Comprehensive Phosphoproteomic Analysis of Pepper Fruit Development Provides Insight into Plant Signaling Transduction
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phosphoproteomic Profiling
2.2. Characteristics of the Phosphorylation Sites
2.3. Identification of Differentially Expressed Phosphoproteins (DEPPs) at Different Stages
2.4. Motif Analysis of Lysine Phosphorylated Peptides
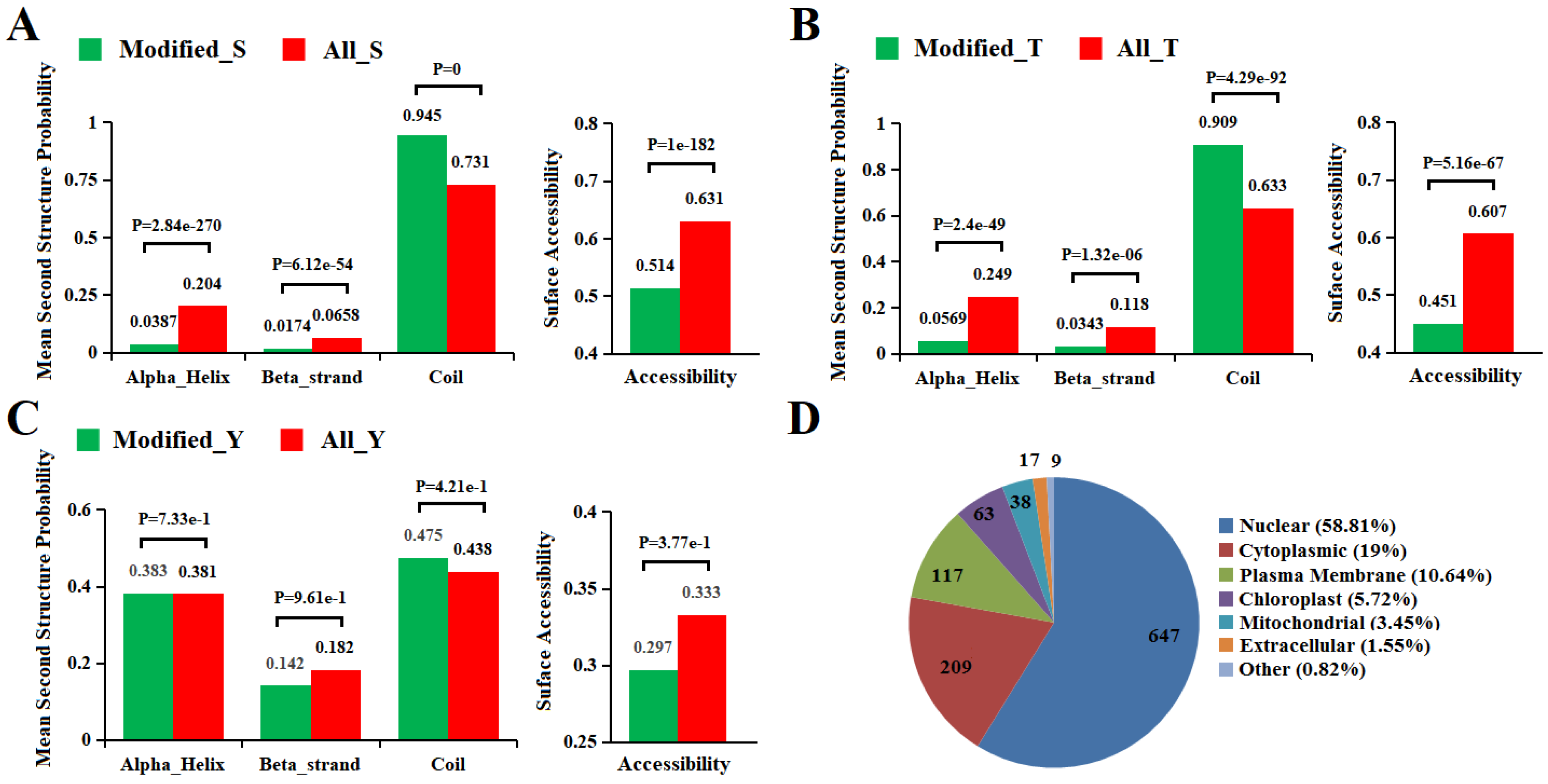
2.5. Secondary Structure and Subcellular Localization Analysis
2.6. Top 20 Most Abundant GO Term Analysis of DEPPs
2.7. Top 20 Most Abundant KEGG Analysis of DEPPs
2.8. The 120 Kinase Analysis in Phosphorylated Proteome
2.9. Plant Hormone Signal Transduction in Phosphorylated Proteome
3. Materials and Methods
3.1. Plant Growth and Sampling
3.2. Protein Extraction and FASP Digestion
3.3. TMT Labeling and Enrichment of Phosphorylated Peptides by the TiO2 Beads
3.4. HPLC and LC-MS/MS Analysis
3.5. Data Analysis
3.6. Gene Ontology (GO) and KEGG Annotation
3.7. Motif Prediction
3.8. Accession Numbers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| ABA | Abscisic acid |
| Br | Breaking |
| CDPKs | Calcium-dependent protein kinases |
| CKs | Casein kinases |
| CDKs | Cyclin-depenent kinases |
| DEPPs | Differentially expressed phosphoproteins |
| IMG | Immature Green |
| MG | Mature Green |
| MR | Mature Red |
| MAPKs | Mitogen-activated protein kinases |
| MAPKKs | Mitogen-activated protein kinase kinases |
| MAPKKKs | Mitogen-activated protein kinase kinase kinases |
| PLKs | Serine/threonine kinases |
| PEPCK | Photophosphorylpyruvate carboxylase |
| pSer | Phosphoserine |
| pThr | Phosphothreonine |
| pTyr | Phosphotyrosine |
| RPKs | Receptor protein kinases |
| TMT | Tandem Mass Tag |
References
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S. PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Aza-González, C.; Núñez-Palenius, H.G.; Ochoa-Alejo, N. Molecular biology of capsaicinoid biosynthesis in chili pepper (Capsicum spp.). Plant Cell Rep. 2011, 30, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Howard, L.; Talcott, S.; Brenes, C.; Villalon, B. Changes in phytochemical and antioxidant activity of selected pepper cultivars (Capsicum species) as influenced by maturity. J. Agric. Food Chem. 2000, 48, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Mikulic-Petkovsek, M.; Rescic, J.; Schmitzer, V.; Stampar, F.; Slatnar, A.; Koron, D.; Veberic, R. Changes in fruit quality parameters of four Ribes species during ripening. Food Chem. 2015, 173, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Martínez-López, L.A.; Ochoa-Alejo, N.; Martínez, O. Dynamics of the chili pepper transcriptome during fruit development. BMC Genom. 2014, 15, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.M.; Kim, S.; Lee, J.Y.; Yoo, E.Y.; Cho, M.C.; Cho, M.R.; Kim, B.D.; Bahk, Y.Y. A differentially expressed proteomic analysis in placental tissues in relation to pungency during the pepper fruit development. Proteomics 2006, 6, 5248–5259. [Google Scholar] [CrossRef]
- Kersten, B.; Agrawal, G.K.; Iwahashi, H.; Rakwal, R. Plant phosphoproteomics: A long road ahead. Proteomics 2006, 6, 5517–5528. [Google Scholar] [CrossRef]
- Hunter, T. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 2007, 28, 730–738. [Google Scholar] [CrossRef]
- Wan, J.; Kang, S.; Tang, C.; Yan, J.; Ren, Y.; Liu, J.; Gao, X.; Banerjee, A.; Ellis, L.B.; Li, T. Meta-prediction of phosphorylation sites with weighted voting and restricted grid search parameter selection. Nucleic Acids Res. 2008, 36, e22. [Google Scholar] [CrossRef] [Green Version]
- Peuchen, E.H.; Cox, O.F.; Sun, L.; Hebert, A.S.; Coon, J.J.; Champion, M.M.; Dovichi, N.J.; Huber, P.W. Phosphorylation dynamics dominate the regulated proteome during early Xenopus development. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Wu, M.; Pei, W.; Li, H.; Li, X.; Zhang, J.; Yu, J.; Yu, S. Quantitative phosphoproteomic profiling of fiber differentiation and initiation in a fiberless mutant of cotton. BMC Genom. 2014, 15, 466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.-H.; Sladek, T.L. Novel phosphorylated forms of E2F-1 transcription factor bind to the retinoblastoma protein in cells overexpressing an E2F-1 cDNA. Biochem. Biophys. Res. Commun. 1997, 232, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Dephoure, N.; Gould, K.L.; Gygi, S.P.; Kellogg, D.R. Mapping and analysis of phosphorylation sites: A quick guide for cell biologists. Mol. Biol. Cell 2013, 24, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Ong, S.-E.; Grønborg, M.; Steen, H.; Jensen, O.N.; Pandey, A. Analysis of protein phosphorylation using mass spectrometry: Deciphering the phosphoproteome. Trends biotechnol. 2002, 20, 261–268. [Google Scholar] [CrossRef]
- Meyer, L.J.; Gao, J.; Xu, D.; Thelen, J.J. Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol. 2012, 159, 517–528. [Google Scholar] [CrossRef] [Green Version]
- Bi, Y.-D.; Wang, H.-X.; Lu, T.-C.; Li, X.-H.; Shen, Z.; Chen, Y.-B.; Wang, B.-C. Large-scale analysis of phosphorylated proteins in maize leaf. Planta 2011, 233, 383–392. [Google Scholar] [CrossRef]
- Facette, M.R.; Shen, Z.; Björnsdóttir, F.R.; Briggs, S.P.; Smith, L.G. Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. Plant Cell 2013, 25, 2798–2812. [Google Scholar] [CrossRef] [Green Version]
- Lv, D.-W.; Ge, P.; Zhang, M.; Cheng, Z.-W.; Li, X.-H.; Yan, Y.-M. Integrative network analysis of the signaling cascades in seedling leaves of bread wheat by large-scale phosphoproteomic profiling. J. Proteome Res. 2014, 13, 2381–2395. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Thelen, J.J. Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol. Cell. Proteom. 2006, 5, 2044–2059. [Google Scholar] [CrossRef] [Green Version]
- Chitteti, B.R.; Peng, Z. Proteome and phosphoproteome dynamic change during cell dedifferentiation in Arabidopsis. Proteomics 2007, 7, 1473–1500. [Google Scholar] [CrossRef]
- Chao, Q.; Liu, X.-Y.; Mei, Y.-C.; Gao, Z.-F.; Chen, Y.-B.; Qian, C.-R.; Hao, Y.-B.; Wang, B.-C. Light-regulated phosphorylation of maize phosphoenolpyruvate carboxykinase plays a vital role in its activity. Plant Mol. Biol. 2014, 85, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Medzihradszky, M.; Bindics, J.; Ádám, É.; Viczián, A.; Klement, É.; Lorrain, S.; Gyula, P.; Mérai, Z.; Fankhauser, C.; Medzihradszky, K.F. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 2013, 25, 535–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anguenot, R.; Nguyen-Quoc, B.; Yelle, S.; Michaud, D. Protein phosphorylation and membrane association of sucrose synthase in developing tomato fruit. Plant Physiol. Biochem. 2006, 44, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Stulemeijer, I.J.; Joosten, M.H.; Jensen, O.N. Quantitative phosphoproteomics of tomato mounting a hypersensitive response reveals a swift suppression of photosynthetic activity and a differential role for hsp90 isoforms. J. Proteome Res. 2009, 8, 1168–1182. [Google Scholar] [CrossRef] [PubMed]
- Elagamey, E.; Narula, K.; Sinha, A.; Aggarwal, P.R.; Ghosh, S.; Chakraborty, N.; Chakraborty, S. Extracellular matrix proteome and phosphoproteome of potato reveals functionally distinct and diverse canonical and non-canonical proteoforms. Proteomes 2016, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Lv, J.; Zhang, Z.; Li, H.; Yang, B.; Chen, W.; Dai, X.; Li, X.; Yang, S.; Liu, L. Integrative transcriptome and proteome analysis identifies major metabolic pathways involved in pepper fruit development. J. Proteome Res. 2019, 18, 982–994. [Google Scholar] [CrossRef]
- Cieśla, J.; Frączyk, T.; Rode, W. Phosphorylation of basic amino acid residues in proteins: Important but easily missed. Acta Biochim. Pol. 2011, 58. [Google Scholar] [CrossRef] [Green Version]
- Reiland, S.; Messerli, G.; Baerenfaller, K.; Gerrits, B.; Endler, A.; Grossmann, J.; Gruissem, W.; Baginsky, S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009, 150, 889–903. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, N.; Nakagami, H.; Mochida, K.; Daudi, A.; Tomita, M.; Shirasu, K.; Ishihama, Y. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 2008, 4, 193. [Google Scholar] [CrossRef]
- Lv, D.-W.; Li, X.; Zhang, M.; Gu, A.-Q.; Zhen, S.-M.; Wang, C.; Li, X.-H.; Yan, Y.-M. Large-scale phosphoproteome analysis in seedling leaves of Brachypodium distachyon L. BMC Genom. 2014, 15, 375. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.-X.; Zhen, S.-M.; Liu, Y.-L.; Yan, X.; Zhang, M.; Yan, Y.-M. In vivo phosphoproteome characterization reveals key starch granule-binding phosphoproteins involved in wheat water-deficit response. BMC Plant Biol. 2017, 17, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, J.; Chen, Y.; Hu, Y.; Deng, Y.; Liu, Y.; Li, G.; Zou, W.; Zhao, J. Arabidopsis replication factor C4 is critical for DNA replication during the mitotic cell cycle. Plant J. 2018, 94, 288–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.A.; Marzahn, M.R.; O’Donnell, M.; Bloom, L.B. Replication factor C is a more effective proliferating cell nuclear antigen (PCNA) opener than the checkpoint clamp loader, Rad24-RFC. J. Biol. Chem. 2012, 287, 2203–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Deng, Y.; Li, G.; Zhao, J. Replication factor C 1 (RFC 1) is required for double-strand break repair during meiotic homologous recombination in A rabidopsis. Plant J. 2013, 73, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Twyffels, L.; Gueydan, C.; Kruys, V. Shuttling SR proteins: More than splicing factors. FEBS J. 2011, 278, 3246–3255. [Google Scholar] [CrossRef]
- Melcák, I.; Cermanová, S.T.; Jirsová, K.; Koberna, K.; Malınsky, J.; Raška, I. Nuclear pre-mRNA compartmentalization: Trafficking of released transcripts to splicing factor reservoirs. Mol. Biol. Cell 2000, 11, 497–510. [Google Scholar] [CrossRef] [Green Version]
- Lopato, S.; Kalyna, M.; Dorner, S.; Kobayashi, R.; Krainer, A.R.; Barta, A. atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 1999, 13, 987–1001. [Google Scholar] [CrossRef] [Green Version]
- Hinnebusch, A.G. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006, 31, 553–562. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- van Wijk, K.J.; Friso, G.; Walther, D.; Schulze, W.X. Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 2014, 26, 2367–2389. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Hou, Y.; Wang, Y.; Li, Z.; Zhao, J.; Tong, X.; Lin, H.; Wei, X.; Ao, H.; Zhang, J. A comprehensive proteomic survey of ABA-induced protein phosphorylation in rice (Oryza sativa L.). Int. J. Mol. Sci. 2017, 18, 60. [Google Scholar] [CrossRef]
- Petersen, B.; Petersen, T.N.; Andersen, P.; Nielsen, M.; Lundegaard, C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009, 9, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Qiu, J.; Wang, Y.; Li, Z.; Zhao, J.; Tong, X.; Lin, H.; Zhang, J. A quantitative proteomic analysis of brassinosteroid-induced protein phosphorylation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Yang, M.-K.; Li, C.-Y.; Wang, Y.; Zhang, J.; Wang, D.-B.; Zhang, X.-E.; Ge, F. Phosphoproteomic analysis provides novel insights into stress responses in Phaeodactylum tricornutum, a model diatom. J. Proteome Res. 2014, 13, 2511–2523. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Bentem, S.D.L.F.; Hirt, H. Protein tyrosine phosphorylation in plants: More abundant than expected? Trends Plant Sci. 2009, 14, 71–76. [Google Scholar] [CrossRef]
- Chen, P.; Li, R.; Zhou, R. Comparative phosphoproteomic analysis reveals differentially phosphorylated proteins regulate anther and pollen development in kenaf cytoplasmic male sterility line. Amino Acids 2018, 50, 841–862. [Google Scholar] [CrossRef]
- Zeng, Y.; Pan, Z.; Wang, L.; Ding, Y.; Xu, Q.; Xiao, S.; Deng, X. Phosphoproteomic analysis of chromoplasts from sweet orange during fruit ripening. Physiol. Plant. 2014, 150, 252–270. [Google Scholar] [CrossRef]
- Bögre, L.; Ligterink, W.; Heberle-Bors, E.; Hirt, H. Mechanosensors in plants. Nature 1996, 383, 489–490. [Google Scholar] [CrossRef]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.-E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar]
- Sugiura, R.; Kita, A.; Shimizu, Y.; Shuntoh, H.; Sio, S.O.; Kuno, T. Feedback regulation of MAPK signalling by an RNA-binding protein. Nature 2003, 424, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Kosetsu, K.; Matsunaga, S.; Nakagami, H.; Colcombet, J.; Sasabe, M.; Soyano, T.; Takahashi, Y.; Hirt, H.; Machida, Y. The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 2010, 22, 3778–3790. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Cai, Z.; Guo, Y.; Gan, S. An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 2009, 150, 167–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.-Q.; Oono, K.; Imai, R. Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol. 2002, 129, 1880–1891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, G.K.; Agrawal, S.K.; Shibato, J.; Iwahashi, H.; Rakwal, R. Novel rice MAP kinases OsMSRMK3 and OsWJUMK1 involved in encountering diverse environmental stresses and developmental regulation. Biochem. Biophys. Res. Commun. 2003, 300, 775–783. [Google Scholar] [CrossRef]
- Walton, A.; Stes, E.; De Smet, I.; Goormachtig, S.; Gevaert, K. Plant hormone signalling through the eye of the mass spectrometer. Proteomics 2015, 15, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Bethke, G.; Unthan, T.; Uhrig, J.F.; Pöschl, Y.; Gust, A.A.; Scheel, D.; Lee, J. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 8067–8072. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.-D.; Cho, Y.-H.; Tena, G.; Xiong, Y.; Sheen, J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C 2 H 4 signalling. Nature 2008, 451, 789–795. [Google Scholar] [CrossRef]
- Wen, X.; Zhang, C.; Ji, Y.; Zhao, Q.; He, W.; An, F.; Jiang, L.; Guo, H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012, 22, 1613–1616. [Google Scholar] [CrossRef] [Green Version]
- Alonso, J.M.; Hirayama, T.; Roman, G.; Nourizadeh, S.; Ecker, J.R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 1999, 284, 2148–2152. [Google Scholar] [CrossRef]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, C.; Yoon, G.M.; Shemansky, J.M.; Lin, D.Y.; Ying, Z.I.; Chang, J.; Garrett, W.M.; Kessenbrock, M.; Groth, G.; Tucker, M.L. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 19486–19491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Binder, B.M.; Garrett, W.M.; Tucker, M.L.; Chang, C.; Cooper, B. Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Mol. Biosyst. 2011, 7, 2637–2650. [Google Scholar] [CrossRef] [PubMed]
- Merchante, C.; Alonso, J.M.; Stepanova, A.N. Ethylene signaling: Simple ligand, complex regulation. Curr. Opin. Plant Biol. 2013, 16, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.Y.; Chae, M.J.; Lee, I.S.; Lee, Y.N.; Nam, M.H.; Kim, D.Y.; Byun, M.O.; Yoon, I.S. Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry 2011, 72, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhang, W.; Wang, X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017, 15, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.; Oosten, M.; Zhang, H.; Tao, W.; Zhu, J. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Singal. 2013, 6, rs8. [Google Scholar] [CrossRef]
- Ishikawa, S.; Barrero, J.; Takahashi, F.; Takahashi, F.; Peck, S.; Gubler, F.; Shinozaka, K.; Umezawa, T. Comparative phosphoproteomic analysis of barley embryos with different dormancy during imbibition. Int. J. Mol. Sci. 2019, 20, 451. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Hou, Y.; Tong, X.; Wng, Y.; Lin, H.; Liu, Q.; Zhang, W.; Li, Z.; Nallamilli, B.; Zhang, J. Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol. Biol. 2016, 90, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.-M.; Jia, H.-F.; Li, C.-L.; Dong, Q.-H.; Shen, Y.-Y. FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 2011, 62, 5079–5089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Lu, D.; Luo, J.-J.; Qin, L.; Shen, Y.-Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.-F.; Lu, D.; Sun, J.-H.; Li, C.-L.; Xing, Y.; Qin, L.; Shen, Y.-Y. Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J. Exp. Bot. 2013, 64, 1677–1687. [Google Scholar] [CrossRef] [Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef] [Green Version]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Molina, L.; Chua, N.-H. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- McAlister, G.C.; Huttlin, E.L.; Haas, W.; Ting, L.; Jedrychowski, M.P.; Rogers, J.C.; Kuhn, K.; Pike, I.; Grothe, R.A.; Blethrow, J.D. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 2012, 84, 7469–7478. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Differential Expressed Proteins | MG/IMG | Br/MG | MR/Br |
|---|---|---|---|
| Upregulated | 74 | 17 | 7 |
| Downregulated | 25 | 649 | 416 |
| Total | 99 | 666 | 423 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Lv, J.; Liu, Y.; Wang, J.; Zhang, Z.; Chen, W.; Song, J.; Yang, B.; Tan, F.; Zou, X.; et al. Comprehensive Phosphoproteomic Analysis of Pepper Fruit Development Provides Insight into Plant Signaling Transduction. Int. J. Mol. Sci. 2020, 21, 1962. https://doi.org/10.3390/ijms21061962
Liu Z, Lv J, Liu Y, Wang J, Zhang Z, Chen W, Song J, Yang B, Tan F, Zou X, et al. Comprehensive Phosphoproteomic Analysis of Pepper Fruit Development Provides Insight into Plant Signaling Transduction. International Journal of Molecular Sciences. 2020; 21(6):1962. https://doi.org/10.3390/ijms21061962
Chicago/Turabian StyleLiu, Zhoubin, Junheng Lv, Yuhua Liu, Jing Wang, Zhuqing Zhang, Wenchao Chen, Jingshuang Song, Bozhi Yang, Fangjun Tan, Xuexiao Zou, and et al. 2020. "Comprehensive Phosphoproteomic Analysis of Pepper Fruit Development Provides Insight into Plant Signaling Transduction" International Journal of Molecular Sciences 21, no. 6: 1962. https://doi.org/10.3390/ijms21061962
APA StyleLiu, Z., Lv, J., Liu, Y., Wang, J., Zhang, Z., Chen, W., Song, J., Yang, B., Tan, F., Zou, X., & Ou, L. (2020). Comprehensive Phosphoproteomic Analysis of Pepper Fruit Development Provides Insight into Plant Signaling Transduction. International Journal of Molecular Sciences, 21(6), 1962. https://doi.org/10.3390/ijms21061962





