Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease
Abstract
1. Introduction
2. Results
2.1. Evaluation of ApoB-Derived Peptide Effects on Clinically Isolated Baterial Strains
2.2. Evaluation of ApoB-Derived Peptide Anti-biofilm Activity on Clinically Isolated Baterial Strains
2.2.1. Evaluation of ApoB-Derived Peptide Anti-Biofilm Activity by Microtiter Plate Assay
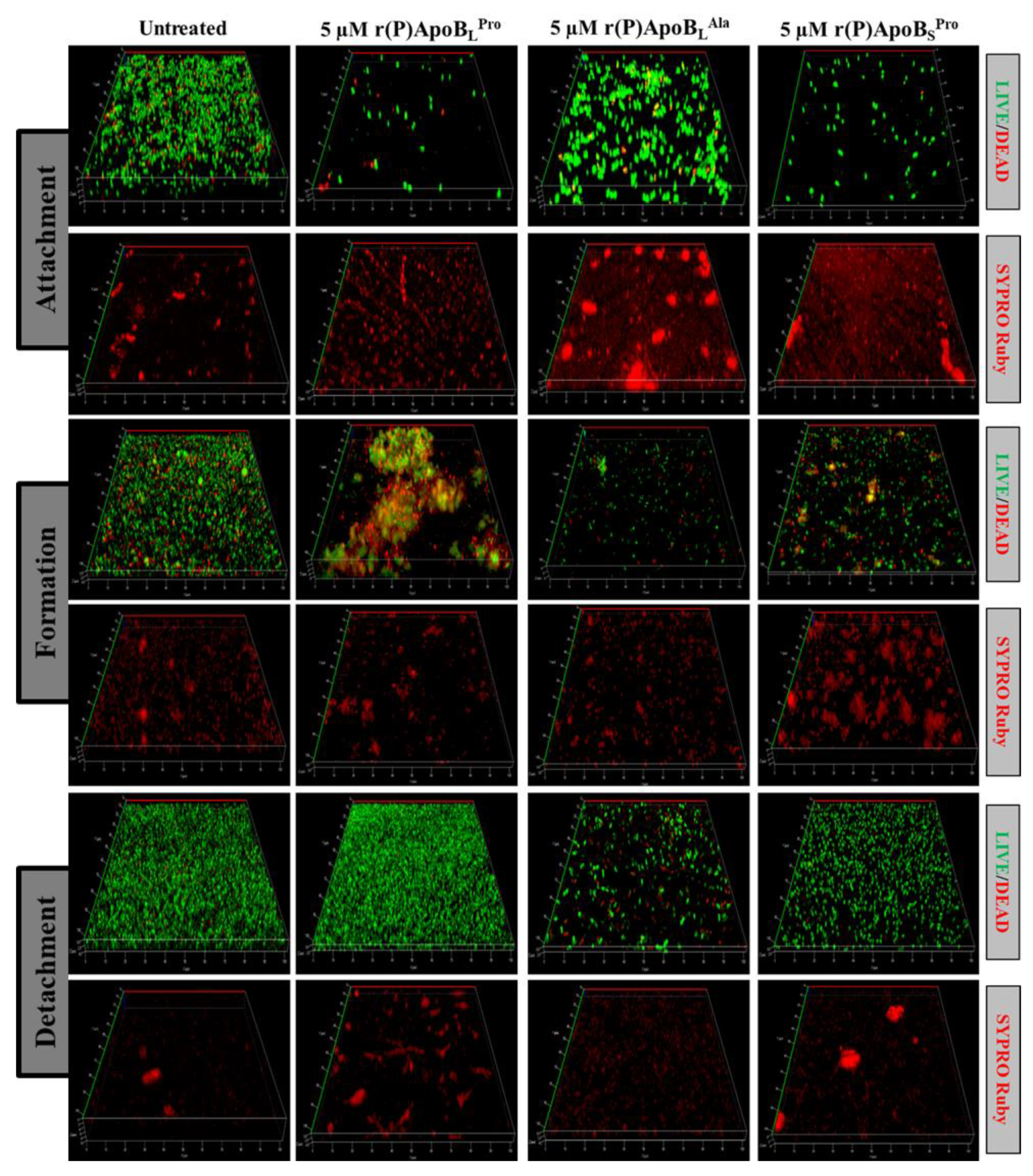
2.2.2. Evaluation of ApoB-Derived Peptides Anti-Biofilm Activity by Laser Scanning Confocal Microscopy
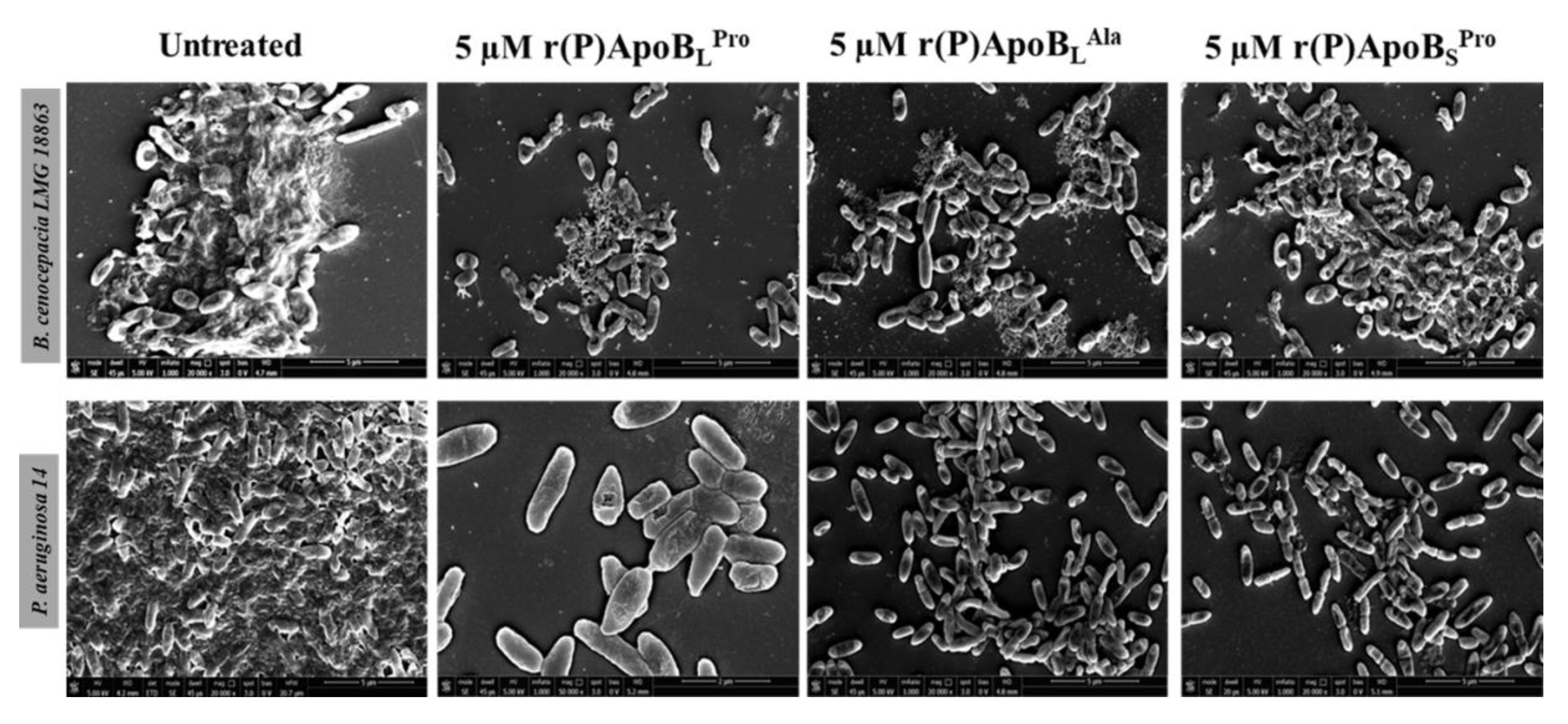
2.2.3. Evaluation of ApoB-Derived Peptides Anti-Biofilm Activity by Scanning Electron Microscopy
2.3. Combinatorial Therapeutic Approach
2.4. Evaluation of Peptide Biocompatibility
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Recombinant Production of ApoB-Derived Peptides
4.3. Bacterial Strains and Growth Conditions
4.4. Eukaryotic Cells and Growth Conditions
4.5. Cell Viability Assays
4.6. Antimicrobial Activity Assays
4.7. Anti-Biofilm Activity by Crystal Violet Assay
4.8. Anti-Biofilm Activity by CLSM Analyses
4.9. Anti-Biofilm Activity by Scanning Electron Microscopy
4.10. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CF | Cystic Fibrosis |
| MDR | Multidrug Resistance |
| ApoB | Apolipoprotein B |
| HDPs | Host Defense Peptides |
| CFTR | Cystic fibrosis transmembrane conductance regulator |
| AMPs | Antimicrobial peptides |
| MIC | Minimum inhibitory concentration |
| MHB | Mueller Hinton Broth |
| CLSM | Confocal laser scanning microscopy |
| SEM | Scanning electron microscopy |
| PI | Propidium iodide |
| LPS | Lipopolysaccharide |
References
- Farrell, P.M. The prevalence of cystic fibrosis in the European Union. J. Cyst. Fibros. 2008, 7, 450–453. [Google Scholar] [CrossRef]
- Evans, C.M.; Koo, J.S. Airway mucus: The good, the bad, the sticky. Pharmacol Ther. 2009, 121, 332–348. [Google Scholar] [CrossRef]
- Chace, K.V.; Flux, M.; Sachdev, G.P. Comparison of physicochemical properties of purified mucus glycoproteins isolated from respiratory secretions of cystic fibrosis and asthmatic patients. Biochemistry 1985, 24, 7334–7341. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.R.; Durie, P.R. What is cystic fibrosis? N. Engl. J. Med. 2002, 347, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Hansen, C.R.; Høiby, N. Respiratory bacterial infections in cystic fibrosis. Curr. Opin. Pulm. Med. 2013, 19, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Rudkjøbing, V.B.; Thomsen, T.R.; Alhede, M.; Kragh, K.N.; Nielsen, P.H.; Johansen, U.R.; Givskov, M.; Høiby, N.; Bjarnsholt, T. The microorganisms in chronically infected end-stage and non-end-stage cystic fibrosis patients. FEMS Immunol. Med. Microbiol. 2012, 65, 236–244. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Jensen, P.Ø.; Fiandaca, M.J.; Pedersen, J.; Hansen, C.R.; Andersen, C.B.; Pressler, T.; Givskov, M.; Høiby, N. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 2009, 44, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.; Ahn, D.; Cohen, T.; Prince, A. Innate Immune Signaling Activated by MDR Bacteria in the Airway. Physio. Rev. 2016, 96, 19–53. [Google Scholar] [CrossRef]
- Blanc, D.S.; Petignat, C.; Janin, B.; Bille, J.; Francioli, P. Frequency and molecular diversity of Pseudomonas aeruginosa upon admission and during hospitalization: A prospective epidemiologic study. Clin. Microbiol. Infect. 1998, 4, 242–247. [Google Scholar] [CrossRef]
- Sadikot, R.T.; Blackwell, T.S.; Christman, J.W.; Prince, A.S. Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 1209–1223. [Google Scholar] [CrossRef]
- Larrosa, M.; Truchado, P.; Espin, J.C.; Tomas-Barberan, F.A.; Allende, A.; Garcia-Conesa, M.T. Evaluation of Pseudomonas aeruginosa (PAO1) adhesion to human alveolar epithelial cells A549 using SYTO 9 dye. Mol. Cell Probes 2012, 26, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Hirakata, Y.; Izumikawa, K.; Yamaguchi, T.; Igimi, S.; Furuya, N.; Maesaki, S.; Tomono, K.; Yamada, Y.; Kohno, S.; Yamaguchi, K.; et al. Adherence to and penetration of human intestinal Caco-2 epithelial cell monolayers by Pseudomonas aeruginosa. Infect. Immun. 1998, 66, 1748–1751. [Google Scholar] [CrossRef]
- Pier, G.B.; Grout, M.; Zaidi, T.S. Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc. Natl. Acad. Sci. USA 1997, 94, 12088–12093. [Google Scholar] [CrossRef] [PubMed]
- Pier, G.B.; Grout, M.; Zaidi, T.S.; Olsen, J.C.; Johnson, L.G.; Yankaskas, J.R.; Goldberg, J.B. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science 1996, 271, 64–67. [Google Scholar] [CrossRef]
- Esen, M.; Grassme, H.; Riethmuller, J.; Riehle, A.; Fassbender, K.; Gulbins, E. Invasion of human epithelial cells by Pseudomonas aeruginosa involves src-like tyrosine kinases p60Src and p59Fyn. Infect. Immun. 2001, 69, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Darling, K.E.; Evans, T.J. Effects of nitric oxide on Pseudomonas aeruginosa infection of epithelial cells from a human respiratory cell line derived from a patient with cystic fibrosis. Infect. Immun. 2003, 71, 2341–2349. [Google Scholar] [CrossRef]
- Brinch, K.S.; Frimodt-Moller, N.; Hoiby, N.; Kristensen, H.H. Influence of antidrug antibodies on plectasin efficacy and pharmacokinetics. Antimicrob. Agents Chemother. 2009, 53, 4794–4800. [Google Scholar] [CrossRef]
- Chi, E.; Mehl, T.; Nunn, D.; Lory, S. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infect. Immun. 1991, 59, 822–828. [Google Scholar] [CrossRef]
- Drenkard, E.; Ausubel, F.M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 2002, 416, 740–743. [Google Scholar] [CrossRef]
- Scoffone, V.C.; Chiarelli, L.R.; Trespidi, G.; Mentasti, M.; Riccardi, G.; Buroni, S. Burkholderia cenocepacia Infections in Cystic Fibrosis Patients: Drug Resistance and Therapeutic Approaches. Front. Microbiol. 2017, 8, 1592. [Google Scholar] [CrossRef]
- Alexander, B.D.; Petzold, E.W.; Reller, L.B.; Palmer, S.M.; Davis, R.D.; Woods, C.W.; Lipuma, J.J. Survival after lung transplantation of cystic fibrosis patients infected with Burkholderia cepacia complex. Am. J. Transplant. 2008, 8, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, C.; Keshavjee, S. Lung transplantation for cystic fibrosis: An update. Expert Rev. Respir. Med. 2016, 10, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Mercer, D.K.; O’Neil, D.A. Peptides as the next generation of anti-infectives. Future Med. Chem. 2013, 5, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Hancock, R.B. Peptide design for antimicrobial and immunomodulatory applications. Biopolymers 2013, 100, 572–583. [Google Scholar] [CrossRef]
- Cruz, J.; Ortiz, C.; Guzman, F.; Fernandez-Lafuente, R.; Torres, R. Antimicrobial peptides: Promising compounds against pathogenic microorganisms. Curr. Med. Chem. 2014, 21, 2299–2321. [Google Scholar] [CrossRef]
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1999, 1462, 11–28. [Google Scholar] [CrossRef]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Cao, H.; Ke, T.; Liu, R.; Yu, J.; Dong, C.; Cheng, M.; Huang, J.; Liu, S. Identification of a Novel Proline-Rich Antimicrobial Peptide from Brassica napus. PLoS ONE 2015, 10, e0137414. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Vineeth Kumar, T.V.; Sanil, G. A Review of the Mechanism of Action of Amphibian Antimicrobial Peptides Focusing on Peptide-Membrane Interaction and Membrane Curvature. Curr. Protein. Pept. Sci. 2017, 18, 1263–1272. [Google Scholar] [PubMed]
- Shahrour, H.; Ferrer-Espada, R.; Dandache, I.; Bárcena-Varela, S.; Sánchez-Gómez, S.; Chokr, A.; Martinez-de-Tejada, G. AMPs as Anti-biofilm Agents for Human Therapy and Prophylaxis. Adv. Exp. Med. Biol. 2019, 1117, 257–279. [Google Scholar] [PubMed]
- Gaglione, R.; Dell’Olmo, E.; Bosso, A.; Chino, M.; Pane, K.; Ascione, F.; Itri, F.; Caserta, S.; Amoresano, A.; Lombardi, A.; et al. Novel human bioactive peptides identified in Apolipoprotein B: Evaluation of their therapeutic potential. Biochem. Pharmacol. 2017, 130, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Pane, K.; Durante, L.; Crescenzi, O.; Cafaro, V.; Pizzo, E.; Varcamonti, M.; Zanfardino, A.; Izzo, V.; Di Donato, A.; Notomista, E. Antimicrobial potency of cationic antimicrobial peptides can be predicted from their amino acid composition: Application to the detection of “cryptic” antimicrobial peptides. J. Theor. Biol. 2017, 419, 254–265. [Google Scholar] [CrossRef]
- Gaglione, R.; Cesaro, A.; Dell’Olmo, E.; Della Ventura, B.; Casillo, A.; Di Girolamo, R.; Velotta, R.; Notomista, E.; Veldhuizen, E.J.A.; Corsaro, M.M.; et al. Effects of human antimicrobial cryptides identified in apolipoprotein B depend on specific features of bacterial strains. Sci. Rep. 2019, 9, 6728. [Google Scholar] [CrossRef]
- Gaglione, R.; Pane, K.; Dell’Olmo, E.; Cafaro, V.; Pizzo, E.; Olivieri, G.; Notomista, E.; Arciello, A. Cost-effective production of recombinant peptides in Escherichia coli. N. Biotechnol. 2019, 51, 39–48. [Google Scholar] [CrossRef]
- Pane, K.; Cafaro, V.; Avitabile, A.; Torres, M.T.; Vollaro, A.; De Gregorio, E.; Catania, M.R.; Di Maro, A.; Bosso, A.; Gallo, G.; et al. Identification of Novel Cryptic Multifunctional Antimicrobial Peptides from the Human Stomach Enabled by a Computational-Experimental Platform. ACS Synth. Biol. 2018, 7, 2105–2115. [Google Scholar] [CrossRef]
- Zanfardino, A.; Bosso, A.; Gallo, G.; Pistorio, V.; Di Napoli, M.; Gaglione, R.; Dell’Olmo, E.; Varcamonti, M.; Notomista, E.; Arciello, A.; et al. Human apolipoprotein E as a reservoir of cryptic bioactive peptides: The case of ApoE 133-167. J. Pept. Sci. 2018, 24, e3095. [Google Scholar] [CrossRef]
- Pizzo, E.; Pane, K.; Bosso, A.; Landi, N.; Ragucci, S.; Russo, R.; Gaglione, R.; Torres, M.D.T.; de la Fuente-Nunez, C.; Arciello, A.; et al. Novel bioactive peptides from PD-L1/2, a type 1 ribosome inactivating protein from Phytolacca dioica L. Evaluation of their antimicrobial properties and anti-biofilm activities. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1425–1435. [Google Scholar] [CrossRef]
- Pizzo, E.; Cafaro, V.; Di Donato, A.; Notomista, E. Cryptic Antimicrobial Peptides: Identification Methods and Current Knowledge of their Immunomodulatory Properties. Curr. Pharm. Des. 2018, 24, 1054–1066. [Google Scholar] [CrossRef]
- Gaglione, R.; Pirone, L.; Farina, B.; Fusco, S.; Smaldone, G.; Aulitto, M.; Dell’Olmo, E.; Roscetto, E.; Del Gatto, A.; Fattorusso, R.; et al. Insights into the anticancer properties of the first antimicrobial peptide from Archaea. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2155–2164. [Google Scholar] [CrossRef] [PubMed]
- Kasetty, G.; Papareddy, P.; Kalle, M.; Rydengård, V.; Walse, B.; Svensson, B.; Mörgelin, M.; Malmsten, M.; Schmidtchen, A. The C-terminal sequence of several human serine proteases encodes host defense functions. J. Innate Immun. 2011, 3, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Huang, C.M.; Nakatsuji, T.; Thiboutot, D.; Kang, S.A.; Monestier, M.; Gallo, R.L. Histone H4 is a major component of the antimicrobial action of human sebocytes. J. Invest. Dermatol. 2009, 129, 2489–2496. [Google Scholar] [CrossRef] [PubMed]
- Beck, W.H.; Adams, C.P.; Biglang-Awa, I.M.; Patel, A.B.; Vincent, H.; Haas-Stapleton, E.J.; Weers, P.M. Apolipoprotein A-I binding to anionic vesicles and lipopolysaccharides: Role for lysine residues in antimicrobial properties. Biochim. Biophys. Acta 2013, 1828, 1503–1510. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Langton Hewer, S.C.; Smyth, A.R. Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis. Cochrane Database Syst. Rev. 2014, 11, CD004197. [Google Scholar]
- Sanchez, Z.; Tani, A.; Kimbara, K. Extensive reduction of cell viability and enhanced matrix production in Pseudomonas aeruginosa PAO1 flow biofilms treated with a D-amino acid mixture. Appl. Environ. Microbiol. 2013, 79, 1396–1399. [Google Scholar] [CrossRef]
- Bechinger, B.; Gorr, S.U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef]
- De Soyza, A.; Ellis, C.D.; Khan, C.M.A.; Corris, P.A.; Demarco De Hormaeche, R. Burkholderia cenocepacia lipopolysaccharide, lipid A, and proinflammatory activity. Am. J. Respir. Crit. Care Med. 2004, 170, 70–77. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Reynolds, C.M.; Trent, M.S.; Bishop, R.E. Lipid A Modification Systems in Gram-Negative Bacteria. Annu. Rev. Biochem. 2007, 76, 295–329. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 29–30. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef] [PubMed]
- Pletzer, D.; Hancock, R.E.W. Antibiofilm peptides: Potential as broadspectrum agents. J. Bacteriol. 2016, 198, 2572–2578. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.C.; de la Fuente-Núñez, C.; Hancock, R.E. Peptide IDR-1018: Modulating the immune system and targeting bacterial biofilms to treat antibiotic-resistant bacterial infections. J. Pept. Sci. 2015, 21, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, T.; Stone, T.A.; Glibowicka, M.; Adams, C.; Yau, Y.; Ahmadi, S.; Bear, C.E.; Grasemann, H.; Waters, V.; Deber, C.M. Activity of a novel antimicrobial peptide against Pseudomonas aeruginosa biofilms. Sci. Rep. 2018, 8, 14728. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.; Roussel, V.; Pestel-Caron, M.; Barreau, M.; Caron, F.; Bouffartigues, E.; Chevalier, S.; Etienne, M. Understanding Ciprofloxacin Failure in Pseudomonas aeruginosa Biofilm: Persister Cells Survive Matrix Disruption. Front. Microbiol. 2019, 10, 2603. [Google Scholar] [CrossRef]
- Reffuveille, F.; De La Fuente-Nú̃nez, C.; Mansour, S.; Hancock, R.E.W. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 2014, 58, 5363–5371. [Google Scholar] [CrossRef]
- Dosler, S.; Karaaslan, E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef]






| MIC100 (μM) | |||
|---|---|---|---|
| r(P)ApoBLPro | r(P)ApoBLAla | r(P)ApoBSPro | |
| P. aeruginosa RP 73 | 10–20 | 5–10 | 20–40 |
| P. aeruginosa 14 | >40 | >40 | >40 |
| P. aeruginosa AA2 | >40 | >40 | >40 |
| P. aeruginosa KK 27 | 20–40 | 10–20 | 20–40 |
| Burkholderia cenocepacia LMG 18863 | >40 | >40 | >40 |
| Burkholderia multivorans LMG 17582 | 10–20 | 10–20 | 20–40 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaglione, R.; Cesaro, A.; Dell’Olmo, E.; Di Girolamo, R.; Tartaglione, L.; Pizzo, E.; Arciello, A. Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease. Int. J. Mol. Sci. 2020, 21, 2049. https://doi.org/10.3390/ijms21062049
Gaglione R, Cesaro A, Dell’Olmo E, Di Girolamo R, Tartaglione L, Pizzo E, Arciello A. Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease. International Journal of Molecular Sciences. 2020; 21(6):2049. https://doi.org/10.3390/ijms21062049
Chicago/Turabian StyleGaglione, Rosa, Angela Cesaro, Eliana Dell’Olmo, Rocco Di Girolamo, Luca Tartaglione, Elio Pizzo, and Angela Arciello. 2020. "Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease" International Journal of Molecular Sciences 21, no. 6: 2049. https://doi.org/10.3390/ijms21062049
APA StyleGaglione, R., Cesaro, A., Dell’Olmo, E., Di Girolamo, R., Tartaglione, L., Pizzo, E., & Arciello, A. (2020). Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease. International Journal of Molecular Sciences, 21(6), 2049. https://doi.org/10.3390/ijms21062049