Overexpression and Biochemical Characterization of an Endo-α-1,4-polygalacturonase from Aspergillus nidulans in Pichia pastoris
Abstract
1. Introduction
2. Results and Discussion
2.1. Sequence Alignment and Phylogenetic Analysis of AnEPG with Other GH 28 endo-PGs
2.2. Overexpression of AnEPG in P. pastoris
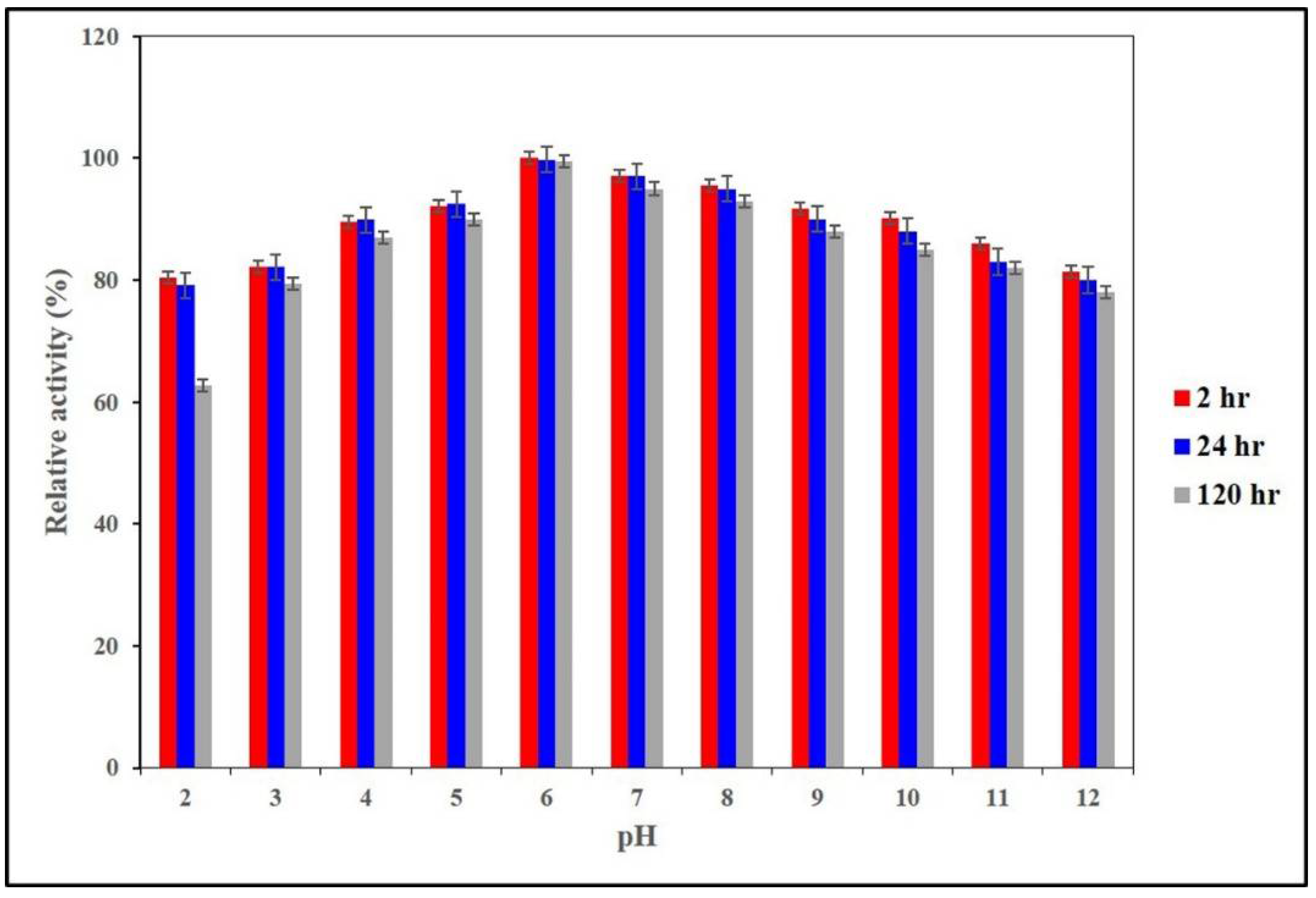
2.3. Determination of pH Optima and pH Stability of AnEPG
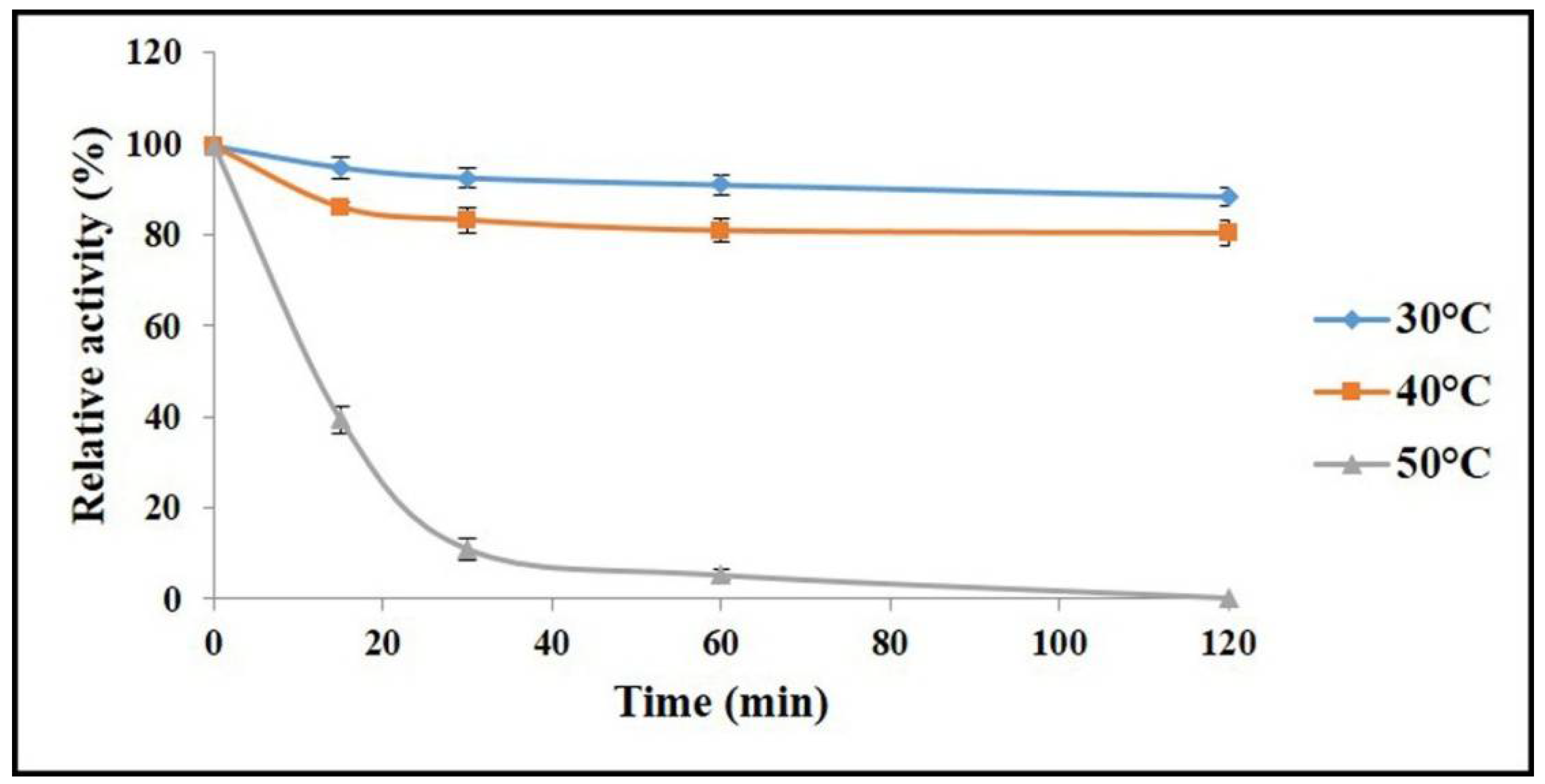
2.4. Determination of Optimal Temperature and Thermal Stability of AnEPG
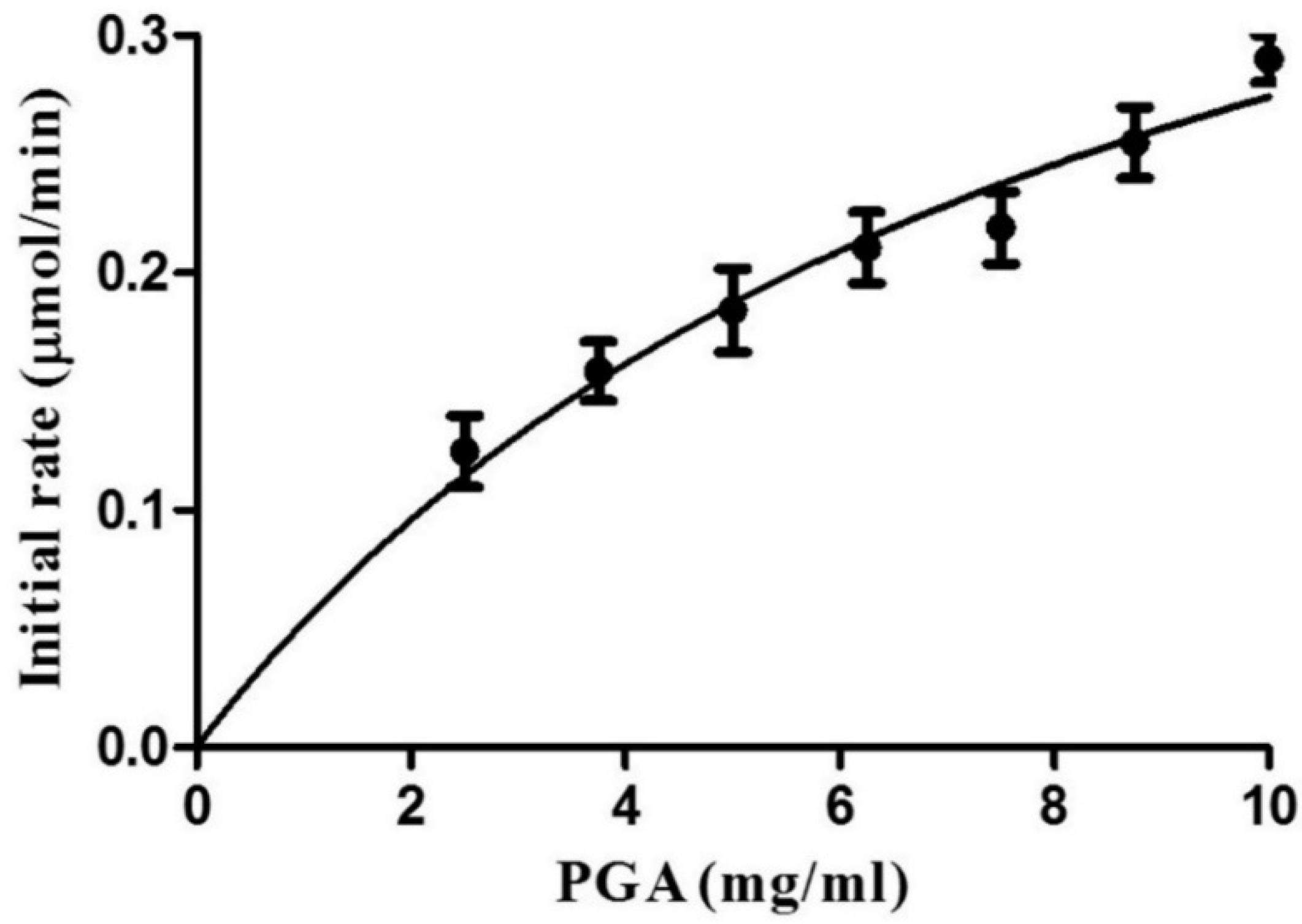
2.5. Determination of Kinetic Parameters of AnEPG
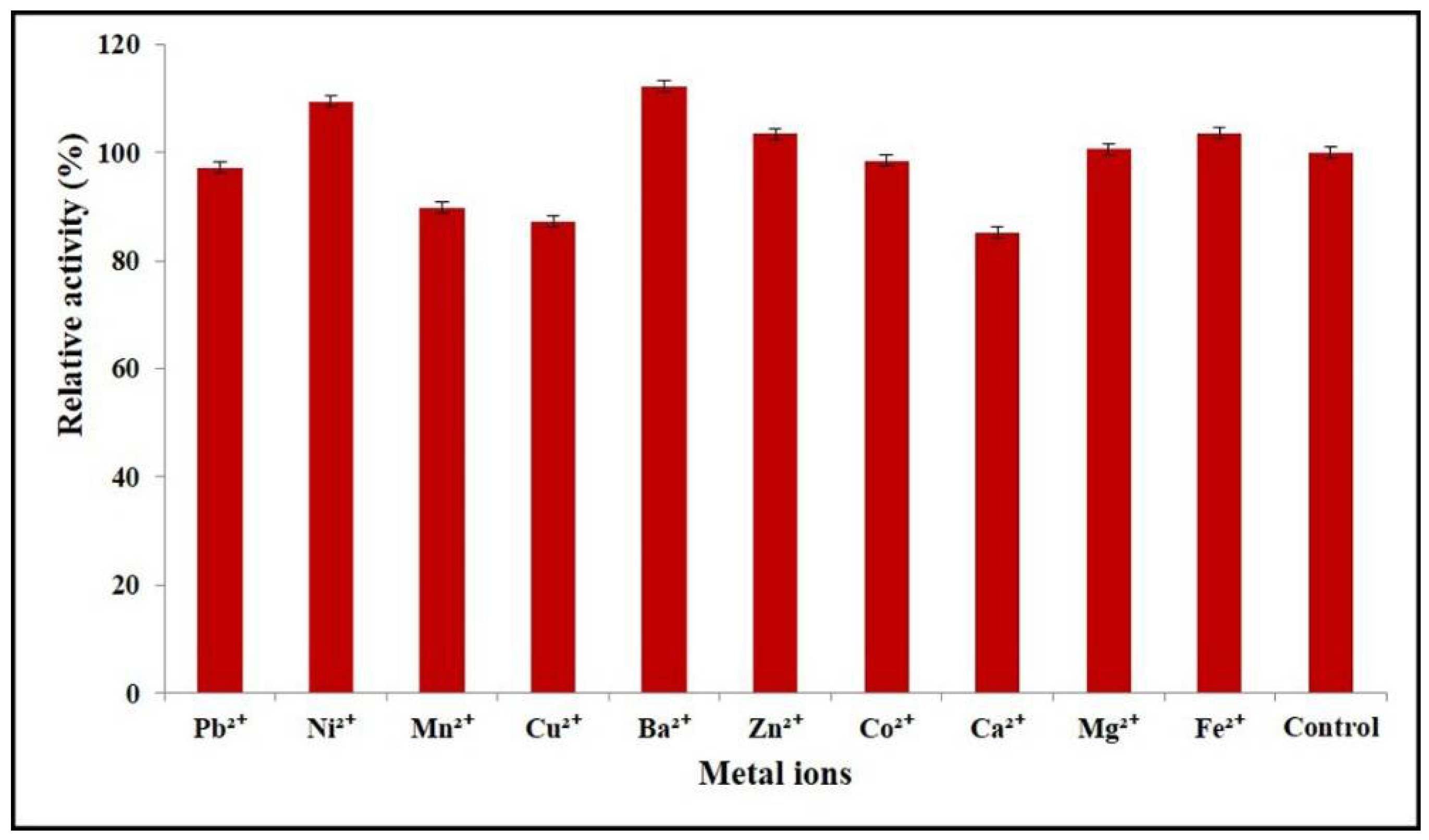
2.6. Effects of Divalent Metal Ions on Enzyme Activity
2.7. Analysis of Hydrolysis Products of PGA by AnEPG
3. Materials and Methods
3.1. Materials
3.2. Bacterial Strains, Plasmids, and Media
3.3. Protein Overexpression
3.4. Enzyme Activity Assay
3.5. Determination of Optimal pH and pH Stability
3.6. Determination of Optimal Temperature and Thermal Stability
3.7. Determination of Kinetic Parameters
3.8. Effects of Divalent Metal Ions on Enzyme Activity
3.9. Thin Layer Chromatography Analysis of Hydrolysis Products of PGA by AnEPG
3.10. Sequence Analysis
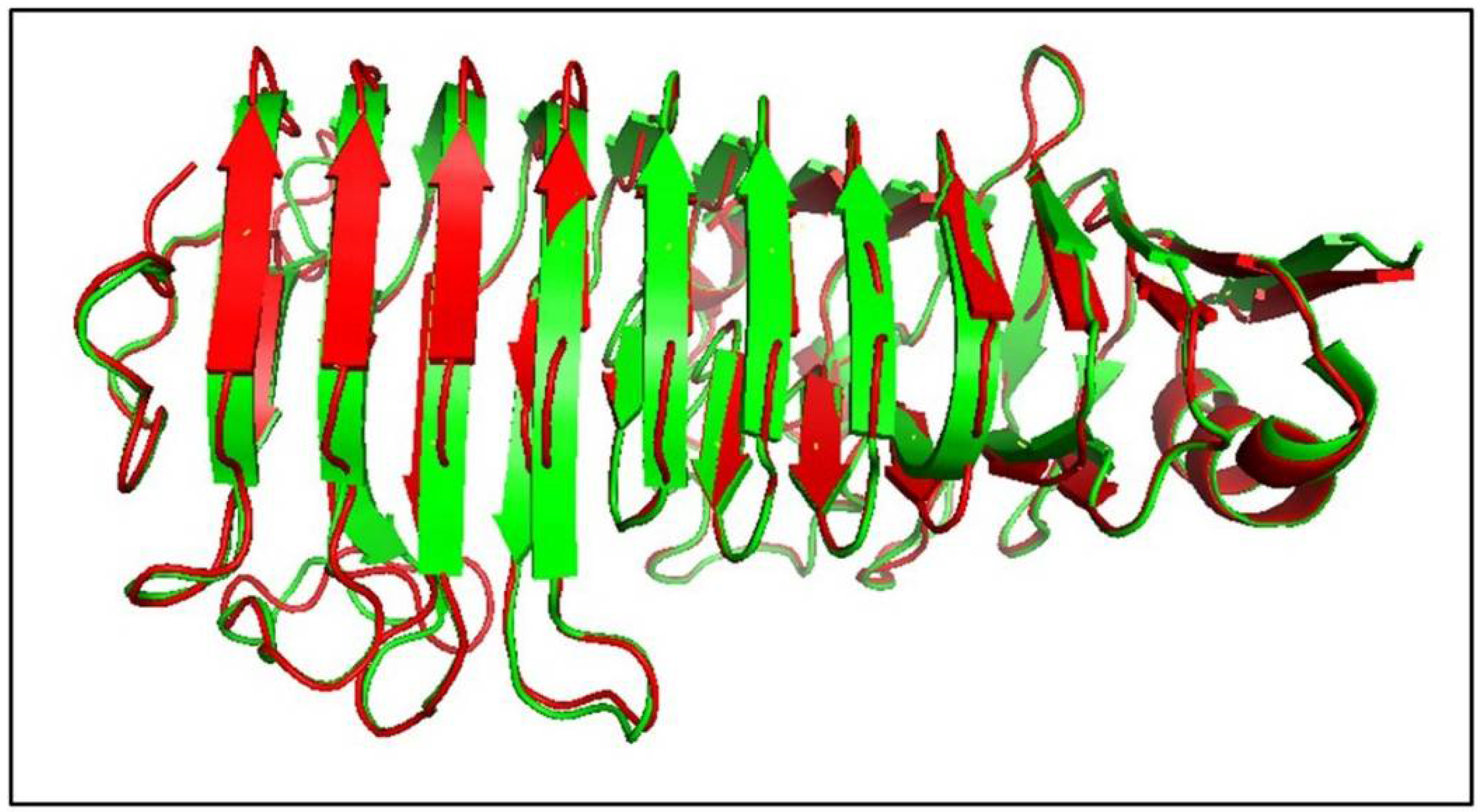
3.11. Structure Modelling of AnEPG
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AnEPG | endo-α-1,4-polygalacturonase from Aspergillus nidulans |
| PGA | polygalacturonic acid |
| HG | homogalacturonan |
| CAZy | carbohydrate-active enzymes database |
| ORFs | open reading frames |
| C-score | confidence score |
| TM-score | template modeling score |
| RMSD | the root-mean-square deviation |
| CMC | Carboxymethyl Cellulose |
| FGSC | Fungal Genetics Stock Center |
| YPD | yeast extract peptone dextrose |
| BMGY | buffered complex glycerol medium |
| BMMY | buffered complex methanol medium |
| DNS | 3,5-dinitrosalicylic acid |
| TLC | thin-layer chromatography |
References
- Harholt, J.; Suttangkakul, A.; Vibe Scheller, H. Biosynthesis of pectin. Plant Physiol. 2010, 153, 384–395. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Willats, W.G.; McCartney, L.; Mackie, W.; Knox, J.P. Pectin: Cell biology and prospects for functional analysis. Plant Mol. Biol. 2001, 47, 9–27. [Google Scholar] [CrossRef]
- Van den Brink, J.; de Vries, R.P. Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbial. Biotechnol. 2011, 91, 1477–1492. [Google Scholar] [CrossRef]
- Jayani, R.S.; Saxena, S.; Gupta, R. Microbial pectinolytic enzymes: A review. Process Biochem. 2005, 40, 2931–2944. [Google Scholar] [CrossRef]
- Sharma, N.; Rathore, M.; Sharma, M. Microbial pectinase: Sources, characterization and applications. Rev. Environ. Sci. Bio Technol. 2013, 12, 45–60. [Google Scholar] [CrossRef]
- Khan, M.; Nakkeeran, E.; Umesh-Kumar, S. Potential application of pectinase in developing functional foods. Annu. Rev. Food Sci. Technol. 2013, 4, 21–34. [Google Scholar] [CrossRef]
- Anand, G.S.Y.; Dubey, A.K.; Yadav, D. Recent developments in biotechnology. In Molecular Biology of Microbial Polygalacturonases: A Review; Studium Press: Delhi, India, 2014. [Google Scholar]
- Pařenicová, L.; Benen, J.A.; Kester, H.C.; Visser, J. pgaE encodes a fourth member of the endopolygalacturonase gene family from Aspergillus niger. Eur. J. Biochem. 1998, 251, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, X.; Guo, M.; Xu, Q.; Cao, Y.; Qiao, D.; Cao, Y.; Xu, H. Secretory expression and characterization of an acidic endo-polygalacturonase from Aspergillus niger SC323 in Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 2015, 25, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Qin, X.; Gao, L.; Han, B.; Zhang, D.; Li, J.; Huang, H.; Zhang, W. Efficient expression of an acidic endo-polygalacturonase from Aspergillus niger and its application in juice production. J. Agric. Food Chem. 2017, 65, 2730–2736. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.; Katsuragi, T.; Terashita, T.; Yoshikawa, K.; SAKAI, T. Purification and characterization of an endo-polygalacturonase from Aspergillus awamori. Biosci. Biotechnol. Biochem. 2000, 64, 1729–1732. [Google Scholar] [CrossRef] [PubMed]
- Nakkeeran, E.; Umesh-Kumar, S.; Subramanian, R. Aspergillus carbonarius polygalacturonases purified by integrated membrane process and affinity precipitation for apple juice production. Bioresour. Technol. 2011, 102, 3293–3297. [Google Scholar] [CrossRef] [PubMed]
- Abdulrachman, D.; Thongkred, P.; Kocharin, K.; Nakpathom, M.; Somboon, B.; Narumol, N.; Champreda, V.; Eurwilaichitr, L.; Suwanto, A.; Nimchua, T. Heterologous expression of Aspergillus aculeatus endo-polygalacturonase in Pichia pastoris by high cell density fermentation and its application in textile scouring. BMC Biotechnol. 2017, 17, 15. [Google Scholar] [CrossRef]
- Whitehead, M.; Shieh, M.; Cleveland, T.; Cary, J.; Dean, R. Isolation and characterization of polygalacturonase genes (pecA and pecB) from Aspergillus flavus. Appl. Environ. Microbiol. 1995, 61, 3316–3322. [Google Scholar] [CrossRef]
- Cheng, Z.; Chen, D.; Lu, B.; Wei, Y.; Xian, L.; Li, Y.; Luo, Z.; Huang, R. A novel acid-stable endo-polygalacturonase from Penicillium oxalicum CZ1028: Purification, characterization, and application in the beverage industry. J. Microbiol. Biotechnol. 2016, 26, 989–998. [Google Scholar] [CrossRef]
- Yuan, P.; Meng, K.; Huang, H.; Shi, P.; Luo, H.; Yang, P.; Yao, B. A novel acidic and low-temperature-active endo-polygalacturonase from Penicillium sp. CGMCC 1669 with potential for application in apple juice clarification. Food Chem. 2011, 129, 1369–1375. [Google Scholar] [CrossRef]
- Sassi, A.H.; Tounsi, H.; Trigui-Lahiani, H.; Bouzouita, R.; Romdhane, Z.B.; Gargouri, A. A low-temperature polygalacturonase from P. occitanis: Characterization and application in juice clarification. Int. J. Biol. Macromol. 2016, 91, 158–164. [Google Scholar] [CrossRef]
- Pan, X.; Li, K.; Ma, R.; Shi, P.; Huang, H.; Yang, P.; Meng, K.; Yao, B. Biochemical characterization of three distinct polygalacturonases from Neosartorya fischeri P1. Food Chem. 2015, 188, 569–575. [Google Scholar] [CrossRef]
- Yang, J.; Luo, H.; Li, J.; Wang, K.; Cheng, H.; Bai, Y.; Yuan, T.; Fan, Y.; Yao, B. Cloning, expression and characterization of an acidic endo-polygalacturonase from Bispora sp. MEY-1 and its potential application in juice clarification. Process Biochem. 2011, 46, 272–277. [Google Scholar] [CrossRef]
- Tu, T.; Meng, K.; Bai, Y.; Shi, P.; Luo, H.; Wang, Y.; Yang, P.; Zhang, Y.; Zhang, W.; Yao, B. High-yield production of a low-temperature-active polygalacturonase for papaya juice clarification. Food Chem. 2013, 141, 2974–2981. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Tu, T.; Zhang, D.; Ma, R.; You, S.; Wang, X.; Yao, B.; Luo, H.; Xu, B. Two acidic, thermophilic GH28 polygalacturonases from Talaromyces leycettanus JCM 12802 with application potentials for grape juice clarification. Food Chem. 2017, 237, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Meng, K.; Huang, H.; Luo, H.; Bai, Y.; Ma, R.; Su, X.; Shi, P.; Yang, P.; Wang, Y. Molecular characterization of a thermophilic endo-polygalacturonase from Thielavia arenaria XZ7 with high catalytic efficiency and application potential in the food and feed industries. J. Agric. Food Chem. 2014, 62, 12686–12694. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.-J.; Wortman, J.R.; Batzoglou, S.; Lee, S.-I.; Baştürkmen, M.; Spevak, C.C.; Clutterbuck, J. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef]
- De Vries, R.; Van Grieken, C.; VanKuyk, P.; Wosten, H. The value of genome sequences in the rapid identification of novel genes encoding specific plant cell wall degrading enzymes. Curr. Genom. 2005, 6, 157–187. [Google Scholar] [CrossRef]
- Coutinho, P.M.; Andersen, M.R.; Kolenova, K.; Benoit, I.; Gruben, B.S.; Trejo-Aguilar, B.; Visser, H.; van Solingen, P.; Pakula, T.; Seiboth, B. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet. Biol. 2009, 46, S161–S169. [Google Scholar] [CrossRef]
- Bauer, S.; Vasu, P.; Persson, S.; Mort, A.J.; Somerville, C.R. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. USA 2006, 103, 11417–11422. [Google Scholar] [CrossRef]
- Contesini, F.J.; Liberato, M.V.; Rubio, M.V.; Calzado, F.; Zubieta, M.P.; Riaño-Pachón, D.M.; Squina, F.M.; Bracht, F.; Skaf, M.S.; Damasio, A.R. Structural and functional characterization of a highly secreted α-l-arabinofuranosidase (GH62) from Aspergillus nidulans grown on sugarcane bagasse. Biochim. Biophys. Acta Proteins Proteomics 2017, 1865, 1758–1769. [Google Scholar] [CrossRef]
- van Santen, Y.; Benen, J.A.; Schröter, K.-H.; Kalk, K.H.; Armand, S.; Visser, J.; Dijkstra, B.W. 1.68-Å crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J. Biol. Chem. 1999, 274, 30474–30480. [Google Scholar] [CrossRef]
- Bonivento, D.; Pontiggia, D.; Matteo, A.D.; Fernandez-Recio, J.; Salvi, G.; Tsernoglou, D.; Cervone, F.; Lorenzo, G.D.; Federici, L. Crystal structure of the endopolygalacturonase from the phytopathogenic fungus Colletotrichum lupini and its interaction with polygalacturonase-inhibiting proteins. Proteins Struct. Funct. Bioinf. 2008, 70, 294–299. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Lee, S.; Shin, W. The X-ray structure of Aspergillus aculeatus polygalacturonase and a modeled structure of the polygalacturonase-octagalacturonate complex. J. Mol. Biol. 2001, 311, 863–878. [Google Scholar] [CrossRef] [PubMed]
- Daly, R.; Hearn, M.T. Expression of heterologous proteins in Pichia pastoris: A useful experimental tool in protein engineering and production. J. Mol. Recognit. Interdiscip. J. 2005, 18, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Anand, G.; Yadav, S.; Yadav, D. Purification and characterization of polygalacturonase from Aspergillus fumigatus MTCC 2584 and elucidating its application in retting of Crotalaria juncea fiber. 3 Biotech 2016, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Zhang, P.; Zhang, Y.; Liu, Z.; Zhang, X.; Li, Z.; Li, J.-J.; Du, Y. Overexpression and Biochemical Characterization of an Endo-α-1,4-polygalacturonase from Aspergillus nidulans in Pichia pastoris. Int. J. Mol. Sci. 2020, 21, 2100. https://doi.org/10.3390/ijms21062100
Xu H, Zhang P, Zhang Y, Liu Z, Zhang X, Li Z, Li J-J, Du Y. Overexpression and Biochemical Characterization of an Endo-α-1,4-polygalacturonase from Aspergillus nidulans in Pichia pastoris. International Journal of Molecular Sciences. 2020; 21(6):2100. https://doi.org/10.3390/ijms21062100
Chicago/Turabian StyleXu, Hua, Pengfei Zhang, Yuchen Zhang, Zebin Liu, Xuebing Zhang, Zhimin Li, Jian-Jun Li, and Yuguang Du. 2020. "Overexpression and Biochemical Characterization of an Endo-α-1,4-polygalacturonase from Aspergillus nidulans in Pichia pastoris" International Journal of Molecular Sciences 21, no. 6: 2100. https://doi.org/10.3390/ijms21062100
APA StyleXu, H., Zhang, P., Zhang, Y., Liu, Z., Zhang, X., Li, Z., Li, J.-J., & Du, Y. (2020). Overexpression and Biochemical Characterization of an Endo-α-1,4-polygalacturonase from Aspergillus nidulans in Pichia pastoris. International Journal of Molecular Sciences, 21(6), 2100. https://doi.org/10.3390/ijms21062100





