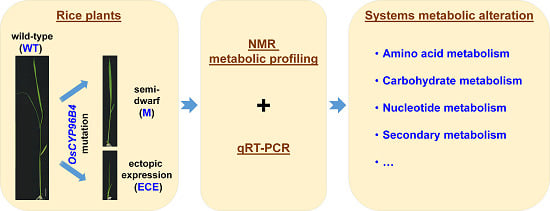
Systems Metabolic Alteration in a Semi-Dwarf Rice Mutant Induced by OsCYP96B4 Gene Mutation
Abstract
1. Introduction
2. Results
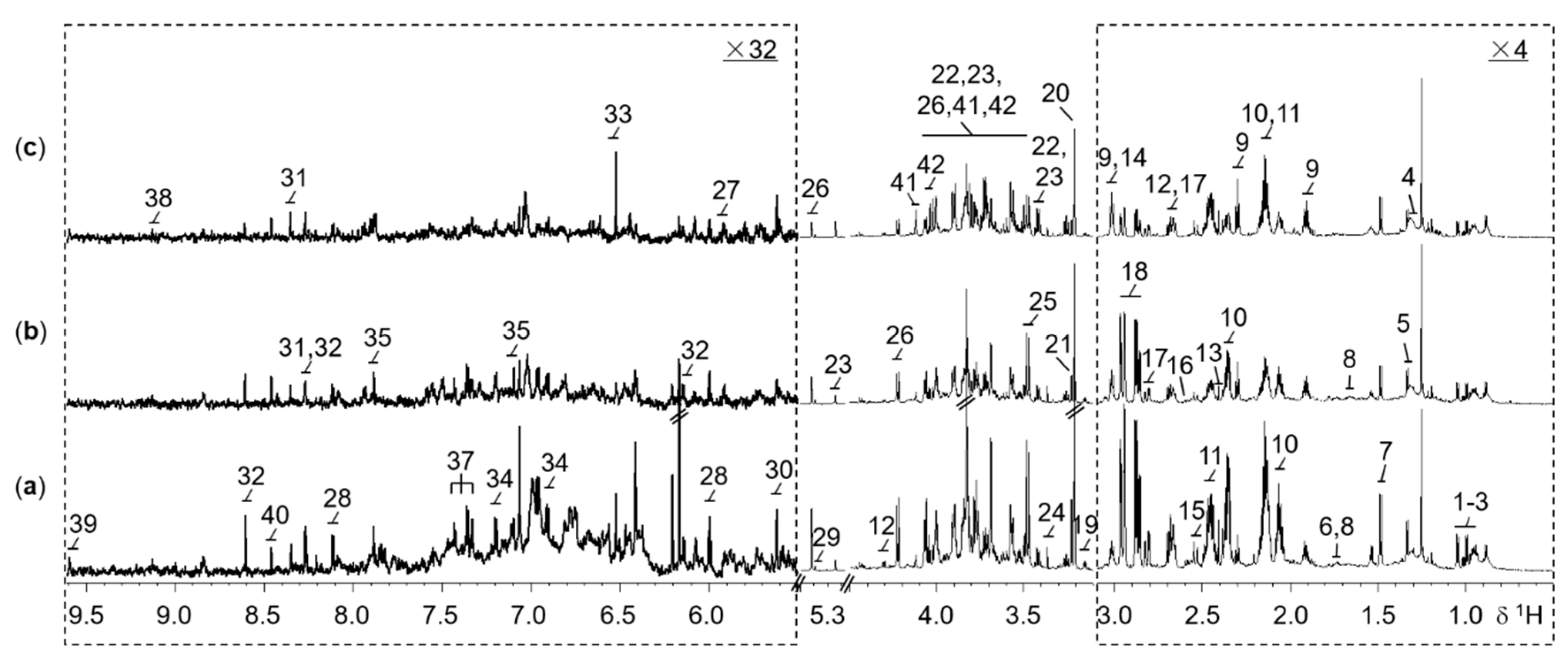
2.1. Metabolite Profiling in Wild-Type and Mutant Rice
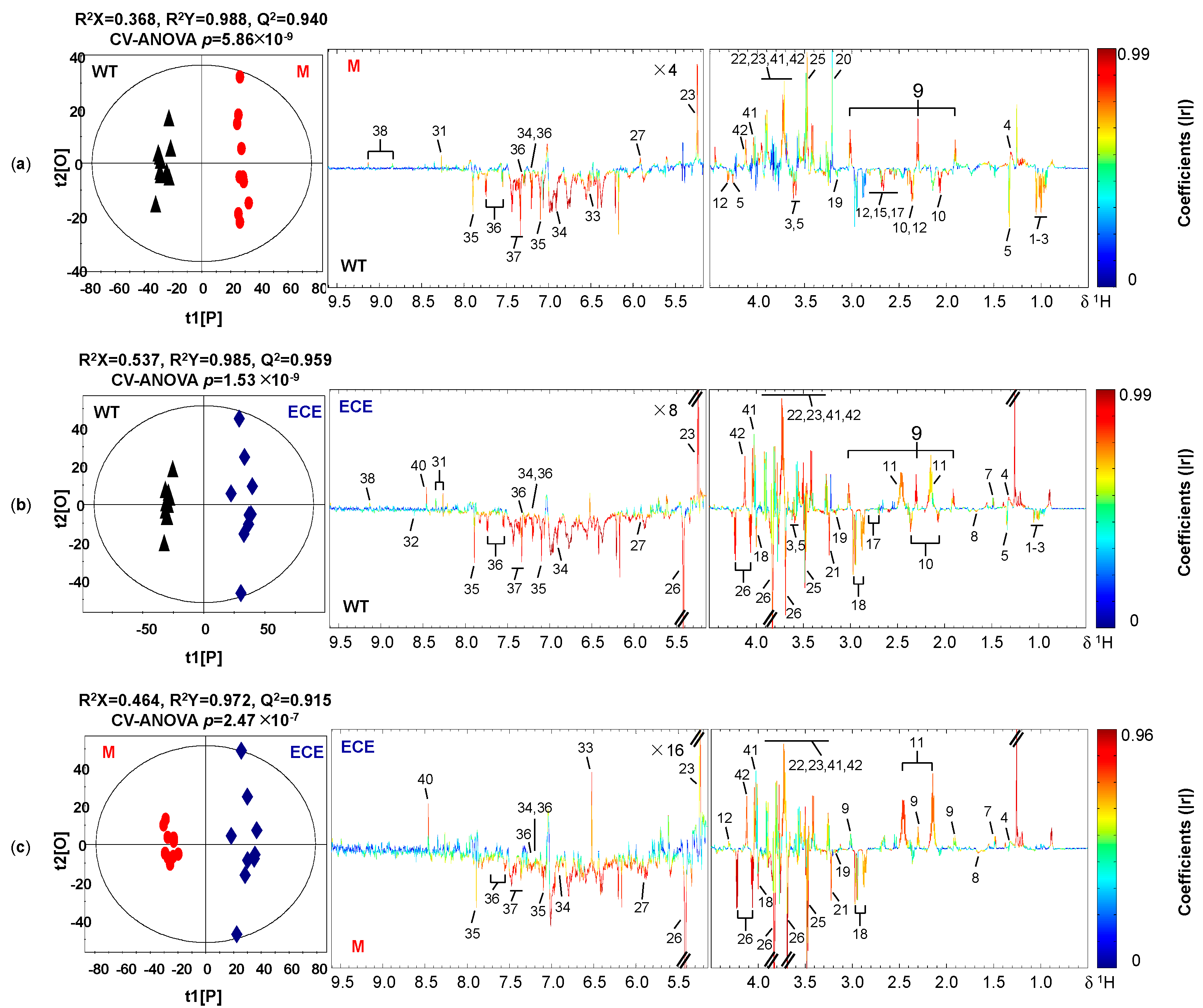
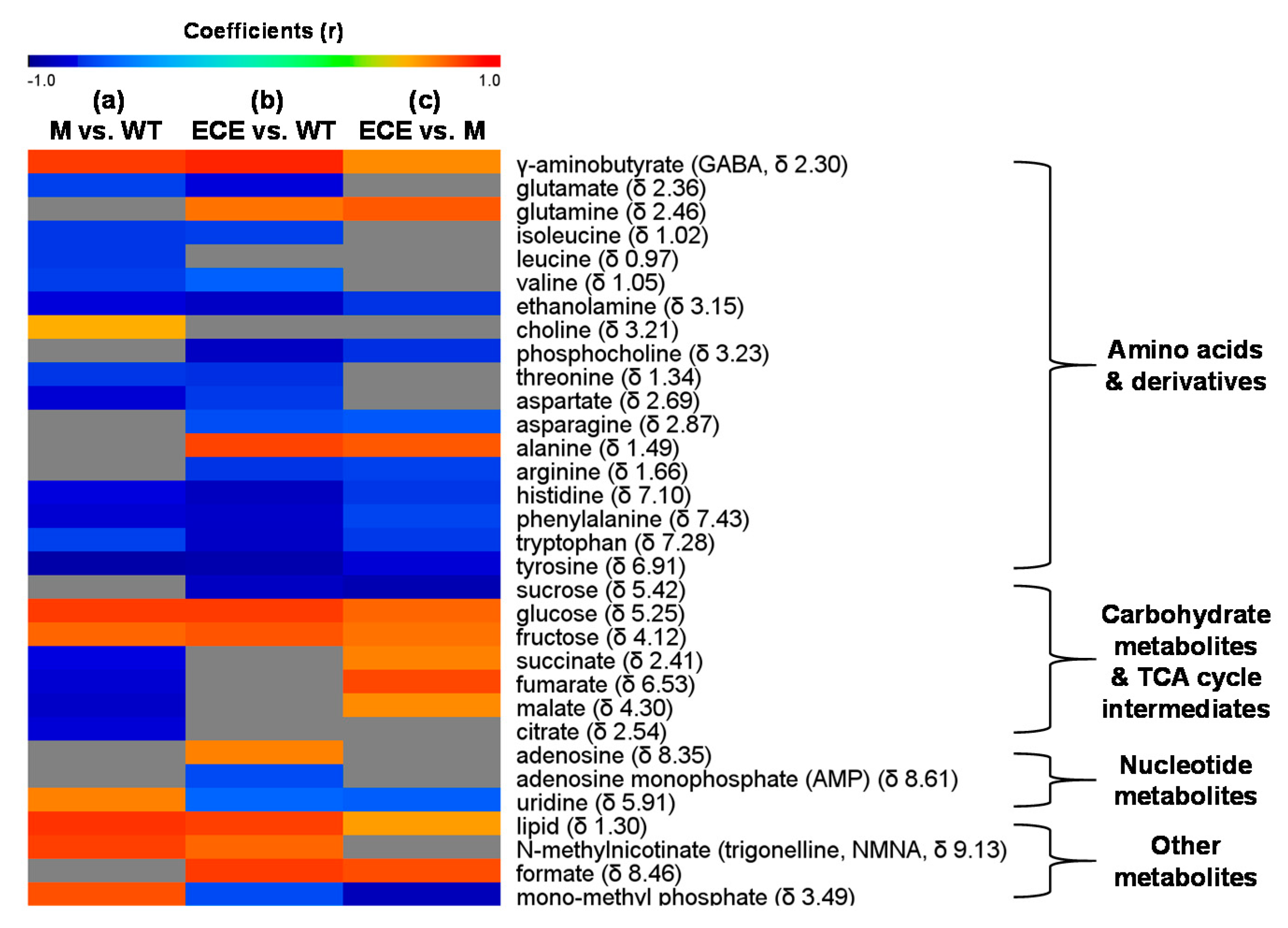
2.2. OsCYP96B4 Gene Mutation Induced Metabolic Changes in Mutant Rice
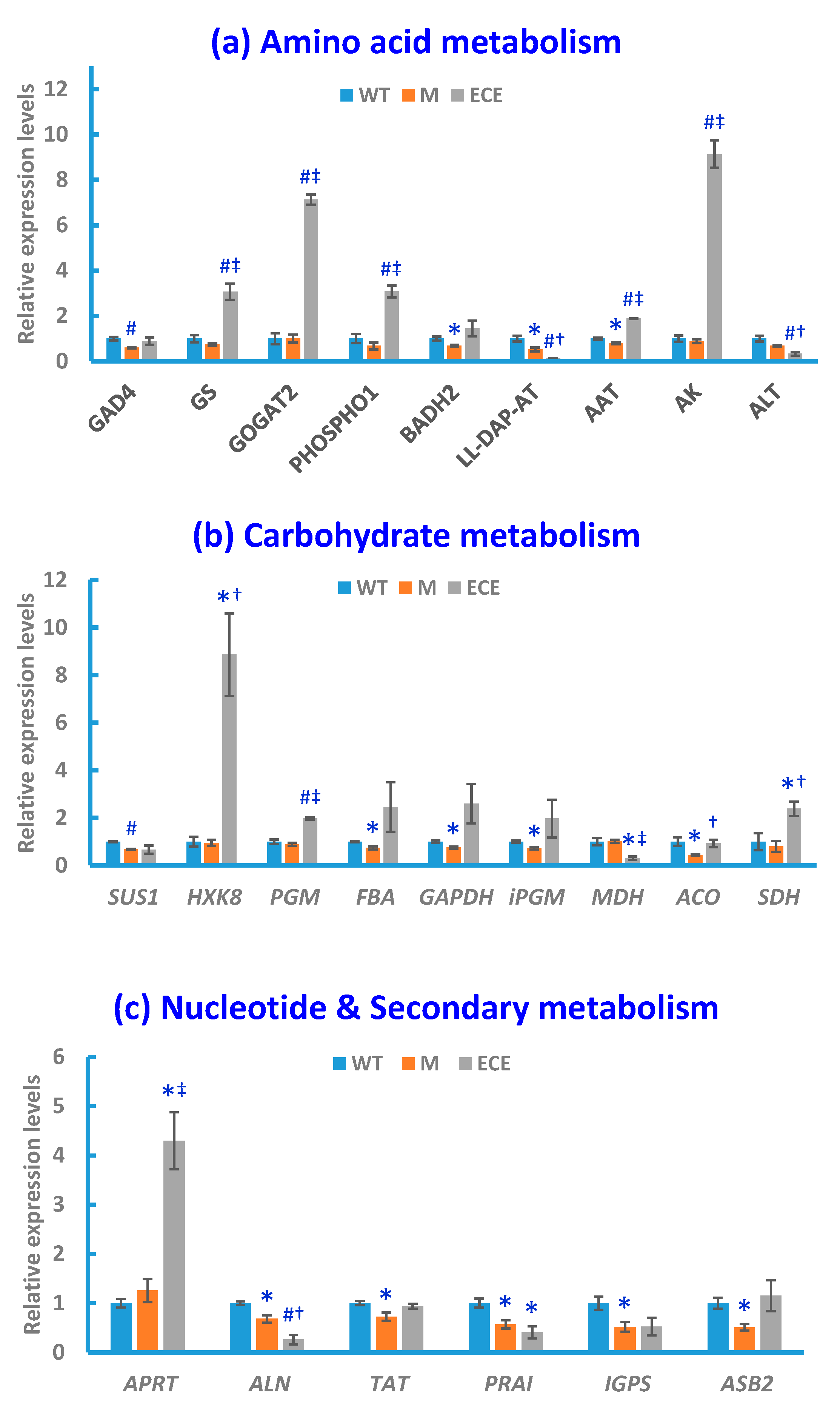
2.3. Gene Expression Analysis
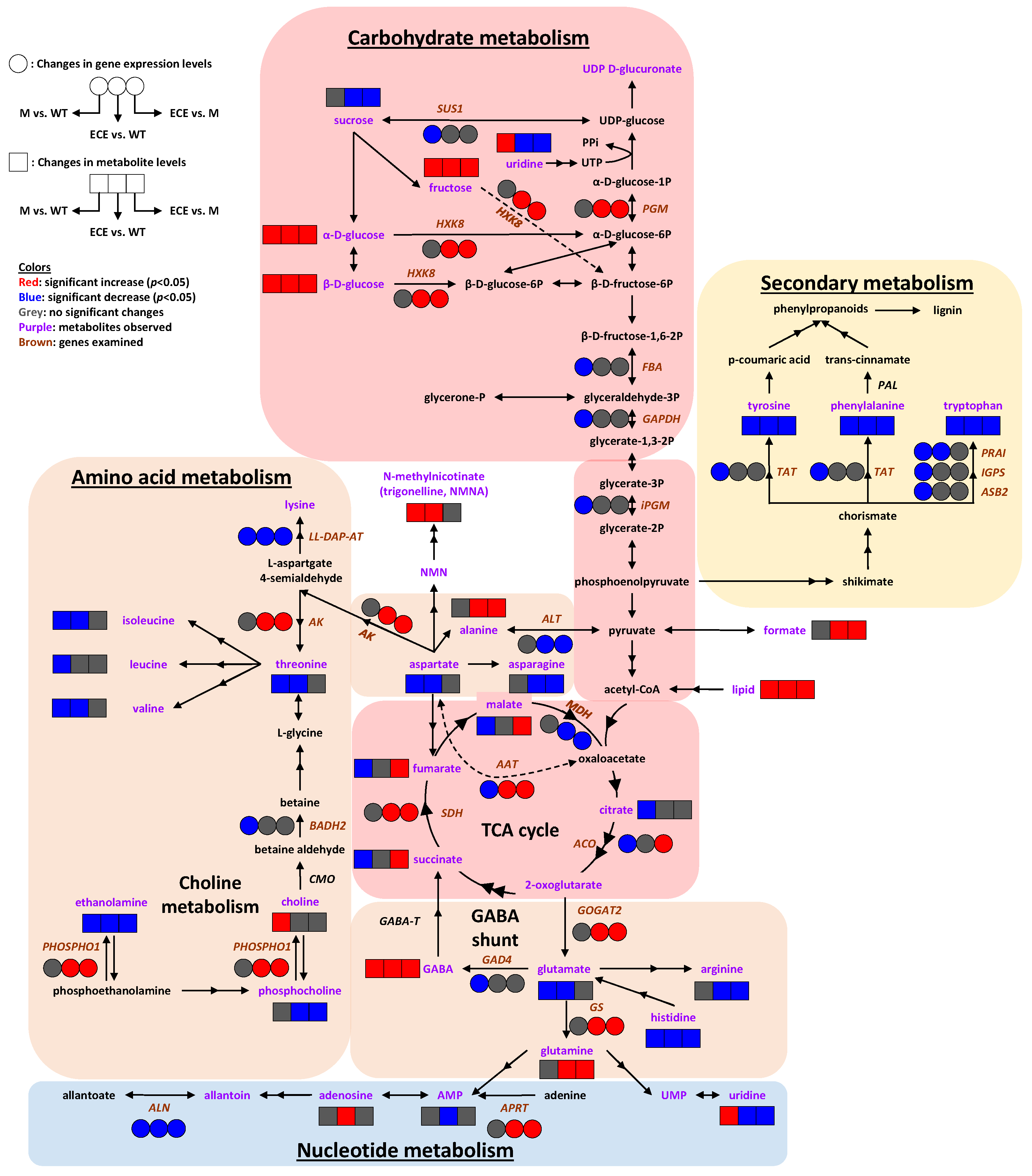
3. Discussion
3.1. Amino Acid Metabolism
3.2. Carbohydrate Metabolism
3.3. Nucleotide Metabolism
3.4. Secondary Metabolism
3.5. Other Metabolism
4. Materials and Methods
4.1. Plant Materials
4.2. Metabolome Analysis
4.2.1. Chemicals
4.2.2. Plant Extraction and Sample Preparation
4.2.3. NMR Spectroscopy
4.2.4. Multivariate Data Analysis
4.3. RNA Extraction and qRT-PCR Analysis
4.4. Metabolic Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Ds | Dissociator |
| ECE | Ectopic expression |
| GABA | γ-aminobutyrate |
| M | Mutant |
| NMR | Nuclear magnetic resonance |
| OPLS-DA | Orthogonal projection to latent structure discriminant analysis |
| OsCYP96B4 | Oryza sativa Cytochrome P450 96B4 |
| PCA | Principal component analysis |
| PLS-DA | Projection to latent structure discriminant analysis |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| TCA | Tricarboxylic acid |
| WT | Wild type |
References
- Yu, J.; Hu, S.N.; Wang, J.; Wong, G.K.S.; Li, S.G.; Liu, B.; Deng, Y.J.; Dai, L.; Zhou, Y.; Zhang, X.Q.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S. Green revolution: Preparing for the 21st century. Genome 1999, 42, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Morinaka, Y.; Sakamoto, T.; Inukai, Y.; Agetsuma, M.; Kitano, H.; Ashikari, M.; Matsuoka, M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 2006, 141, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Berry, P.M.; Sterling, M.; Spink, J.H.; Baker, C.J.; Sylvester-Bradley, R.; Mooney, S.J.; Tams, A.R.; Ennos, A.R. Understanding and reducing lodging in cereals. Adv. Agron. 2004, 84, 217–271. [Google Scholar]
- Ramamoorthy, R.; Jiang, S.Y.; Ramachandran, S. Oryza sativa Cytochrome P450 Family Member OsCYP96B4 Reduces Plant Height in a Transcript Dosage Dependent Manner. PLoS ONE 2011, 6, e28069. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.Q.; Li, S.Y.; Cheng, Z.K.; Li, C.Y. The Rice Semi-Dwarf Mutant sd37, Caused by a Mutation in CYP96B4, Plays an Important Role in the Fine-Tuning of Plant Growth. PLoS ONE 2014, 9, e88068. [Google Scholar] [CrossRef]
- Asano, K.; Hirano, K.; Ueguchi-Tanaka, M.; Angeles-Shim, R.B.; Komura, T.; Satoh, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Genet. Genomics 2009, 281, 223–231. [Google Scholar] [CrossRef]
- Tong, J.P.; Han, Z.S.; Han, A.N.; Liu, X.J.; Zhang, S.Y.; Fu, B.Y.; Hu, J.; Su, J.P.; Li, S.Q.; Wang, S.J.; et al. Sdt97: A Point Mutation in the 5 Untranslated Region Confers Semidwarfism in Rice. G3-Genes Genomes Genet. 2016, 6, 1491–1502. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.Y.; Guo, W.Z. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, H.F.; Wang, X.C.; Qiu, Y.J.; Tian, L.H.; Qi, X.Q.; Qu, L. Cytochrome P450 family member CYP96B5 hydroxylates alkanes to primary alcohols and is involved in rice leaf cuticular wax synthesis. New Phytol. 2020, 225, 2094–2107. [Google Scholar] [CrossRef]
- Wang, X.L.; Cheng, Z.J.; Zhao, Z.C.; Gan, L.; Qin, R.Z.; Zhou, K.N.; Ma, W.W.; Zhang, B.C.; Wang, J.L.; Zhai, H.Q.; et al. BRITTLE SHEATH1 encoding OsCYP96B4 is involved in secondary cell wall formation in rice. Plant Cell Rep. 2016, 35, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Tamiru, M.; Undan, J.R.; Takagi, H.; Abe, A.; Yoshida, K.; Undan, J.Q.; Natsume, S.; Uemura, A.; Saitoh, H.; Matsumura, H.; et al. A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 85–99. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics-the link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Connelly, J.; Lindon, J.C.; Holmes, E. Metabonomics: A platform for studying drug toxicity and gene function. Nat. Rev. Drug Discovery 2002, 1, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Kopka, J.; Dormann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef]
- Tang, H.R.; Wang, Y.L. Metabonomics: A revolution in progress. Prog. Biochem. Biophys. 2006, 33, 401–417. [Google Scholar]
- Jacobs, A.; Lunde, C.; Bacic, A.; Tester, M.; Roessner, U. The impact of constitutive heterologous expression of a moss Na+ transporter on the metabolomes of rice and barley. Metabolomics 2007, 3, 307–317. [Google Scholar] [CrossRef]
- Barding, G.A.; Fukao, T.; Beni, S.; Bailey-Serres, J.; Larive, C.K. Differential Metabolic Regulation Governed by the Rice SUB1A Gene during Submergence Stress and Identification of Alanylglycine by H-1 NMR Spectroscopy. J. Proteome Res. 2012, 11, 320–330. [Google Scholar] [CrossRef]
- Chen, F.F.; Zhang, J.T.; Song, X.S.; Yang, J.; Li, H.P.; Tang, H.R.; Liao, Y.C. Combined metabonomic and quantitative real-time PCR analyses reveal systems metabolic changes of Fusarium graminearum induced by Tri5 gene deletion. J. Proteome Res. 2011, 10, 2273–2285. [Google Scholar] [CrossRef]
- Liu, C.X.; Ding, F.; Hao, F.H.; Yu, M.; Lei, H.H.; Wu, X.Y.; Zhao, Z.X.; Guo, H.X.; Yin, J.; Wang, Y.L.; et al. Reprogramming of Seed Metabolism Facilitates Pre-harvest Sprouting Resistance of Wheat. Sci. Rep. 2016, 6, 20593. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Zhang, L.M.; Panigrahi, P.; Dholakia, B.B.; Dewangan, V.; Chavan, S.G.; Kunjir, S.M.; Wu, X.Y.; Li, N.; Rajmohanan, P.R.; et al. Fusarium oxysporum mediates systems metabolic reprogramming of chickpea roots as revealed by a combination of proteomics and metabolomics. Plant Biotechnol. J. 2016, 14, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Hao, F.H.; Hu, J.; Zhang, W.L.; Wan, L.L.; Zhu, L.L.; Tang, H.R.; He, G.C. Revealing different systems responses to brown planthopper infestation for pest susceptible and resistant rice plants with the combined metabonomic and gene-expression analysis. J. Proteome Res. 2010, 9, 6774–6785. [Google Scholar] [CrossRef] [PubMed]
- Villette, C.; Zumsteg, J.; Schaller, H.; Heintz, D. Non-targeted metabolic profiling of BW312 Hordeum vulgare semi dwarf mutant using UHPLC coupled to QTOF high resolution mass spectrometry. Sci. Rep. 2018, 8, 13178. [Google Scholar] [CrossRef]
- Cevallos-Cevallos, J.M.; Jines, C.; Mariduena-Zavala, M.G.; Molina-Miranda, M.J.; Ochoa, D.E.; Flores-Cedeno, J.A. GC-MS metabolite profiling for specific detection of dwarf somaclonal variation in banana plants. Appl. Plant Sci. 2018, 6, e1194. [Google Scholar] [CrossRef]
- Flores, P.; Hernandez, V.; Hellin, P.; Fenoll, J.; Cava, J.; Mestre, T.; Martinez, V. Metabolite profile of the tomato dwarf cultivar Micro-Tom and comparative response to saline and nutritional stresses with regard to a commercial cultivar. J. Sci. Food Agric. 2016, 96, 1562–1570. [Google Scholar] [CrossRef]
- John, K.M.M.; Khan, F.; Luthria, D.L.; Matthews, B.; Garrett, W.M.; Natarajan, S. Proteomic and metabolomic analysis of minimax and Williams 82 soybeans grown under two different conditions. J. Food Biochem. 2017, 41, e12404. [Google Scholar] [CrossRef]
- Blomstedt, C.K.; O’Donnell, N.H.; Bjarnholt, N.; Neale, A.D.; Hamill, J.D.; Moller, B.L.; Gleadow, R.M. Metabolic consequences of knocking out UGT85B1, the gene encoding the glucosyltransferase required for synthesis of dhurrin in Sorghum bicolor (L. Moench). Plant Cell Physiol. 2016, 57, 373–386. [Google Scholar] [CrossRef]
- Fan, W.M.T. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 1996, 28, 161–219. [Google Scholar] [CrossRef]
- Shelp, B.J.; Bown, A.W.; McLean, M.D. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999, 4, 446–452. [Google Scholar] [CrossRef]
- Bouche, N.; Fait, A.; Bouchez, D.; Moller, S.G.; Fromm, H. Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. USA 2003, 100, 6843–6848. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.; Matsukura, C.; Takayama, M.; Asamizu, E.; Ezura, H. Suppression of gamma-Aminobutyric Acid (GABA) Transaminases Induces Prominent GABA Accumulation, Dwarfism and Infertility in the Tomato (Solanum lycopersicum L.). Plant Cell Physiol. 2013, 54, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Akama, K.; Takaiwa, F. C-terminal extension of rice glutamate decarboxylase (OsGAD2) functions as an autoinhibitory domain and overexpression of a truncated mutant results in the accumulation of extremely high levels of GABA in plant cells. J. Exp. Bot. 2007, 58, 2699–2707. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Nian, J.Q.; Xie, Q.J.; Feng, J.; Zhang, F.X.; Jing, H.W.; Zhang, J.; Dong, G.J.; Liang, Y.; Peng, J.L.; et al. Rice Ferredoxin-Dependent Glutamate Synthase Regulates Nitrogen-Carbon Metabolomes and Is Genetically Differentiated between japonica and indica Subspecies. Mol. Plant 2016, 9, 1520–1534. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.X.; Zhao, R.; Li, F.F.; Tang, W.; Han, L.B. Simultaneous Expression of Spinacia oleracea Chloroplast Choline Monooxygenase (CMO) and Betaine Aldehyde Dehydrogenase (BADH) Genes Contribute to Dwarfism in Transgenic Lolium perenne. Plant Mol. Biol. Rep. 2011, 29, 379–388. [Google Scholar] [CrossRef]
- Fan, C.F.; Feng, S.Q.; Huang, J.F.; Wang, Y.T.; Wu, L.M.; Li, X.K.; Wang, L.Q.; Tu, Y.Y.; Xia, T.; Li, J.Y.; et al. AtCesA8-driven OsSUS3 expression leads to largely enhanced biomass saccharification and lodging resistance by distinctively altering lignocellulose features in rice. Biotechnol. Biofuels 2017, 10, 221. [Google Scholar] [CrossRef]
- Huang, P.; Yoshida, H.; Yano, K.; Kinoshita, S.; Kawai, K.; Koketsu, E.; Hattori, M.; Takehara, S.; Huang, J.; Hirano, K.; et al. OsIDD2, a zinc finger and INDETERMINATE DOMAIN protein, regulates secondary cell wall formation. J. Integr. Plant Biol. 2018, 60, 130–143. [Google Scholar] [CrossRef]
- Wu, B.M.; Li, L.; Qiu, T.H.; Zhang, X.; Cui, S.X. Cytosolic APX2 is a pleiotropic protein involved in H2O2 homeostasis, chloroplast protection, plant architecture and fertility maintenance. Plant Cell Rep. 2018, 37, 833–848. [Google Scholar] [CrossRef]
- Beeler, S.; Liu, H.C.; Stadler, M.; Schreier, T.; Eicke, S.; Lue, W.L.; Truernit, E.; Zeeman, S.C.; Chen, J.; Kotting, O. Plastidial NAD-Dependent Malate Dehydrogenase Is Critical for Embryo Development and Heterotrophic Metabolism in Arabidopsis. Plant Physiol. 2014, 164, 1175–1190. [Google Scholar] [CrossRef]
- Itai, R.; Suzuki, K.; Yamaguchi, H.; Nakanishi, H.; Nishizawa, N.K.; Yoshimura, E.; Mori, S. Induced activity of adenine phosphoribosyltransferase (APRT) in iron-deficient barley roots: A possible role for phytosiderophore production. J. Exp. Bot. 2000, 51, 1179–1188. [Google Scholar] [CrossRef][Green Version]
- Casartelli, A.; Melino, V.J.; Baumann, U.; Riboni, M.; Suchecki, R.; Jayasinghe, N.S.; Mendis, H.; Watanabe, M.; Erban, A.; Zuther, E.; et al. Opposite fates of the purine metabolite allantoin under water and nitrogen limitations in bread wheat. Plant Mol.Biol. 2019, 99, 477–497. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.J.; Li, J.; Zou, J.C.; Liang, F.S.; Ye, C.J.; Jin, D.M.; Weng, M.L.; Wang, B. Cloning and characterization of a second form of the rice adenine phosphoribosyl transferase gene (OsAPT2) and its association with TGMS. Plant Mol.Biol. 2006, 60, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, B.; Somerville, C. Positive selection for male-sterile mutants of Arabidopsis lacking adenine phosphoribosyl transferase activity. Plant Physiol. 1988, 86, 1150–1154. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Galili, G. The Biosynthetic Pathways for Shikimate and Aromatic Amino Acids in Arabidopsis thaliana. Arabidopsis Book 2010, 8, e0132. [Google Scholar] [CrossRef]
- Tzin, V.; Galili, G. New Insights into the Shikimate and Aromatic Amino Acids Biosynthesis Pathways in Plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef]
- Yang, D.H.; Chung, B.Y.; Kim, J.S.; Kim, J.H.; Yun, P.Y.; Lee, Y.K.; Lim, Y.P.; Lee, M.C. cDNA cloning and sequence analysis of the rice cinnamate-4-hydroxylase gene, a cytochrome P450-dependent monooxygenase involved in the general phenylpropanoid pathway. J. Plant Biol. 2005, 48, 311–318. [Google Scholar] [CrossRef]
- Kim, J.I.; Dolan, W.L.; Anderson, N.A.; Chapple, C. Indole Glucosinolate Biosynthesis Limits Phenylpropanoid Accumulation in Arabidopsis thaliana. Plant Cell 2015, 27, 1529–1546. [Google Scholar] [CrossRef]
- Ahmad, I.; Kamran, M.; Ali, S.; Bilegjargal, B.; Cai, T.; Ahmad, S.; Meng, X.P.; Su, W.N.; Liu, T.N.; Han, Q.F. Uniconazole application strategies to improve lignin biosynthesis, lodging resistance and production of maize in semiarid regions. Field Crop. Res. 2018, 222, 66–77. [Google Scholar] [CrossRef]
- Wang, C.; Hu, D.; Liu, X.B.; She, H.Z.; Ruan, R.W.; Yang, H.; Yi, Z.L.; Wu, D.Q. Effects of uniconazole on the lignin metabolism and lodging resistance of culm in common buckwheat (Fagopyrum esculentum M.). Field Crop. Res. 2015, 180, 46–53. [Google Scholar] [CrossRef]
- Nishikubo, N.; Araki, T.; Kajita, S.; Kuroda, K.; Kitano, H.; Katayama, Y. Specific accumulation of polysaccharide-linked hydroxycinnamoyl esters in the cell walls of irregularly shaped and collapsed internode parenchyma cells of the dwarf rice mutant Fukei 71. Plant Cell Physiol. 2000, 41, 776–784. [Google Scholar] [CrossRef]
- Kim, Y.S.; Park, S.; Kang, K.; Lee, K.; Back, K. Tyramine accumulation in rice cells caused a dwarf phenotype via reduced cell division. Planta 2011, 233, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Back, K. Melatonin-deficient rice plants show a common semidwarf phenotype either dependent or independent of brassinosteroid biosynthesis. J. Pineal Res. 2019, 66, e12537. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Jiang, L.M.; Huang, J.; Wang, Y.L.; Tang, H.R. Metabonomic analysis reveals the CCl4-induced systems alterations for multiple rat organs. J. Proteome Res. 2012, 11, 3848–3859. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lee, S.C.; Ng, T.C. Pharmacometabonomics Analysis Reveals Serum Formate and Acetate Potentially Associated with Varying Response to Gemcitabine-Carboplatin Chemotherapy in Metastatic Breast Cancer Patients. J. Proteome Res. 2018, 17, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.M.; Zhao, X.J.; Huang, C.Y.; Lei, H.H.; Tang, H.R.; Wang, Y.L. Dynamic changes in metabolic profiles of rats subchronically exposed to mequindox. Mol. Biosyst. 2014, 10, 2914–2922. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, J.; Li, R.; Fang, Z.-M.; Zhu, X.-H.; Yi, X.; Lan, H.; Wei, X.; Jiang, D.-S. Disturbed energy and amino acid metabolism with their diagnostic potential in mitral valve disease revealed by untargeted plasma metabolic profiling. Metabolomics 2019, 15, 57. [Google Scholar] [CrossRef]
- Jiang, L.M.; Huang, J.; Wang, Y.L.; Tang, H.R. Eliminating the dication-induced intersample chemical-shift variations for NMR-based biofluid metabonomic analysis. Analyst 2012, 137, 4209–4219. [Google Scholar] [CrossRef]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Lindgren, F.; Hansen, B.; Karcher, W.; Sjostrom, M.; Eriksson, L. Model validation by permutation tests: Applications to variable selection. J. Chemom. 1996, 10, 521–532. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS® models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Cloarec, O.; Dumas, M.E.; Trygg, J.; Craig, A.; Barton, R.H.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal. Chem. 2005, 77, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Pryor, R.J.; Wittwer, C.T. Real-time polymerase chain reaction and melting curve analysis. In Methods in Molecular Biology; Lo, Y.M.D., Chiu, R.W.K., Chan, K.C.A., Eds.; 999 Riverview Dr, Ste 208; Humana Press Inc.: Totowa, NJ, USA, 2006; Volume 336, pp. 19–32. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Ramamoorthy, R.; Ramachandran, S.; Kumar, P.P. Systems Metabolic Alteration in a Semi-Dwarf Rice Mutant Induced by OsCYP96B4 Gene Mutation. Int. J. Mol. Sci. 2020, 21, 1924. https://doi.org/10.3390/ijms21061924
Jiang L, Ramamoorthy R, Ramachandran S, Kumar PP. Systems Metabolic Alteration in a Semi-Dwarf Rice Mutant Induced by OsCYP96B4 Gene Mutation. International Journal of Molecular Sciences. 2020; 21(6):1924. https://doi.org/10.3390/ijms21061924
Chicago/Turabian StyleJiang, Limiao, Rengasamy Ramamoorthy, Srinivasan Ramachandran, and Prakash P. Kumar. 2020. "Systems Metabolic Alteration in a Semi-Dwarf Rice Mutant Induced by OsCYP96B4 Gene Mutation" International Journal of Molecular Sciences 21, no. 6: 1924. https://doi.org/10.3390/ijms21061924
APA StyleJiang, L., Ramamoorthy, R., Ramachandran, S., & Kumar, P. P. (2020). Systems Metabolic Alteration in a Semi-Dwarf Rice Mutant Induced by OsCYP96B4 Gene Mutation. International Journal of Molecular Sciences, 21(6), 1924. https://doi.org/10.3390/ijms21061924