Transcriptional Profiling of Advanced Urothelial Cancer Predicts Prognosis and Response to Immunotherapy
Abstract
:1. Introduction
2. Results
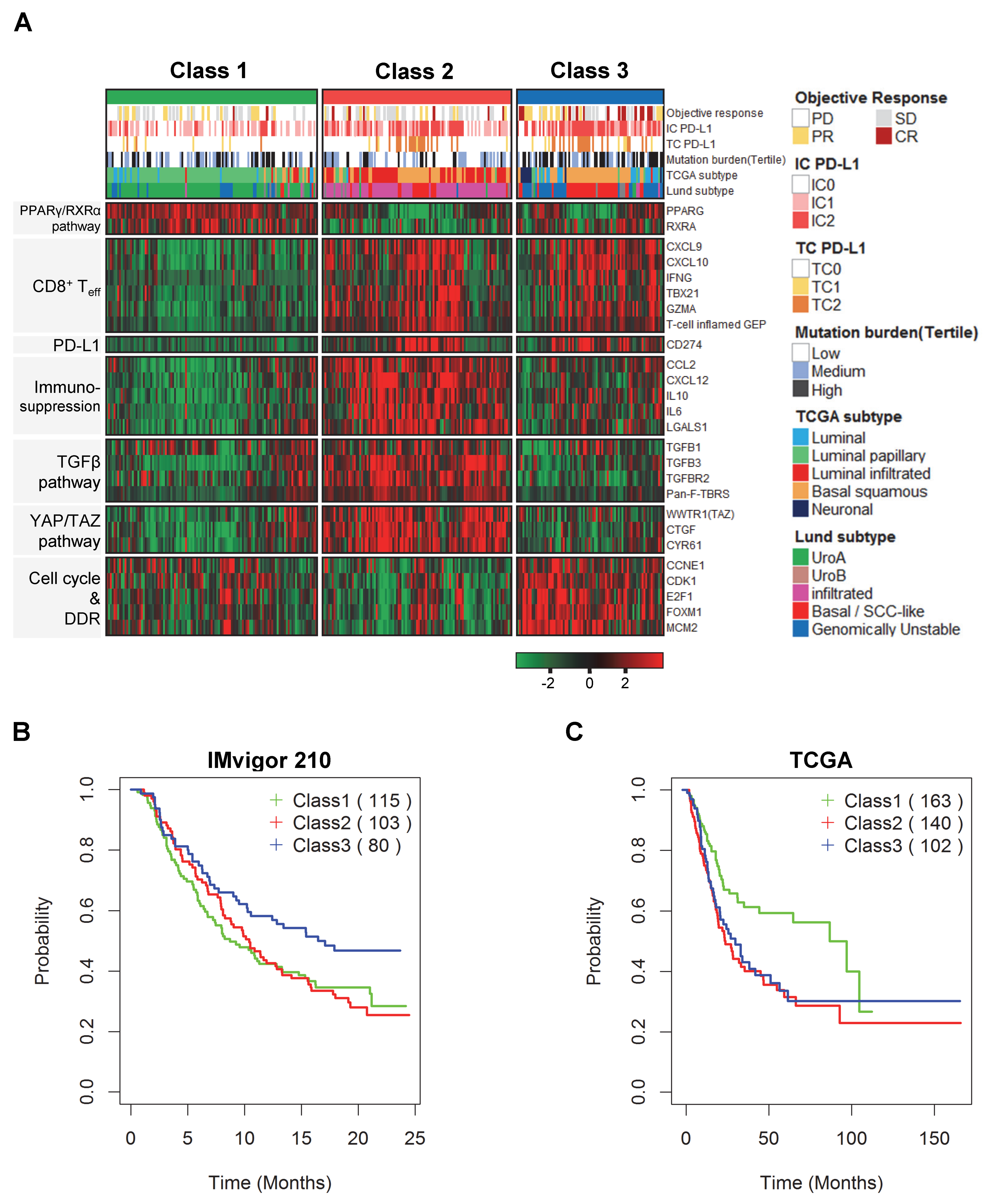
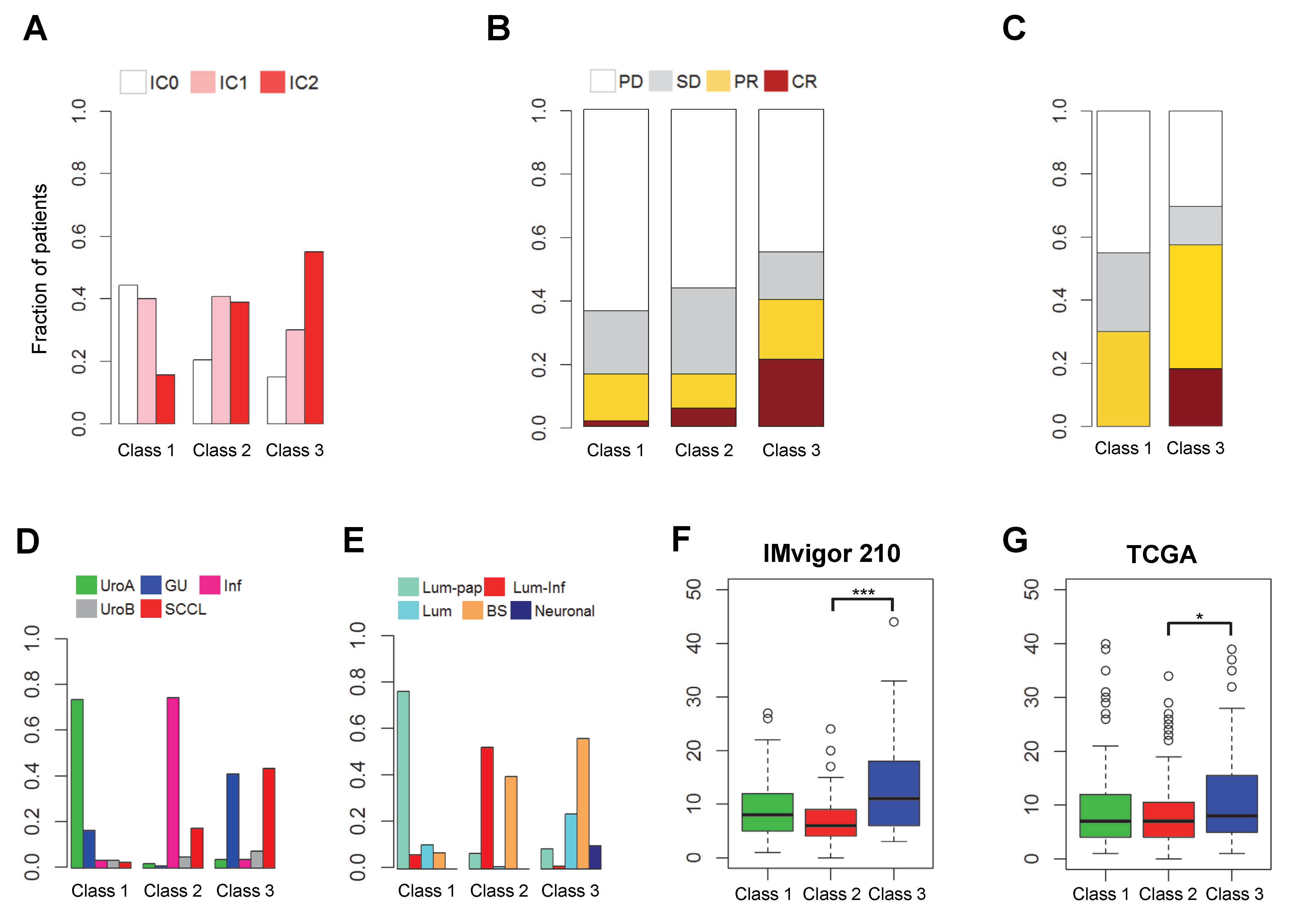
2.1. Discovery of Distinct Three Subtypes and Clinical Characteristics
2.2. Biological Insight into the Newly Identified Subtypes
2.3. Prognostic Impact Based on Unsupervised Clustering Analysis
2.4. Comparison of Clinical Outcomes in the Three Subgroups
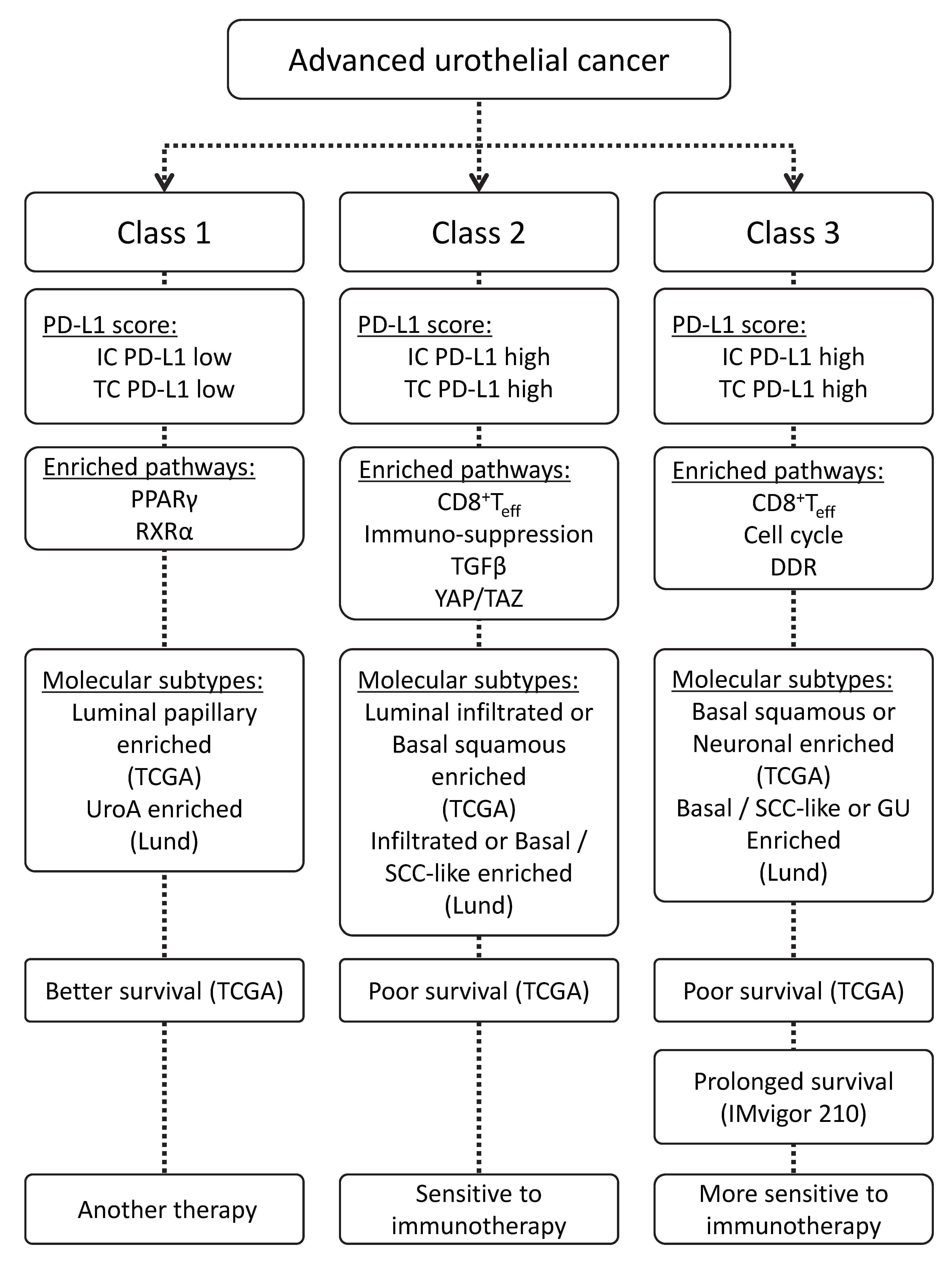
2.5. Schematic Diagram of the Characteristics of Advanced Urothelial Cancer
3. Discussion
4. Materials and Methods
4.1. Patients and Gene Expression Data
4.2. Gene Expression Analysis
4.3. Statistical Analysis and Data Visualization
4.4. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanli, O.; Dobruch, J.; Knowles, M.A.; Burger, M.; Alemozaffar, M.; Nielsen, M.E.; Lotan, Y. Bladder cancer. Nat. Rev. Dis. Prim 2017, 3, 17022. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Hoffman-Censits, J.; Powles, T.; van der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y.; et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef] [Green Version]
- Balar, A.V.; Galsky, M.D.; Rosenberg, J.E.; Powles, T.; Petrylak, D.P.; Bellmunt, J.; Loriot, Y.; Necchi, A.; Hoffman-Censits, J.; Perez-Gracia, J.L.; et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: A single-arm, multicentre, phase 2 trial. Lancet 2017, 389, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Fakhrejahani, F.; Tomita, Y.; Maj-Hes, A.; Trepel, J.B.; De Santis, M.; Apolo, A.B. Immunotherapies for bladder cancer: A new hope. Curr. Opin. Urol. 2015, 25, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Sjodahl, G.; Eriksson, P.; Liedberg, F.; Hoglund, M. Molecular classification of urothelial carcinoma: Global mRNA classification versus tumour-cell phenotype classification. J. Pathol. 2017, 242, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2018, 174, 1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, B.N.; Kim, S.K.; Mun, J.Y.; Choi, Y.D.; Leem, S.H.; Chu, I.S. Identification of an immunotherapy-responsive molecular subtype of bladder cancer. EBioMedicine 2019, 50, 238–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamoun, A.; de Reynies, A.; Allory, Y.; Sjodahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kwiatkowski, D.; McConkey, D.J.; Meeks, J.J.; Freeman, S.S.; Bellmunt, J.; Getz, G.; Lerner, S.P. The Cancer Genome Atlas Expression Subtypes Stratify Response to Checkpoint Inhibition in Advanced Urothelial Cancer and Identify a Subset of Patients with High Survival Probability. Eur. Urol. 2019, 75, 961–964. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Puyang, X.; Jeremy Wu, Z.; Seiler, R.; Furman, C.; Oo, H.Z.; Seiler, M.; Irwin, S.; Subramanian, V.; Julie Joshi, J.; et al. Evasion of immunosurveillance by genomic alterations of PPARgamma/RXRalpha in bladder cancer. Nat. Commun. 2017, 8, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Gong, Y.; Saci, A.; Szabo, P.M.; Martini, A.; Necchi, A.; Siefker-Radtke, A.; Pal, S.; Plimack, E.R.; Sfakianos, J.P.; et al. Fibroblast Growth Factor Receptor 3 Alterations and Response to PD-1/PD-L1 Blockade in Patients with Metastatic Urothelial Cancer. Eur. Urol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Tian, Y.; Cao, C.; Niu, G. The Emerging Role of YAP/TAZ in Tumor Immunity. Mol. Cancer Res. 2019, 17, 1777–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, M.Y.; Seier, K.; Ostrovnaya, I.; Regazzi, A.M.; Kania, B.E.; Moran, M.M.; Cipolla, C.K.; Bluth, M.J.; Chaim, J.; Al-Ahmadie, H.; et al. Alterations in DNA Damage Response and Repair Genes as Potential Marker of Clinical Benefit From PD-1/PD-L1 Blockade in Advanced Urothelial Cancers. J. Clin. Oncol. 2018, 36, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Mouw, K.W.; Goldberg, M.S.; Konstantinopoulos, P.A.; D’Andrea, A.D. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov. 2017, 7, 675–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, S.-W.; Jang, I.-H.; Kim, S.-K.; Nam, J.-K.; Leem, S.-H.; Chu, I.-S. Transcriptional Profiling of Advanced Urothelial Cancer Predicts Prognosis and Response to Immunotherapy. Int. J. Mol. Sci. 2020, 21, 1850. https://doi.org/10.3390/ijms21051850
Baek S-W, Jang I-H, Kim S-K, Nam J-K, Leem S-H, Chu I-S. Transcriptional Profiling of Advanced Urothelial Cancer Predicts Prognosis and Response to Immunotherapy. International Journal of Molecular Sciences. 2020; 21(5):1850. https://doi.org/10.3390/ijms21051850
Chicago/Turabian StyleBaek, Seung-Woo, In-Hwan Jang, Seon-Kyu Kim, Jong-Kil Nam, Sun-Hee Leem, and In-Sun Chu. 2020. "Transcriptional Profiling of Advanced Urothelial Cancer Predicts Prognosis and Response to Immunotherapy" International Journal of Molecular Sciences 21, no. 5: 1850. https://doi.org/10.3390/ijms21051850
APA StyleBaek, S. -W., Jang, I. -H., Kim, S. -K., Nam, J. -K., Leem, S. -H., & Chu, I. -S. (2020). Transcriptional Profiling of Advanced Urothelial Cancer Predicts Prognosis and Response to Immunotherapy. International Journal of Molecular Sciences, 21(5), 1850. https://doi.org/10.3390/ijms21051850





