Peripheral Circulating Exosomal miRNAs Potentially Contribute to the Regulation of Molecular Signaling Networks in Aging
Abstract
1. Introduction
2. Results
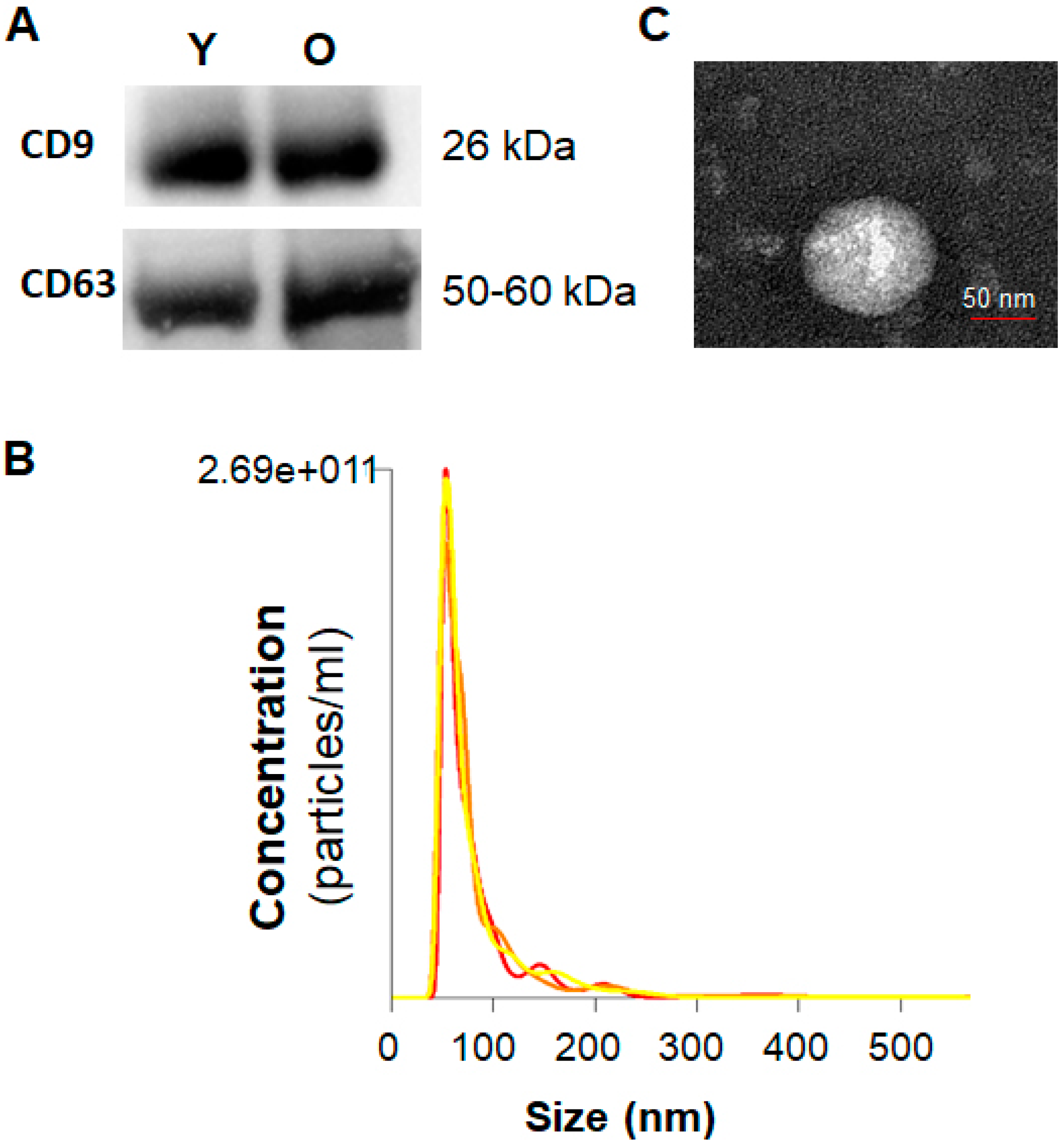
2.1. Characterization of Serum Exosomes
2.2. Differentially Expressed RNAs in Serum Exosomes with Age
2.3. Identification of miRNA-Targeted mRNAs
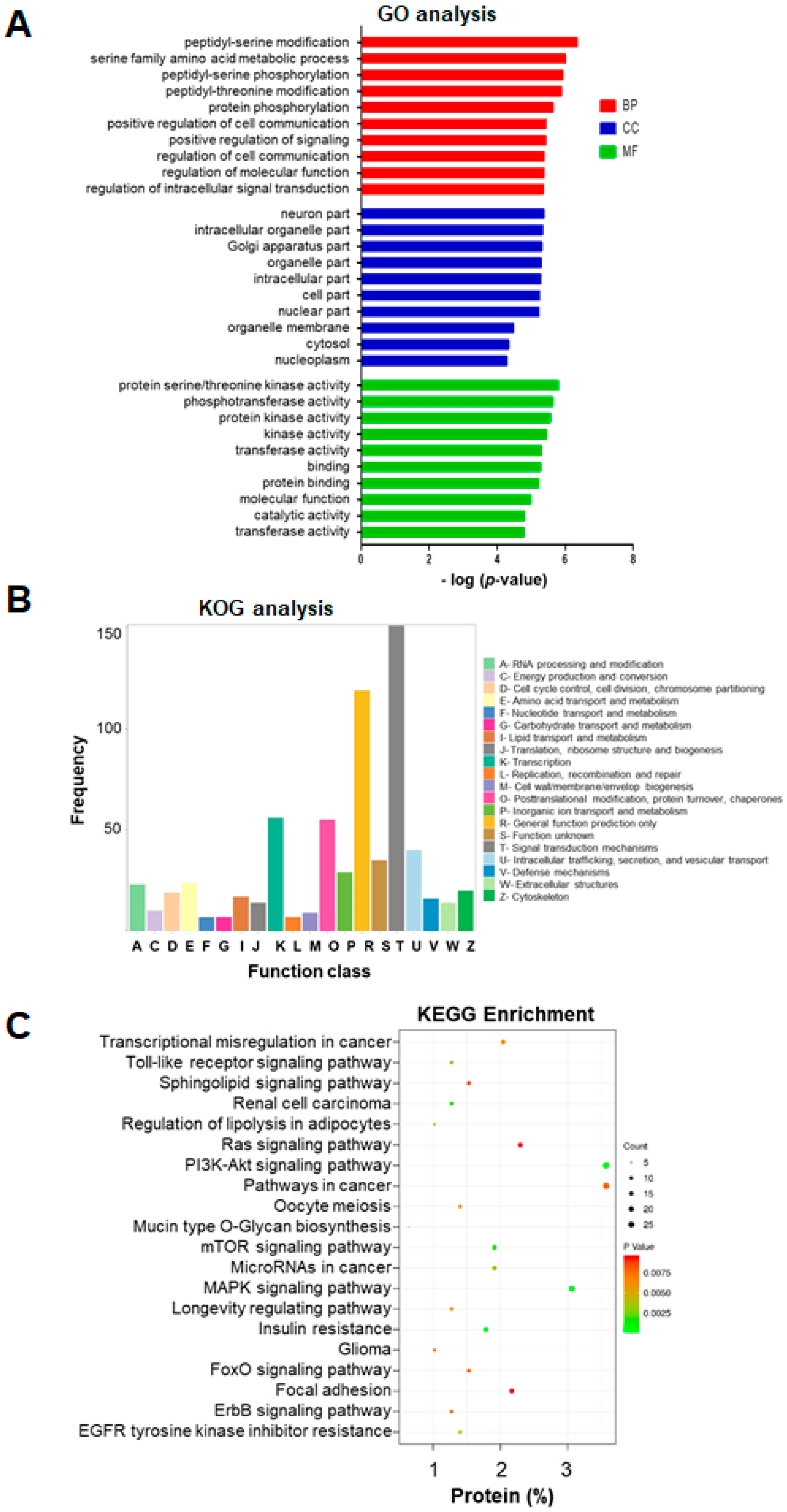
2.4. GO Enrichment Analysis of miRNA-Targeted mRNAs
2.5. KOG and KEGG Enrichment and Analyses
2.6. Analysis of Pathways and Interaction Networks
3. Discussion
4. Materials and Methods
4.1. Isolation of Serum Exosomes
4.2. Characterization of Serum Exosomes
4.3. Western Blotting
4.4. Isolation of Total RNA fromEexosomes and Next-Generation RNA Sequencing
4.5. Data Processing
4.6. Bioinformatics Analysis
4.7. Gene Ontology (GO) and Pathway Enrichment Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tinetti, M.E.; Speechley, M.; Ginter, S.F. Risk factors for falls among elderly persons living in the community. N. Engl. J. Med. 1988, 319, 1701–1707. [Google Scholar] [CrossRef]
- Hebert, L.E.; Scherr, P.A.; Bienias, J.L.; Bennett, D.A.; Evans, D.A. Alzheimer disease in the us population: Prevalence estimates using the 2000 census. Arch. Neurol. 2003, 60, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Jin, K. Modern biological theories of aging. Aging Dis. 2010, 1, 72–74. [Google Scholar] [PubMed]
- Jin, K. A microcirculatory theory of aging. Aging Dis. 2019, 10, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Conboy, I.M.; Conboy, M.J.; Wagers, A.J.; Girma, E.R.; Weissman, I.L.; Rando, T.A. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 2005, 433, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Brack, A.S.; Conboy, M.J.; Roy, S.; Lee, M.; Kuo, C.J.; Keller, C.; Rando, T.A. Increased wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 2007, 317, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Loffredo, F.; Steinhauser, M.L.; Jay, S.M.; Gannon, J.; Pancoast, J.R.; Yalamanchi, P.; Sinha, M.; Dall’Osso, C.; Khong, D.; Shadrach, J.L.; et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 2013, 153, 828–839. [Google Scholar] [CrossRef]
- Zhang, H.; Cherian, R.; Jin, K. Systemic milieu and age-related deterioration. Geroscience 2019, 41, 275–284. [Google Scholar] [CrossRef]
- Sebastiani, P.; Thyagarajan, B.; Sun, F.; Schupf, N.; Newman, A.B.; Montano, M.; Perls, T. Biomarker signatures of aging. Aging Cell 2017, 16, 329–338. [Google Scholar] [CrossRef]
- Xia, X.; Chen, W.; McDermott, J.; Han, J.J. Molecular and phenotypic biomarkers of aging. F1000Research 2017, 6, 860. [Google Scholar] [CrossRef]
- Urbanelli, L.; Magini, A.; Buratta, S.; Brozzi, A.; Sagini, K.; Polchi, A.; Tancini, B.; Emiliani, C. Signaling pathways in exosomes biogenesis, secretion and fate. Genes 2013, 4, 152–170. [Google Scholar] [CrossRef] [PubMed]
- Zomer, A.; Vendrig, T.; Hopmans, E.S.; van Eijndhoven, M.; Middeldorp, J.M.; Pegtel, D.M. Exosomes: Fit to deliver small rna. Commun. Integr. Biol. 2010, 3, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kim, M.S.; Jia, B.; Yan, J.; Zuniga-Hertz, J.P.; Han, C.; Cai, D. Hypothalamic stem cells control ageing speed partly through exosomal mirnas. Nature 2017, 548, 52–57. [Google Scholar] [CrossRef]
- Lee, B.R.; Kim, J.H.; Choi, E.S.; Cho, J.H.; Kim, E. Effect of young exosomes injected in aged mice. Int. J. Nanomed. 2018, 13, 5335–5345. [Google Scholar] [CrossRef] [PubMed]
- Pulliam, L.; Sun, B.; Mustapic, M.; Chawla, S.; Kapogiannis, D. Plasma neuronal exosomes serve as biomarkers of cognitive impairment in hiv infection and alzheimer’s disease. J. Neurovirol. 2019, 25, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Dalvi, P.; Abadjian, L.; Tang, N.; Pulliam, L. Blood neuron-derived exosomes as biomarkers of cognitive impairment in hiv. AIDS 2017, 31, F9–F17. [Google Scholar] [CrossRef]
- Rani, A.; O’Shea, A.; Ianov, L.; Cohen, R.A.; Woods, A.J.; Foster, T.C. Mirna in circulating microvesicles as biomarkers for age-related cognitive decline. Front. Aging Neurosci. 2017, 9, 323. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of sirna to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Matsumoto, J.; Stewart, T.; Banks, W.A.; Zhang, J. The transport mechanism of extracellular vesicles at the blood-brain barrier. Curr. Pharm. Des. 2017, 23, 6206–6214. [Google Scholar] [CrossRef]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in danio rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef]
- Qu, M.; Lin, Q.; Huang, L.; Fu, Y.; Wang, L.; He, S.; Fu, Y.; Yang, S.; Zhang, Z.; Zhang, L.; et al. Dopamine-loaded blood exosomes targeted to brain for better treatment of parkinson’s disease. J. Control Release 2018, 287, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Eitan, E.; Green, J.; Bodogai, M.; Mode, N.A.; Bæk, R.; Jørgensen, M.M.; Freeman, D.W.; Witwer, K.W.; Zonderman, A.B.; Biragyn, A.; et al. Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci. Rep. 2017, 7, 1342. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microrna targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Bartel, D.P. Micrornas: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.L.; Plotkin, J.; Venø, M.T.; Von Schimmelmann, M.; Feinberg, P.; Mann, S.; Handler, A.; Kjems, J.; Surmeier, D.J.; O’Carroll, D.; et al. Microrna-128 governs neuronal excitability and motor behavior in mice. Science 2013, 342, 1254–1258. [Google Scholar] [CrossRef]
- Pedersen, M.E.; Snieckute, G.; Kagias, K.; Nehammer, C.; Multhaupt, H.A.; Couchman, J.R.; Pocock, R. An epidermal microrna regulates neuronal migration through control of the cellular glycosylation state. Science 2013, 341, 1404–1408. [Google Scholar] [CrossRef]
- Gomez, G.G.; Volinia, S.; Croce, C.M.; Zanca, C.; Li, M.; Emnett, R.; Gutmann, D.H.; Brennan, C.; Furnari, F.B.; Cavenee, W.K. Suppression of microrna-9 by mutant egfr signaling upregulates foxp1 to enhance glioblastoma tumorigenicity. Cancer Res. 2014, 74, 1429–1439. [Google Scholar] [CrossRef]
- Mushtaq, G.; Greig, N.H.; Anwar, F.; A Zamzami, M.; Choudhry, H.; Shaik, M.M.; Tamargo, I.A.; Kamal, M.A. Mirnas as circulating biomarkers for alzheimer’s disease and parkinson’s disease. Med. Chem. 2016, 12, 217–225. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free micrornas in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Sheinerman, K.S.; Tsivinsky, V.G.; Abdullah, L.; Crawford, F.; Umansky, S.R. Plasma microrna biomarkers for detection of mild cognitive impairment: Biomarker validation study. Aging 2013, 5, 925–938. [Google Scholar] [CrossRef]
- Cossetti, C.; Iraci, N.; Mercer, T.R.; Leonardi, T.; Alpi, E.; Drago, D.; Alfaro-Cervello, C.; Saini, H.K.; Davis, M.; Schaeffer, J.; et al. Extracellular vesicles from neural stem cells transfer ifn-gamma via ifngr1 to activate stat1 signaling in target cells. Mol. Cell 2014, 56, 193–204. [Google Scholar] [CrossRef]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine 2011, 7, 780–788. [Google Scholar] [CrossRef]
- Feng, L.-B.; Pang, X.-M.; Zhang, L.; Li, J.; Huang, L.-G.; Su, S.-Y.; Zhou, X.; Li, S.-H.; Xiang, H.-Y.; Chen, C.-Y.; et al. Microrna involvement in mechanism of endogenous protection induced by fastigial nucleus stimulation based on deep sequencing and bioinformatics. BMC Med. Genom. 2015, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. Kegg: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Hatse, S.; Brouwers, B.; Dalmasso, B.S.; Laenen, A.; Kenis, C.; Schöffski, P.; Wildiers, H. Circulating micrornas as easy-to-measure aging biomarkers in older breast cancer patients: Correlation with chronological age but not with fitness/frailty status. PLoS ONE 2014, 9, e110644. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Huang, R.; Diao, M.; Li, L.; Cui, X. Integrative analysis of microrna and mrna expression profiles in fetal rat model with anorectal malformation. PeerJ 2018, 6, e5774. [Google Scholar] [CrossRef]
- Kim, J.-H.; Lee, B.-R.; Choi, E.-S.; Lee, K.-M.; Choi, S.-K.; Cho, J.H.; Jeon, W.B.; Kim, E. Reverse expression of aging-associated molecules through transfection of mirnas to aged mice. Mol. Ther. Nucleic Acids 2017, 6, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.P.; Buric, I.; George-Pandeth, A.; Flurkey, K.; Harrison, D.E.; Yuan, R.; Peters, L.; Kuchel, G.A.; Melzer, D.; Harries, L.W. Micrornas mir-203-3p, mir-664-3p and mir-708-5p are associated with median strain lifespan in mice. Sci. Rep. 2017, 7, 44620. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, C.; Wang, M.; Su, L.; Qu, Y.; Li, J.; Yu, B.; Yan, M.; Yu, Y.; Liu, B.; et al. Decrease of mir-202-3p expression, a novel tumor suppressor, in gastric cancer. PLoS ONE 2013, 8, e69756. [Google Scholar] [CrossRef]
- Dluzen, D.F.; Noren Hooten, N.; De, S.; Wood, W.H., 3rd; Zhang, Y.; Becker, K.G.; Zonderman, A.B.; Tanaka, T.; Ferrucci, L.; Evans, M.K. Extracellular rna profiles with human age. Aging Cell 2018, 17, e12785. [Google Scholar] [CrossRef]
- Tsai, H.P.; Huang, S.F.; Li, C.F.; Chien, H.T.; Chen, S.C. Differential microrna expression in breast cancer with different onset age. PLoS ONE 2018, 13, e0191195. [Google Scholar] [CrossRef] [PubMed]
- Noren Hooten, N.; Fitzpatrick, M.; Wood, W.H., 3rd; De, S.; Ejiogu, N.; Zhang, Y.; Mattison, J.A.; Becker, K.G.; Zonderman, A.B.; Evans, M.K. Age-related changes in microrna levels in serum. Aging 2013, 5, 725–740. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Abdelmohsen, K.; Gorospe, M.; Ejiogu, N.; Zonderman, A.B.; Evans, M.K. Microrna expression patterns reveal differential expression of target genes with age. PLoS ONE 2010, 5, e10724. [Google Scholar] [CrossRef] [PubMed]
- Inukai, S.; de Lencastre, A.; Turner, M.; Slack, F. Novel micrornas differentially expressed during aging in the mouse brain. PLoS ONE 2012, 7, e40028. [Google Scholar] [CrossRef] [PubMed]
- Hara, N.; Kikuchi, M.; Miyashita, A.; Hatsuta, H.; Saito, Y.; Kasuga, K.; Murayama, S.; Ikeuchi, T.; Kuwano, R. Serum microrna mir-501-3p as a potential biomarker related to the progression of alzheimer’s disease. Acta Neuropathol. Commun. 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Kovanda, A.; Leonardis, L.; Zidar, J.; Koritnik, B.; Dolenc-Grošelj, L.; Kovacic, S.R.; Curk, T.; Rogelj, B. Differential expression of micrornas and other small rnas in muscle tissue of patients with als and healthy age-matched controls. Sci. Rep. 2018, 8, 5609. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.Y.; Kim, D.H.; Lee, E.K.; Chung, K.W.; Chung, S.; Lee, B.; Seo, A.Y.; Chung, J.H.; Jung, Y.S.; Im, E.; et al. Redefining chronic inflammation in aging and age-related diseases: Proposal of the senoinflammation concept. Aging Dis. 2019, 10, 367–382. [Google Scholar] [CrossRef]
- Lafferty-Whyte, K.; Cairney, C.J.; Jamieson, N.B.; Oien, K.A.; Keith, W.N. Pathway analysis of senescence-associated mirna targets reveals common processes to different senescence induction mechanisms. Biochim. Biophys. Acta 2009, 1792, 341–352. [Google Scholar] [CrossRef]
- Carlson, M.E.; Silva, H.S.; Conboy, I.M. Aging of signal transduction pathways, and pathology. Exp. Cell Res. 2008, 314, 1951–1961. [Google Scholar] [CrossRef]
- Newgard, C.B.; Pessin, J.E. Recent progress in metabolic signaling pathways regulating aging and life span. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2014, 69, S21–S27. [Google Scholar] [CrossRef]
- Pan, H.; Finkel, T. Key proteins and pathways that regulate lifespan. J. Biol. Chem. 2017, 292, 6452–6460. [Google Scholar] [CrossRef] [PubMed]
- Dharap, A.; Vemuganti, R. Ischemic pre-conditioning alters cerebral micrornas that are upstream to neuroprotective signaling pathways. J. Neurochem. 2010, 113, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.M.; da Silva Xavier, G.; Dawe, H.R.; Rutter, G.A.; Harries, L.W. Increased expression of mir-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia 2014, 57, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Danielson, K.M.; Benton, M.C.; Ziegler, O.; Shah, R.; Stubbs, R.S.; Das, S.; Macartney-Coxson, D. Mirna signatures of insulin resistance in obesity. Obesity 2017, 25, 1734–1744. [Google Scholar] [CrossRef]
- Tatar, M.; Bartke, A.; Antebi, A. The endocrine regulation of aging by insulin-like signals. Science 2003, 299, 1346–1351. [Google Scholar] [CrossRef]
- Markowska, A.L.; Mooney, M.; Sonntag, W.E. Insulin-like growth factor-1 ameliorates age-related behavioral deficits. Neuroscience 1998, 87, 559–569. [Google Scholar] [CrossRef]
- Vijg, J.; Campisi, J. Puzzles, promises and a cure for ageing. Nature 2008, 454, 1065–1071. [Google Scholar] [CrossRef]
- Holzenberger, M.; Dupont, J.; Ducos, B.; Leneuve, P.; Géloën, A.; Even, P.; Cervera, P.; Le Bouc, Y. Igf-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 2003, 421, 182–187. [Google Scholar] [CrossRef]
- Kennedy, B.K.; Lamming, D.W. The mechanistic target of rapamycin: The grand conductor of metabolism and aging. Cell Metab. 2016, 23, 990–1003. [Google Scholar] [CrossRef]
- Henderson, S.T.; Bonafe, M.; Johnson, T.E. Daf-16 protects the nematode caenorhabditis elegans during food deprivation. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2006, 61, 444–460. [Google Scholar] [CrossRef]
- Hansen, M.; Taubert, S.; Crawford, D.; Libina, N.; Lee, S.J.; Kenyon, C. Lifespan extension by conditions that inhibit translation in caenorhabditis elegans. Aging Cell 2007, 6, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Syntichaki, P.; Troulinaki, K.; Tavernarakis, N. Eif4e function in somatic cells modulates ageing in caenorhabditis elegans. Nature 2007, 445, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of lifespan in drosophila by modulation of genes in the tor signaling pathway. Curr. Biol. 2004, 14, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009, 460, 392–395. [Google Scholar] [CrossRef]
- Miller, R.A.; Harrison, D.E.; Astle, C.M.; Fernandez, E.; Flurkey, K.; Han, M.; Javors, M.A.; Li, X.; Nadon, N.L.; Nelson, J.F.; et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell 2014, 13, 468–477. [Google Scholar] [CrossRef]
- Greer, E.L.; Brunet, A. Signaling networks in aging. J. Cell Sci. 2008, 121, 407–412. [Google Scholar] [CrossRef]
- Tait, I.S.; Li, Y.; Lu, J. Pten, longevity and age-related diseases. Biomedicines 2013, 1, 17–48. [Google Scholar] [CrossRef]
- Iwanami, A.; Cloughesy, T.F.; Mischel, P.S. Striking the balance between pten and pdk1: It all depends on the cell context. Genes Dev. 2009, 23, 1699–1704. [Google Scholar] [CrossRef]




| ID | p-Value | Fold Change | Expression Level (Old vs. Young) |
|---|---|---|---|
| rno-miR-101b-3p | 0.0295919 | 3.0300921 | Up |
| rno-miR-122-5p | 0.005130112 | 10.50495019 | Up |
| rno-miR-133a-3p | 0.000973173 | −3.631787956 | Down |
| rno-miR-143-3p | 0.033831212 | 2.469145377 | Up |
| rno-miR-145-5p | 0.032740298 | 17.04524832 | Up |
| rno-miR-150-3p | 0.016694482 | −3.404719964 | Down |
| rno-miR-181c-5p | 0.006634803 | −2.79454293 | Down |
| rno-miR-187-3p | 0.001 | 64 | Up |
| rno-miR-194-5p | 0.042745612 | 16.8112814 | Up |
| rno-miR-199a-5p | 0.009166599 | 3.06058214 | Up |
| rno-miR-202-3p | 0.001 | 64 | Up |
| rno-miR-203b-3p | 0.03954997 | 9.395112765 | Up |
| rno-miR-378a-3p | 0.043871243 | 1.797380304 | Up |
| rno-miR-450b-5p | 0.001 | 64 | Up |
| rno-miR-483-3p | 0.001 | −64 | Down |
| rno-miR-489-3p | 0.001 | −64 | Down |
| rno-miR-501-3p | 0.001 | 64 | Up |
| rno-miR-511-5p | 0.001 | 64 | Up |
| rno-miR-598-3p | 0.001 | 64 | Up |
| Ingenuity Canonical Pathways | −log (p-Value) | Related miRNA | Target Genes | Full Name |
|---|---|---|---|---|
| Insulin receptor signaling | 4.48 | miR-378a-3p | PDK1 | 3-phosphoinositide-dependent protein kinase 1 |
| miR-187-3p | INSR | insulin receptor | ||
| miR-202-3p | 4E-BP1 | Eukaryotic translation initiation factor 4E-binding protein 1 | ||
| miR-101b-3p | AKT | RAC-alpha serine/threonine-protein kinase | ||
| miR-199a-5p | STX4 | Syntaxin-4 | ||
| miR-203b-3p | PTEN | phosphatase and tensin homolog deleted on chromosome | ||
| mTOR signaling | 3.77 | miR-378a-3p | PDK1 | 3-phosphoinositide-dependent protein kinase 1 |
| miR-187-3p | INSR | insulin receptor | ||
| miR-202-3p | eIF4E-BP1 | Eukaryotic translation initiation factor 4E-binding protein 1 | ||
| miR-101b-3p | AKT | RAC-alpha serine/threonine-protein kinase | ||
| miR-194-5p | RAC | Aryl-hydrocarbon-interacting protein-like 1 | ||
| AMPK signaling | 2.84 | miR-378a-3p | PDK1 | phosphoinositide-dependent kinase-1 |
| miR-187-3p | INSR | insulin receptor | ||
| miR-202-3p | eIF4E-BP1 | Eukaryotic translation initiation factor 4E-binding protein 1 | ||
| miR-101b-3p | AKT | RAC-alpha serine/threonine-protein kinase | ||
| eNOS signaling | 2.37 | miR-378a-3p | PDK1 | phosphoinositide-dependent kinase-1 |
| miR-187-3p | CASP8 | caspase-8 | ||
| miR-101b-3p | AKT | RAC-alpha serine/threonine-protein kinase | ||
| miR-122-5p | CAT1 | cationic amino acid transporter 1 | ||
| miR-143-3p | CASP8 | caspase-8 | ||
| IGF-1 signaling | 2.94 | miR-378a-3p | PDK1, IGF-1R | phosphoinositide-dependent kinase-1, Insulin-like growth factor 1 receptor |
| miR-202-3p | IGF-1R | Insulin-like growth factor 1 receptor | ||
| miR-101b-3p | AKT | RAC-alpha serine/threonine-protein kinase | ||
| miR-145-5p | IGF-1R, PXN | insulin-like growth factor 1 receptor, Paxillin | ||
| miR-199a-5p | PXN | paxillin | ||
| PTEN signaling | 3.31 | miR-378a-3p | PDK1 | phosphoinositide-dependent kinase-1 |
| miR-101b-3p | BCL2L11, AKT | bcl-2-like protein 11, RAC-alpha serine/threonine-protein kinase | ||
| miR-203b-3p | PTEN | phosphatase and tensin homolog deleted on chromosome | ||
| miR-532-5p | NF-ƙB | Nuclear factor NF-kappa-B | ||
| p53 signaling | 2.13 | miR-202-3p | MDM4 | protein Mdm4 |
| miR-101b-3p | AKT | RAC-alpha serine/threonine-protein kinase | ||
| miR-203b-3p | PTEN | phosphatase and tensin homolog deleted on chromosome | ||
| miR-532-5p | MDM4, Slug | Protein Mdm4, Zinc finger protein SNAI2 | ||
| miR-450b-5p | MDM4 | Protein Mdm4 | ||
| miR-194-5p | PAI-1 | Glia-derived nexin | ||
| miR-199a-5p | HIPK2 | Homeodomain-interacting protein kinase 2 | ||
| miR-143-3p | HIPK2, Chk2 | Homeodomain-interacting protein kinase 2, Serine/threonine-protein kinase Chk2 | ||
| Integrin signaling | 4.13 | miR-378a-3p | PARVIN-α | parvin alpha |
| miR-101b-3p | ASAP1, PARVIN-α, AKT | arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1, parvin alpha, RAC-alpha serine/threonine-protein kinase | ||
| miR-145-5p | PXN, CRKL | Paxillin, Crk-like protein | ||
| miR-122-5p | PDGFβ | Platelet-derived growth factor subunit B | ||
| miR-203b-3p | PTEN | phosphatase and tensin homolog deleted on chromosome | ||
| miR-598-3p | PARVIN-α | parvin alpha | ||
| miR-199a-5p | PXN | Paxillin | ||
| Growth hormone signaling | 2.44 | miR-378a-3p | PDK1 | phosphoinositide-dependent kinase-1 |
| miR-202-3p | IGF-1R | Insulin-like growth factor 1 receptor | ||
| miR-145-5p | IGF-1R | Insulin-like growth factor 1 receptor |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Jin, K. Peripheral Circulating Exosomal miRNAs Potentially Contribute to the Regulation of Molecular Signaling Networks in Aging. Int. J. Mol. Sci. 2020, 21, 1908. https://doi.org/10.3390/ijms21061908
Zhang H, Jin K. Peripheral Circulating Exosomal miRNAs Potentially Contribute to the Regulation of Molecular Signaling Networks in Aging. International Journal of Molecular Sciences. 2020; 21(6):1908. https://doi.org/10.3390/ijms21061908
Chicago/Turabian StyleZhang, Hongxia, and Kunlin Jin. 2020. "Peripheral Circulating Exosomal miRNAs Potentially Contribute to the Regulation of Molecular Signaling Networks in Aging" International Journal of Molecular Sciences 21, no. 6: 1908. https://doi.org/10.3390/ijms21061908
APA StyleZhang, H., & Jin, K. (2020). Peripheral Circulating Exosomal miRNAs Potentially Contribute to the Regulation of Molecular Signaling Networks in Aging. International Journal of Molecular Sciences, 21(6), 1908. https://doi.org/10.3390/ijms21061908





