway to ascorbic acid in plants. J. Biol. Chem. 2007, 282, 18879–18885. [Google Scholar] [CrossRef] [PubMed]
- Lisko, K.A.; Torres, R.; Harris, R.S.; Belisle, M.; Vaughan, M.M.; Jullian, B.; Chevone, B.I.; Mendes, P.; Nessler, C.L.; Lorence, A. Elevating vitamin C content via overexpression of myo-inositol oxygenase and L-gulono-1,4-lactone oxidase in Arabidopsis leads to enhanced biomass and tolerance to abiotic stresses. Vitr. Cell. Dev. Biol. Plant 2013, 49, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Radzio, J.A.; Lorence, A.; Chevone, B.I.; Nessler, C.L. L-Gulono-1,4-lactone oxidase expression rescues vitamin C-deficient Arabidopsis (vtc) mutants. Plant Mol. Biol. 2003, 53, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Nessler, C.L. Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Mol. Breed. 2000, 6, 73–78. [Google Scholar] [CrossRef]
- Upadhyaya, C.P.; Akula, N.; Young, K.E.; Chun, S.C.; Kim, D.H.; Park, S.W. Enhanced ascorbic acid accumulation in transgenic potato confers tolerance to various abiotic stresses. Biotechnol. Lett. 2010, 32, 321–330. [Google Scholar]
- Lim, M.Y.; Pulla, R.K.; Park, J.M.; Harn, C.H.; Jeong, B.R. Over-expression of L-gulono-γ-lactone oxidase (GLOase) gene leads to ascorbate accumulation with enhanced abiotic stress tolerance in tomato. Vitr. Cell. Dev. Biol. Plant 2012, 48, 453–461. [Google Scholar] [CrossRef]
- Aboobucker, S.I.; Suza, W.P.; Lorence, A. Characterization of two Arabidopsis L-gulono-1, 4-lactone oxidases, AtGulLO3 and AtGulLO5, involved in ascorbate biosynthesis. React. Oxyg. Species 2017, 4, 389. [Google Scholar] [CrossRef]
- Maruta, T.; Ichikawa, Y.; Mieda, T.; Takeda, T.; Tamoi, M.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. The contribution of Arabidopsis homologs of L-gulono-1, 4-lactone oxidase to the biosynthesis of ascorbic acid. Biosci. Biotechnol. Biochem. 2010, 74, 1494–1497. [Google Scholar] [CrossRef]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef]
- Tóth, S.Z.; Nagy, V.; Puthur, J.T.; Kovács, L.; Garab, G. The physiological role of ascorbate as photosystem II electron donor: Protection against photoinactivation in heat-stressed leaves. Plant Physiol. 2011, 156, 382–392. [Google Scholar] [CrossRef]
- Zhang, W.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol. 2008, 146, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Belgaroui, N.; Lacombe, B.; Rouached, H.; Hanin, M. Phytase overexpression in Arabidopsis improves plant growth under osmotic stress and in combination with phosphate deficiency. Sci. Rep. 2018, 8, 1137. [Google Scholar] [CrossRef] [PubMed]
- Endres, S.; Tenhaken, R. Myoinositol oxygenase controls the level of myoinositol in Arabidopsis, but does not increase ascorbic acid. Plant Physiol. 2009, 149, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Zhang, M.; Zhang, H.; Xiong, H.; Liu, P.; Ali, J.; Li, J.; Li, Z. OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Plant Sci. 2012, 196, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Kanter, U.; Usadel, B.; Guerineau, F.; Li, Y.; Pauly, M.; Tenhaken, R. The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 2005, 221, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Höller, S.; Ueda, Y.; Wu, L.; Wang, Y.; Hajirezaei, M.-R.; Ghaffari, M.-R.; von Wirén, N.; Frei, M. Ascorbate biosynthesis and its involvement in stress tolerance and plant development in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 545–560. [Google Scholar] [CrossRef]
- Ivanov Kavkova, E.; Blöchl, C.; Tenhaken, R. The myo-inositol pathway does not contribute to ascorbic acid synthesis. Plant Biol. 2019, 21, 95–102. [Google Scholar] [CrossRef]
- Agius, F.; González-Lamothe, R.; Caballero, J.L.; Muñoz-Blanco, J.; Botella, M.A.; Valpuesta, V. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003, 21, 177. [Google Scholar] [CrossRef]
- Upadhyaya, C.P.; Young, K.E.; Akula, N.; Kim, H.; Heung, J.J.; Oh, O.M.; Aswath, C.R.; Chun, S.C.; Kim, D.H.; Park, S.W. Over-expression of strawberry D-galacturonic acid reductase in potato leads to accumulation of vitamin C with enhanced abiotic stress tolerance. Plant Sci. 2009, 177, 659–667. [Google Scholar]
- Upadhyaya, C.P.; Venkatesh, J.; Gururani, M.A.; Asnin, L.; Sharma, K.; Ajappala, H.; Park, S.W. Transgenic potato overproducing L-ascorbic acid resisted an increase in methylglyoxal under salinity stress via maintaining higher reduced glutathione level and glyoxalase enzyme activity. Biotechnol. Lett. 2011, 33, 2297. [Google Scholar] [CrossRef]
- Upadhyaya, C.P.; Akula, N.; Kim, H.S.; Jeon, J.H.; Ho, O.M.; Chun, S.C.; Kim, D.H.; Park, S.W. Biochemical analysis of enhanced tolerance in transgenic potato plants overexpressing D-galacturonic acid reductase gene in response to various abiotic stresses. Mol. Breed. 2011, 28, 105–115. [Google Scholar]
- Amaya, I.; Osorio, S.; Martinez-Ferri, E.; Lima-Silva, V.; Doblas, V.G.; Fernández-Muñoz, R.; Fernie, A.R.; Botella, M.A.; Valpuesta, V. Increased antioxidant capacity in tomato by ectopic expression of the strawberry D-galacturonate reductase gene. Biotechnol. J. 2015, 10, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhang, C.; Ye, J.; Hu, T.; Ye, Z.; Li, H.; Zhang, Y. Ectopic expression of FaGalUR leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Growth Regul. 2015, 76, 187–197. [Google Scholar] [CrossRef]
- Lim, M.Y.; Jeong, B.R.; Jung, M.; Harn, C.H. Transgenic tomato plants expressing strawberry D-galacturonic acid reductase gene display enhanced tolerance to abiotic stresses. Plant Biotechnol. Rep. 2016, 10, 105–116. [Google Scholar] [CrossRef]
- Bao, G.; Zhuo, C.; Qian, C.; Xiao, T.; Guo, Z.; Lu, S. Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 2016, 14, 206–214. [Google Scholar] [CrossRef]
- Chen, Z.; Qin, C.; Lin, L.; Zeng, X.; Zhao, Y.; He, S.; Lu, S.; Guo, Z. Overexpression of yeast arabinono-1, 4-lactone oxidase gene (ALO) increases tolerance to oxidative stress and Al toxicity in transgenic tobacco plants. Plant Mol. Biol. Rep. 2015, 33, 806–818. [Google Scholar] [CrossRef]
- Gallie, D.R. The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2012, 64, 433–443. [Google Scholar] [CrossRef]
- Eastmond, P.J. MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 2007, 19, 1376–1387. [Google Scholar] [CrossRef]
- Eltelib, H.; Fujikawa, Y.; Esaka, M. Overexpression of the acerola (Malpighia glabra) monodehydroascorbate reductase gene in transgenic tobacco plants results in increased ascorbate levels and enhanced tolerance to salt stress. S. Afr. J. Bot. 2012, 78, 295–301. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Kawano, N.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Shibahara, T.; Inanaga, S.; Tanaka, K. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 2007, 225, 1255–1264. [Google Scholar] [CrossRef]
- Yin, L.; Wang, S.; Eltayeb, A.E.; Uddin, M.I.; Yamamoto, Y.; Tsuji, W.; Takeuchi, Y.; Tanaka, K. Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 2010, 231, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wu, Q.Y.; Sun, Y.L.; Wang, L.Y.; Yang, X.H.; Meng, Q.W. Overexpression of chloroplastic monodehydroascorbate reductase enhanced tolerance to temperature and methyl viologen-mediated oxidative stresses. Physiol. Plant 2010, 139, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, K.; George, S.; Venkataraman, G.; Parida, A. A salt-inducible chloroplastic monodehydroascorbate reductase from halophyte Avicennia marina confers salt stress tolerance on transgenic plants. Biochimie 2010, 92, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Haroldsen, V.M.; Chi-Ham, C.L.; Kulkarni, S.; Lorence, A.; Bennett, A.B. Constitutively expressed DHAR and MDHAR influence fruit, but not foliar ascorbate levels in tomato. Plant Physiol. Biochem. 2011, 49, 1244–1249. [Google Scholar] [CrossRef]
- Yoshida, S.; Tamaoki, M.; Shikano, T.; Nakajima, N.; Ogawa, D.; Ioki, M.; Aono, M.; Kubo, A.; Kamada, H.; Inoue, Y. Cytosolic dehydroascorbate reductase is important for ozone tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 304–308. [Google Scholar] [CrossRef]
- Hao, Z.; Wang, X.; Zong, Y.; Wen, S.; Cheng, Y.; Li, H. Enzymatic activity and functional analysis under multiple abiotic stress conditions of a dehydroascorbate reductase gene derived from Liriodendron Chinense. Environ. Exp. Bot. 2019, 167, 103850. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, Y.; Chen, W.; Tang, K.; Zhang, L. Increased vitamin C content accompanied by an enhanced recycling pathway confers oxidative stress tolerance in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 400–409. [Google Scholar] [CrossRef]
- Liu, F.; Guo, X.; Yao, Y.; Tang, W.; Zhang, W.; Cao, S.; Han, Y.; Liu, Y. Cloning and function characterization of two dehydroascorbate reductases from kiwifruit (Actinidia chinensis L.). Plant Mol. Biol. Rep. 2016, 34, 815–826. [Google Scholar] [CrossRef]
- Ushimaru, T.; Nakagawa, T.; Fujioka, Y.; Daicho, K.; Naito, M.; Yamauchi, Y.; Nonaka, H.; Amako, K.; Yamawaki, K.; Murata, N. Transgenic Arabidopsis plants expressing the rice dehydroascorbate reductase gene are resistant to salt stress. J. Plant Physiol. 2006, 163, 1179–1184. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, X.; Kim, H.S.; Yang, Y.; Hou, D.; Liang, X.; Kwak, S.-S. Molecular cloning and functional characterization of a sweetpotato chloroplast IbDHAR3 gene in response to abiotic stress. Plant Biotechnol. Rep. 2020, 14, 9–19. [Google Scholar] [CrossRef]
- Chen, Z.; Young, T.E.; Ling, J.; Chang, S.-C.; Gallie, D.R. Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc. Natl. Acad. Sci. USA 2003, 100, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Zhu, C.; Farre, G.; Ramessar, K.; Bassie, L.; Breitenbach, J.; Conesa, D.P.; Ros, G.; Sandmann, G.; Capell, T. Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 7762–7767. [Google Scholar] [CrossRef] [PubMed]
- Goo, Y.-M.; Chun, H.J.; Kim, T.-W.; Lee, C.-H.; Ahn, M.-J.; Bae, S.-C.; Cho, K.-J.; Chun, J.-A.; Chung, C.-H.; Lee, S.-W. Expressional characterization of dehydroascorbate reductase cDNA in transgenic potato plants. J. Plant Biol. 2008, 51, 35–41. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Yamamoto, S.; Habora, M.E.E.; Yin, L.; Tsujimoto, H.; Tanaka, K. Transgenic potato overexpressing Arabidopsis cytosolic AtDHAR1 showed higher tolerance to herbicide, drought and salt stresses. Breed. Sci. 2011, 61, 3–10. [Google Scholar] [CrossRef]
- Qin, A.; Shi, Q.; Yu, X. Ascorbic acid contents in transgenic potato plants overexpressing two dehydroascorbate reductase genes. Mol. Biol. Rep. 2011, 38, 1557–1566. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Kim, I.-S.; Bae, M.-J.; Choe, Y.-H.; Kim, Y.-H.; Park, H.-M.; Kang, H.-G.; Yoon, H.-S. Homologous expression of cytosolic dehydroascorbate reductase increases grain yield and biomass under paddy field conditions in transgenic rice (Oryza sativa L. japonica). Planta 2013, 237, 1613–1625. [Google Scholar] [CrossRef]
- Kwon, S.-Y.; Ahn, Y.-O.; Lee, H.-S.; Kwak, S.-S. Biochemical characterization of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. BMB Rep. 2001, 34, 316–321. [Google Scholar]
- Kwon, S.-Y.; Choi, S.-M.; Ahn, Y.-O.; Lee, H.-S.; Lee, H.-B.; Park, Y.-M.; Kwak, S.-S. Enhanced stress-tolerance of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. J. Plant Physiol. 2003, 160, 347–353. [Google Scholar] [CrossRef]
- Chen, Z.; Gallie, D.R. Increasing tolerance to ozone by elevating foliar ascorbic acid confers greater protection against ozone than increasing avoidance. Plant Physiol. 2005, 138, 1673–1689. [Google Scholar] [CrossRef]
- Lee, Y.-P.; Kim, S.-H.; Bang, J.-W.; Lee, H.-S.; Kwak, S.-S.; Kwon, S.-Y. Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant Cell Rep. 2007, 26, 591–598. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Kawano, N.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Morishima, I.; Shibahara, T.; Inanaga, S.; Tanaka, K. Enhanced tolerance to ozone and drought stresses in transgenic tobacco overexpressing dehydroascorbate reductase in cytosol. Physiol. Plant 2006, 127, 57–65. [Google Scholar] [CrossRef]
- Le Martret, B.; Poage, M.; Shiel, K.; Nugent, G.D.; Dix, P.J. Tobacco chloroplast transformants expressing genes encoding dehydroascorbate reductase, glutathione reductase, and glutathione-S-transferase, exhibit altered anti-oxidant metabolism and improved abiotic stress tolerance. Plant Biotechnol. J. 2011, 9, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, Y.; Li, C.; Yu, X. Enhanced ascorbic acid accumulation through overexpression of dehydroascorbate reductase confers tolerance to methyl viologen and salt stresses in tomato. Czech J. Genet. Plant Breed. 2012, 48, 74–86. [Google Scholar] [CrossRef]
- Qin, A.; Huang, X.; Zhang, H.; Wu, J.; Yang, J.; Zhang, S. Overexpression of PbDHAR2 from Pyrus sinkiangensis in transgenic tomato confers enhanced tolerance to salt and chilling stresses. HortScience 2015, 50, 789–796. [Google Scholar] [CrossRef]
- Zhang, W.; Lorence, A.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. AMR1, an Arabidopsis gene that coordinately and negatively regulates the mannose/l-galactose ascorbic acid biosynthetic pathway. Plant Physiol. 2009, 150, 942–950. [Google Scholar] [CrossRef]
- Ye, J.; Li, W.; Ai, G.; Li, C.; Liu, G.; Chen, W.; Wang, B.; Wang, W.; Lu, Y.; Zhang, J. Genome-wide association analysis identifies a natural variation in basic helix-loop-helix transcription factor regulating ascorbate biosynthesis via D-mannose/L-galactose pathway in tomato. PLoS Genet. 2019, 15, e1008149. [Google Scholar] [CrossRef]
- Cho, K.M.; Nguyen, H.T.K.; Kim, S.Y.; Shin, J.S.; Cho, D.H.; Hong, S.B.; Shin, J.S.; Ok, S.H. CML 10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol. 2016, 209, 664–678. [Google Scholar] [CrossRef]
- Wang, J.; Yu, Y.; Zhang, Z.; Quan, R.; Zhang, H.; Ma, L.; Deng, X.W.; Huang, R. Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 2013, 25, 625–636. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, C.; Shu, W.; Ye, Z.; Li, H.; Zhang, Y. The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 2016, 474, 736–741. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef]
- Laing, W.A.; Martínez-Sánchez, M.; Wright, M.A.; Bulley, S.M.; Brewster, D.; Dare, A.P.; Rassam, M.; Wang, D.; Storey, R.; Macknight, R.C.; et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 2015, 27, 772–786. [Google Scholar] [CrossRef] [PubMed]
- Broad, R.C.; Bonneau, J.P.; Beasley, J.T.; Roden, S.; Philips, J.G.; Baumann, U.; Hellens, R.P.; Johnson, A.A. Genome-wide identification and characterization of the GDP-L-galactose phosphorylase gene family in bread wheat. BMC Plant Biol. 2019, 19, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 2018, 36, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Ye, J.; Tao, P.; Li, H.; Zhang, J.; Zhang, Y.; Ye, Z. The tomato HD-Zip I transcription factor SlHZ24 modulates ascorbate accumulation through positive regulation of the d-mannose/l-galactose pathway. Plant J. 2016, 85, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, L.; Ye, Z.; Li, H. Ascorbic acid accumulation is transcriptionally modulated in high-pigment-1 tomato fruit. Plant Mol. Biol. Rep. 2014, 32, 52–61. [Google Scholar] [CrossRef]
- Wang, S.; Liu, J.; Feng, Y.; Niu, X.; Giovannoni, J.; Liu, Y. Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. Plant J. 2008, 55, 89–103. [Google Scholar] [CrossRef]
- Xing, C.; Liu, Y.; Zhao, L.; Zhang, S.; Huang, X. A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ. 2019, 42, 832–845. [Google Scholar] [CrossRef]
- Ye, J.; Liu, G.; Chen, W.; Zhang, F.; Li, H.; Ye, Z.; Zhang, Y. Knockdown of SlNL33 accumulates ascorbate, enhances disease and oxidative stress tolerance in tomato (Solanum lycopersicum). Plant Growth Regul. 2019, 89, 49–58. [Google Scholar] [CrossRef]
- Gangadhar, B.H.; Yu, J.W.; Sajeesh, K.; Park, S.W. A systematic exploration of high-temperature stress-responsive genes in potato using large-scale yeast functional screening. Mol. Genet. Genom. 2014, 289, 185–201. [Google Scholar] [CrossRef]
- Gangadhar, B.H.; Sajeesh, K.; Venkatesh, J.; Baskar, V.; Abhinandan, K.; Yu, J.W.; Prasad, R.; Mishra, R.K. Enhanced tolerance of transgenic potato plants over-expressing non-specific lipid transfer protein-1 (StnsLTP1) against multiple abiotic stresses. Front. Plant Sci. 2016, 7, 1228. [Google Scholar] [CrossRef]
- Conklin, P.L.; DePaolo, D.; Wintle, B.; Schatz, C.; Buckenmeyer, G. Identification of Arabidopsis VTC3 as a putative and unique dual function protein kinase:: Protein phosphatase involved in the regulation of the ascorbic acid pool in plants. J. Exp. Bot. 2013, 64, 2793–2804. [Google Scholar] [CrossRef]
- Zhu, L.; Guo, J.; Zhu, J.; Zhou, C. Enhanced expression of EsWAX1 improves drought tolerance with increased accumulation of cuticular wax and ascorbic acid in transgenic Arabidopsis. Plant Physiol. Biochem. 2014, 75, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, B.; Yang, T.; Li, H.; Zhang, L.; Zhang, Y.; Zhang, J.; Fei, Z.; Ye, Z. Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J. Exp. Bot. 2007, 58, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chu, Z.; Luo, J.; Zhou, Y.; Cai, Y.; Lu, Y.; Xia, J.; Kuang, H.; Ye, Z.; Ouyang, B. The C2H2 zinc-finger protein SlZF3 regulates AsA synthesis and salt tolerance by interacting with CSN 5B. Plant Biotechnol. J. 2018, 16, 1201–1213. [Google Scholar] [CrossRef] [PubMed]
- Si, X.; Zhang, H.; Wang, Y.; Chen, K.; Gao, C. Manipulating gene translation in plants by CRISPR–Cas9-mediated genome editing of upstream open reading frames. Nat. Protoc. 2020, 15, 338–363. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. Increasing vitamin C content in plant foods to improve their nutritional value—successes and challenges. Nutrients 2013, 5, 3424–3446. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gallie, D.R. The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 2004, 16, 1143–1162. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gallie, D.R. Induction of monozygotic twinning by ascorbic acid in tobacco. PLoS ONE 2012, 7, e39147. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).