Insights into Interactions of Flavanones with Target Human Respiratory Syncytial Virus M2-1 Protein from STD-NMR, Fluorescence Spectroscopy, and Computational Simulations
Abstract
1. Introduction
2. Results and Discussion
2.1. Production of the Recombinant hRSV M2-1 Protein
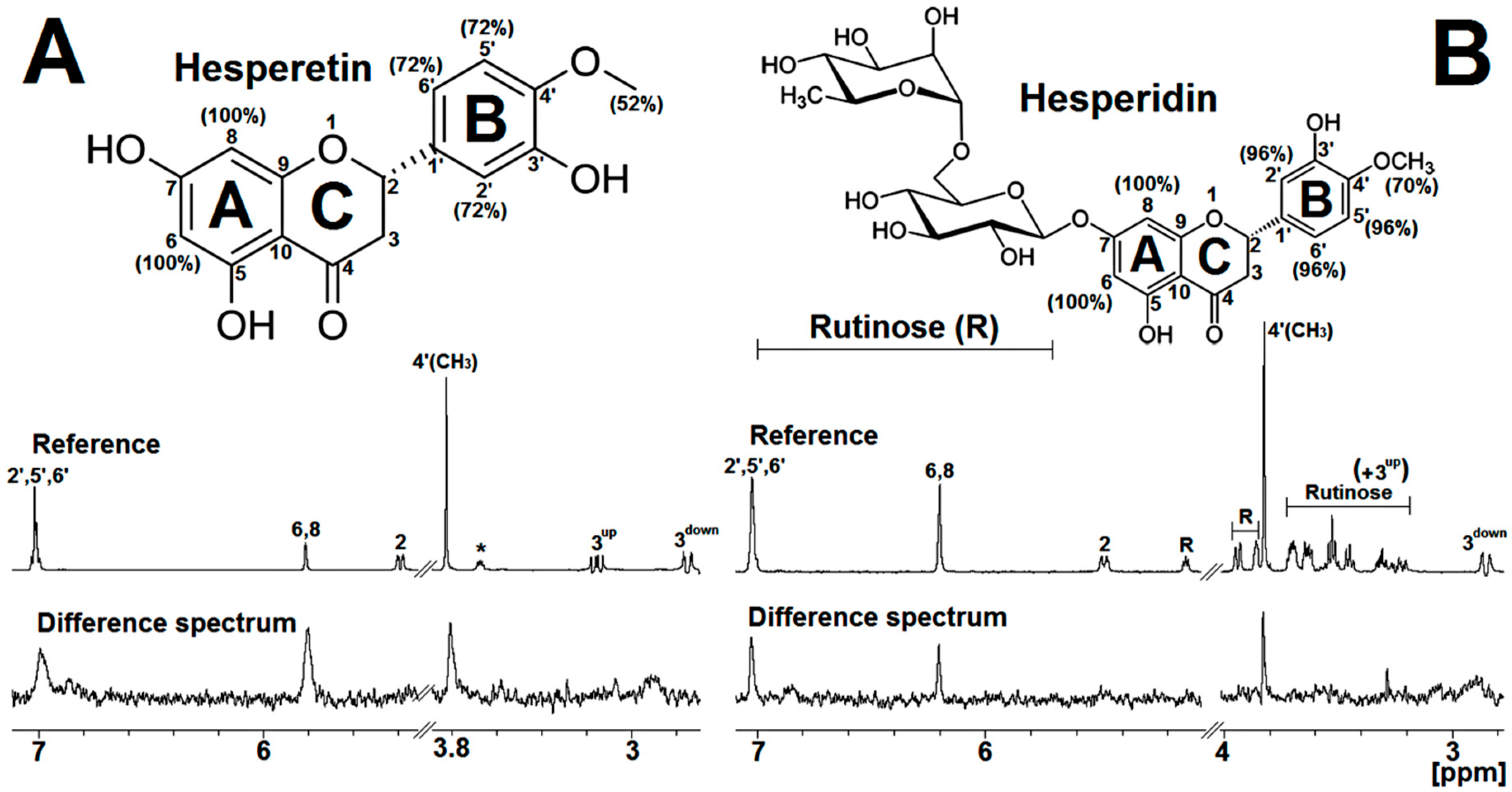
2.2. STD-NMR for Investigating the M2-1/Flavanone Binding
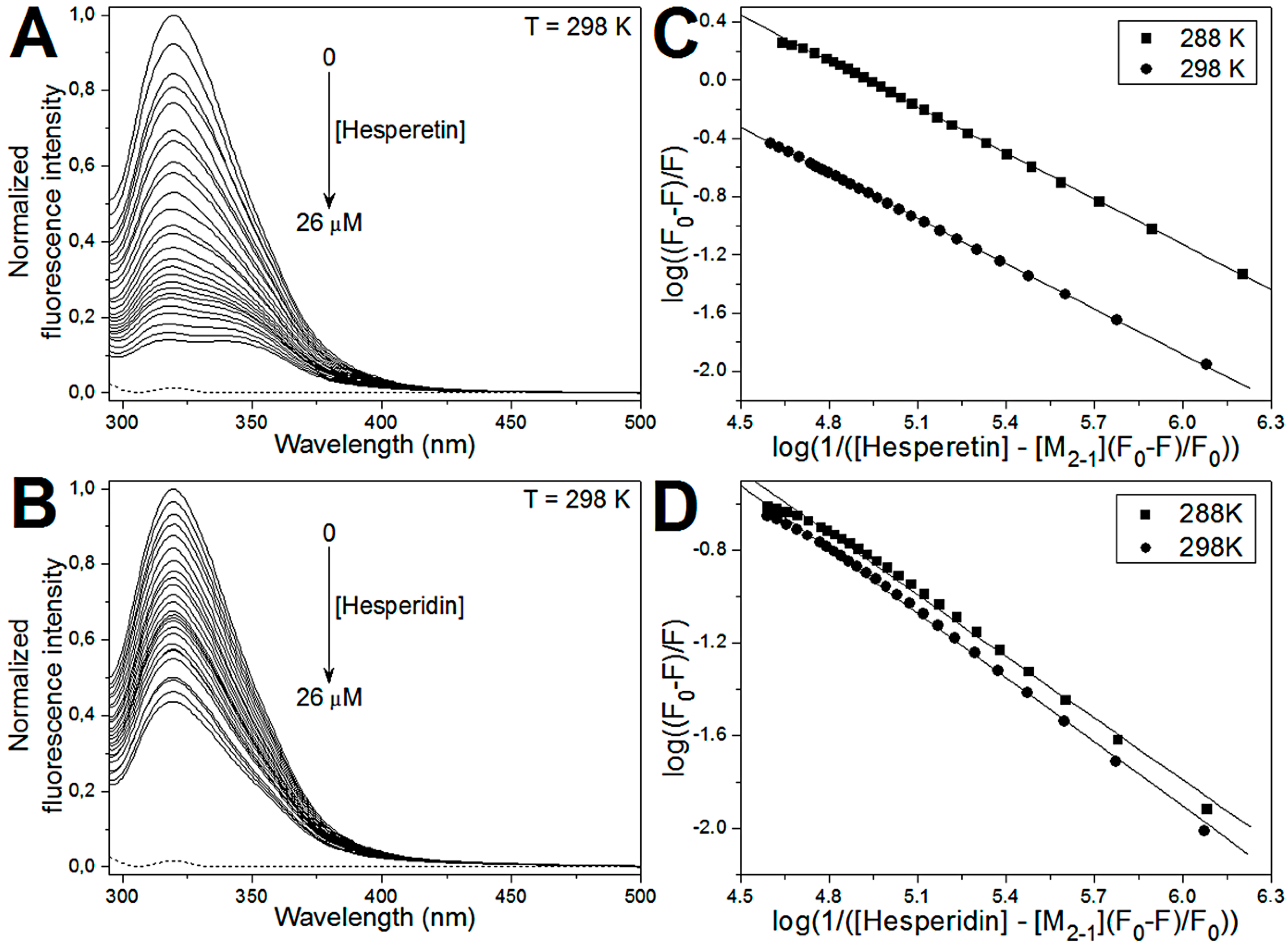
2.3. Fluorescence Quenching of M2-1 by Hesperetin and Hesperidin
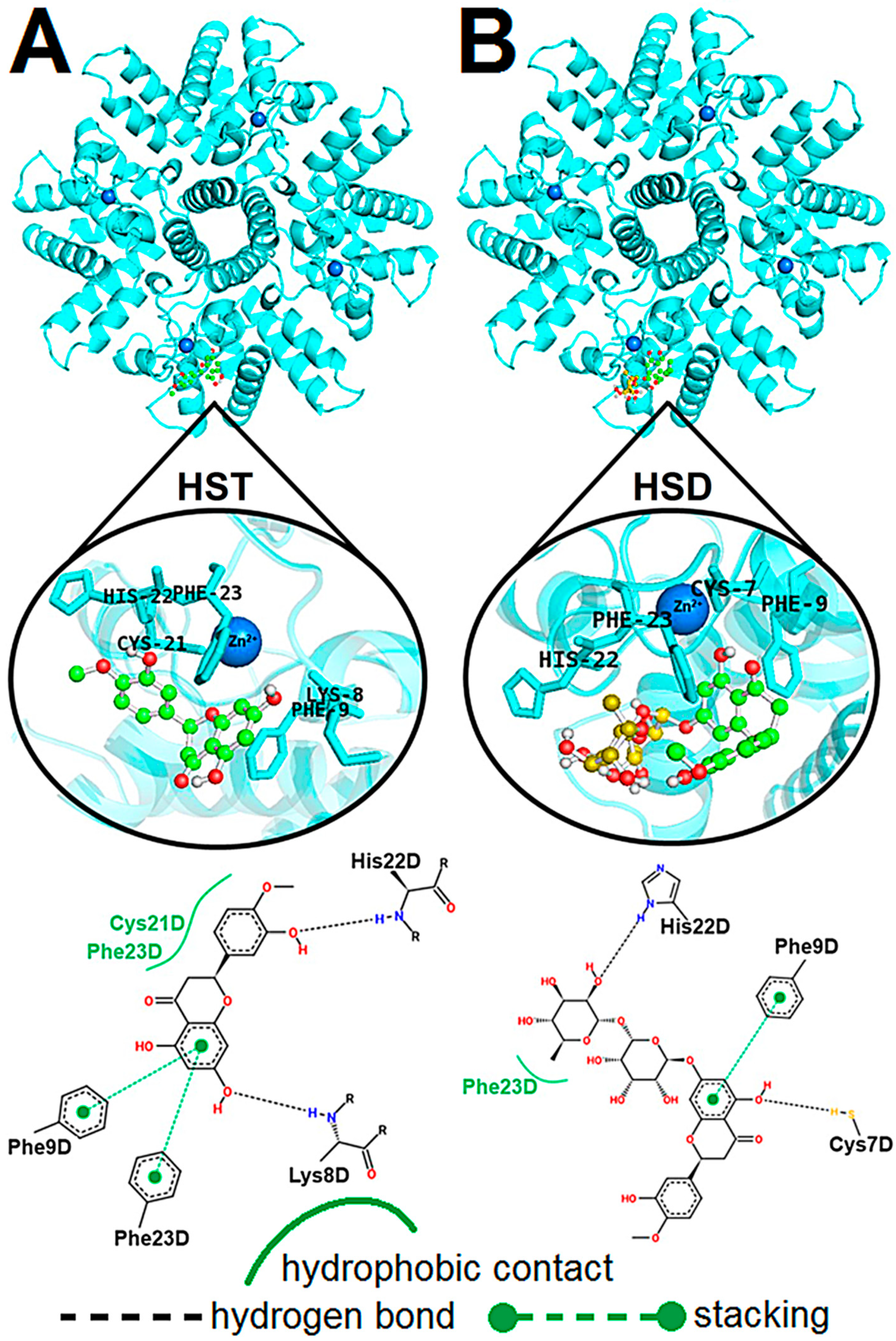
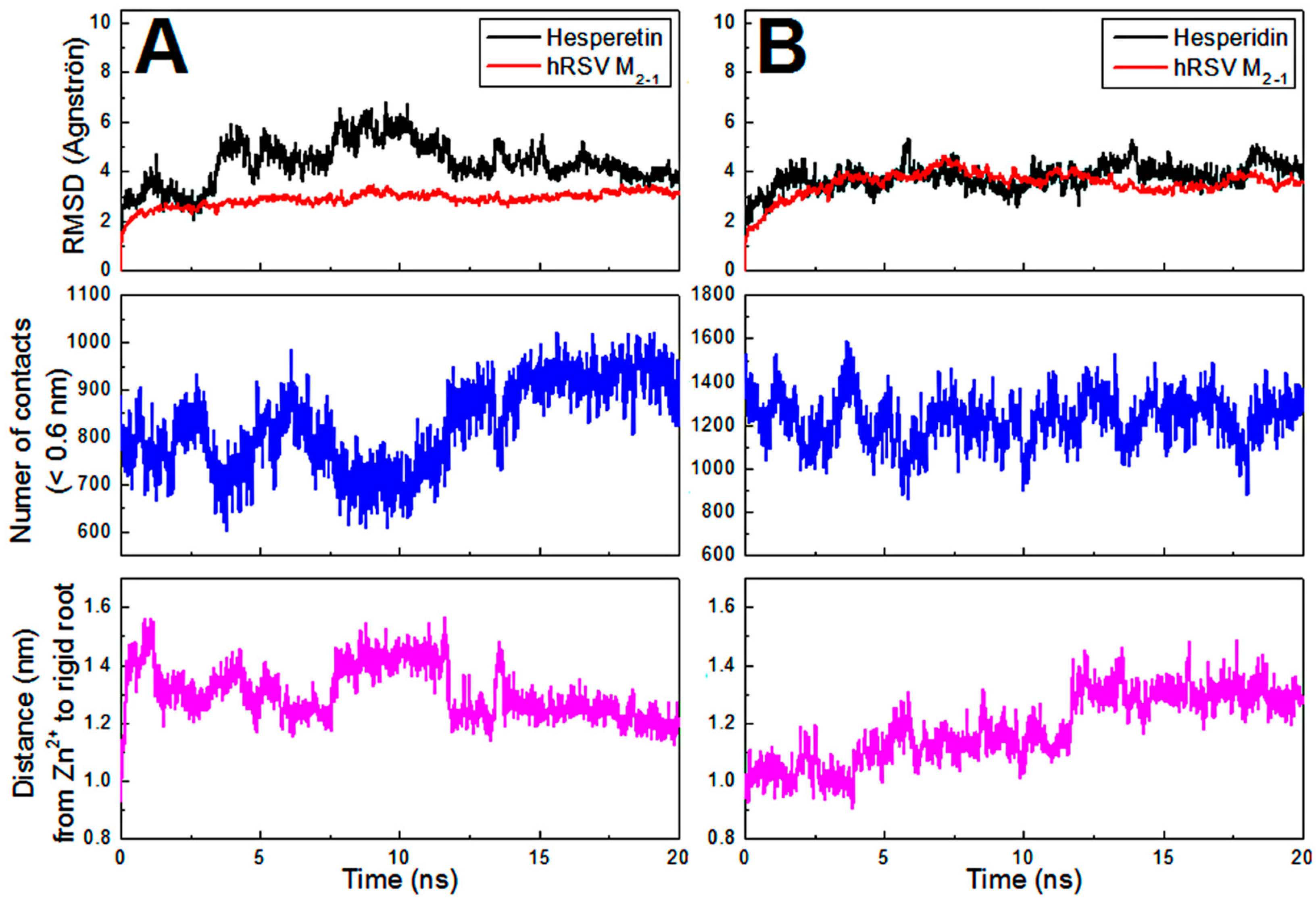
2.4. Computational Approach of the hRSV M2-1/flavonoid Complexes
3. Material and Methods
3.1. Material
3.2. Expression and Purification of the hRSV M2-1 Protein
3.3. Sample Preparation
3.4. Saturation Transfer Difference (STD) by Nuclear Magnetic Resonance (NMR)
3.5. Fluorescence Spectroscopy
3.6. Thermodynamic Analysis
3.7. Molecular Docking
3.8. Molecular Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| hRSV | Human Respiratory Syncytial Virus |
| HST | Hesperetin |
| HSD | Hesperidin |
| AMP | Adenosine monophosphate |
| STD-NMR | Saturation transfer difference by nuclear magnetic resonance |
| ΔH | Enthalpy variation |
| ΔS | Entropy variation |
| ΔG | Gibbs free energy |
| MD | molecular dynamics |
| MM-PBSA | Molecular Mechanics Poisson-Boltzmann Surface Area |
References
- Hall, C.B. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 2015, 344, 1917–1928. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.B.; Weinberg, G.A.; Iwane, M.K.; Blumkin, A.K.; Edwards, K.M.; Staat, M.A.; Auinger, P.; Griffin, M.R.; Poehling, K.A.; Erdman, D.; et al. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 2009, 360, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R.; Hennessey, P.A.; Formica, M.A.; Cox, C.; Walsh, E.E. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 2005, 352, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Whimbey, E.; Ghosh, S. Respiratory syncytial virus infections in immunocompromised adults. Curr. Clin. Top. Infect. Dis. 2000, 20, 232–255. [Google Scholar]
- Santos, N.S.O.; Romanos, M.T.V.; Wigg, M.D. Introdução à Virologia Humana, 2nd ed.; Guanabara Koogan: Rio de Janeiro, Brazil, 2008. [Google Scholar]
- Collins, P.L.; Huang, Y.T.; Wertz, G.W. Identification of a tenth mRNA of respiratory syncytial virus and assignment of polypeptides to the 10 viral genes. J. Virol. 1984, 49, 572–578. [Google Scholar] [CrossRef]
- Collins, P.L.; Hill, M.G.; Camargo, E.; Grosfeld, H.; Chanock, R.M.; Murphy, B.R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 1995, 92, 11563–11567. [Google Scholar]
- Collins, P.L.; Hill, M.G.; Cristina, J.; Grosfeld, H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 1996, 93, 81–85. [Google Scholar] [CrossRef]
- Hardy, R.W.; Wertz, G.W. The product of the respiratory syncytial virus M2 gene ORF1 enhances read through of intergenic junctions during viral transcription. J. Virol. 1998, 72, 520–526. [Google Scholar] [CrossRef]
- Kiss, G.; Holl, J.M.; Williams, G.M.; Alonas, E.; Vanover, D.; Lifland, A.W.; Gudheti, M.; Guerrero-Ferreira, R.C.; Nair, V.; Yi, H.; et al. Structural analysis of respiratory syncytial virus reveals the position of M2-1 between the matrix protein and the ribonucleoprotein complex. J. Virol. 2014, 88, 7602–7617. [Google Scholar] [CrossRef]
- Asenjo, A.; Cuesta, I.; Vivo, A.; Villanueva, N. Phosphorylation of the human respiratory syncytial virus N protein provokes a decrease in viral RNA synthesis. Virus Res. 2012, 163, 396–400. [Google Scholar] [CrossRef]
- Tran, T.L.; Castagné, N.; Dubosclard, V.; Noinville, S.; Koch, E.; Moudjou, M.; Henry, C.; Bernard, J.; Yeo, R.P.; Eléouët, J.F. The respiratory syncytial virus M2-1 protein forms tetramers and interacts with RNA and P in a competitive manner. J. Virol. 2009, 83, 6363–6374. [Google Scholar] [CrossRef] [PubMed]
- Hardy, R.W.; Wertz, G.W. The Cys(3)-His(1) motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 2000, 74, 5880–5885. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanner, S.J.; Ariza, A.; Richard, C.A.; Kyle, H.F.; Dods, R.L.; Blondot, M.L.; Wu, W.; Trincão, J.; Trinh, C.H.; Hiscox, J.A.; et al. Crystal structure of the essential transcription antiterminator M2-1 protein of human respiratory syncytial virus and implications of its phosphorylation. Proc. Natl. Acad. Sci. USA 2014, 111, 1580–1585. [Google Scholar] [CrossRef] [PubMed]
- Corneli, H.M.; Zorc, J.J.; Majahan, P.; Shaw, K.N.; Holubkov, R.; Reeves, S.D.; Ruddy, R.M.; Malik, B.; Nelson, K.A.; Bregstein, J.S.; et al. A multicenter, randomized, controlled trial of dexamethasone for bronchiolitis. Bronchiolitis Study Group of the Pediatric Emergency Care Applied Research Network (PECARN). N. Engl. J. Med. 2007, 357, 331–339. [Google Scholar] [CrossRef]
- Pelletier, A.J.; Mansbach, J.M.; Camargo, C.A. Direct medical costs of bronchiolitis hospitalizations in the United States. Pediatrics 2006, 118, 2418–2423. [Google Scholar] [CrossRef]
- Brady, M.T. Updated guidance for palivizumab prophylaxis among infants and young children at increased risk of hospitalization for respiratory syncytial virus infection. American Academy of Pediatrics Committee on infectious diseases; American Academy of Pediatrics bronchiolitis guidelines committee. Pediatrics 2014, 134, 415–420. [Google Scholar]
- Sidwell, R.W.; Barnard, D.L. Respiratory syncytial virus infections: Recent prospects for control. Antivir. Res. 2006, 71, 379–390. [Google Scholar] [CrossRef]
- Geskey, J.M.; Thomas, N.J.; Brummel, G.L. Palivizumab: A review of its use in the protection of high risk infants against respiratory syncytial virus (RSV). Biologics 2007, 1, 133–143. [Google Scholar]
- Kimura, K.; Mori, S.; Tomita, K.; Ohno, K. Antiviral activity of NMSO3 against respiratory syncytial virus infection in vitro and in vivo. Antivir. Res. 2000, 47, 41–51. [Google Scholar] [CrossRef]
- Cianci, C.; Meanwell, N.; Krystal, M. Antiviral activity and molecular mechanism of an orally active respiratory syncytial virus fusion inhibitor. J. Antimicrob. Chemother. 2005, 55, 289–292. [Google Scholar] [CrossRef]
- Sun, H.; Dong, T.; Zhang, A.; Yang, J.; Yan, G.; Sakurai, T.; Wu, X.; Han, Y.; Wang, X. Pharmacokinetics of hesperetin and naringenin in the Zhi Zhu Wan, a traditional Chinese medicinal formulae, and its pharmacodynamics study. Phytother. Res. 2013, 27, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B.; Mabry, T.J.; Mabry, M. The Flavonoids; Chapman and Hall: London, UK, 1975. [Google Scholar]
- Kuhnau, J. The flavonoids. A class of semi-essential food components: Their role in human nutrition. World Rev. Nutr. Diet. 1976, 24, 117–191. [Google Scholar] [PubMed]
- Parhiz, H.; Roohbakhsh, A.; Soltani, F.; Rezaee, R.; Iranshahi, M. Antioxidant and anti-inflammatory properties of the citrus flavonoids hesperidin and hesperetin: An updated review of their molecular mechanisms and experimental models. Phytother. Res. 2015, 29, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Garg, S.; Zaneveld, L.J.D.; Singla, A.K. Chemistry pharmacology of the citrus bioflavonoid hesperidin. Phytother. Res. 2001, 15, 655–669. [Google Scholar] [CrossRef]
- Iranshahi, M.; Rezaee, R.; Parhiz, H.; Roohbakhsh, A.; Soltani, F. Protective effects of flavonoids against microbes and toxins: The cases of hesperidin and hesperetin. Life Sci. 2015, 137, 125–132. [Google Scholar] [CrossRef]
- Kaul, T.N.; Middleton, E.J.R.; Ogra, P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985, 15, 71–79. [Google Scholar] [CrossRef]
- Liu, Q.; Lu, L.; Hua, M.; Xu, Y.; Xiong, H.; Hou, W.; Yang, Z. Jiawei-Yupingfeng-Tang, a Chinese herbal formula, inhibits respiratory viral infections in vitro and In Vivo. J. Ethnopharmacol. 2013, 150, 521–528. [Google Scholar] [CrossRef]
- Teixeira, T.S.P.; Caruso, I.P.; Lopes, B.R.P.; Regasini, L.O.; Toledo, K.A.; Fossey, M.A.; Souza, F.P. Biophysical characterization of the interaction between M2-1 protein of hRSV and quercetin. Int. J. Biol. Macromol. 2017, 95, 63–71. [Google Scholar] [CrossRef]
- Guimarães, G.C.; Piva, H.R.M.; Araújo, G.C.; Lima, C.S.; Regasini, L.O.; Melo, F.A.; Fossey, M.A.; Caruso, I.P.; Souza, F.P. Binding investigation between M2-1 protein from hRSV and acetylated quercetin derivatives: 1H NMR, fluorescence spectroscopy, and molecular docking. Int. J. Biol. Macromol. 2018, 111, 33–38. [Google Scholar] [CrossRef]
- Cao, H.; Wu, D.; Wang, H.; Xu, M. Effect of the glycosylation of flavonoids on interaction with protein. Spectrochim. Acta A 2009, 75, 972–975. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Hatsumi, A.; Magobei, Y. Thermodynamic characterization of drug binding to human serum albumin by isothermal titration microcalorimetry. J. Pharm. Sci. 1994, 83, 1712–1716. [Google Scholar]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.W.; Olson, A.J. Automated docking using a Larmarckian genetic algorithmand an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Fricker, P.; Gastreich, M.; Rarey, M. Automated generation of structural molecular formulas under constraints. J. Chem. Inf. Comput. Sci. 2004, 44, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Leyrat, C.; Renner, M.; Harlos, K.; Huiskonen, J.T.; Grimes, J.M. Drastic changes in conformational dynamics of the antiterminator M2-1 regulate transcription efficiency in Pneumovirinae. eLife 2014, 3, e02674. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa: A GROMACS tool for high-throughput MMPBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef]
- Tasic, L.; Mandic, B.; Barros, C.H.N.; Cypriano, D.Z.; Stanisic, D.; Schultz, L.G.; Dasilva, L.; Marino, M.A.M.; Queiroz, V.L. Exploring bioactivity of hesperidin, naturally occurring flavanone glycoside, isolated from oranges. In Citrus Fruits: Production, Consumption and Health Benefits; Simmons, D., Ed.; Nova Science Publishers Inc.: New York, NY, USA, 2016; pp. 27–70. [Google Scholar]
- Esperante, S.A.; Chemes, L.B.; Sánchez, I.E.; De Prat-Gay, G. The Respiratory Syncytial Virus Transcription Antiterminator M2-1 is a Highly Stable, Zinc Binding Tetramer with Strong pH-Dependent Dissociation and a Monomeric Unfolding Intermediate. Biochemistry 2011, 50, 8529–8539. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Mayer, M.; Meyer, B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [Google Scholar] [CrossRef]
- Viegas, A.; Manso, J.; Nobrega, F.L.; Cabrita, E.J. Saturation transfer difference (STD) NMR: A simple and fast method for ligand screening and characterization of protein binding. J. Chem. Educ. 2011, 88, 990–994. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 2nd ed.; Kluwer Academic Publishers/Plenum Press: New York, NY, USA, 1999. [Google Scholar]
- Bi, S.; Ding, L.; Tian, Y.; Song, D.; Zhou, X.; Liu, X.; Zhang, H. Investigation of the interaction between flavonoids and human serum albumin. J. Mol. Struc. 2004, 703, 37–45. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schimidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schiegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R. Gaussian 09, Revision A.02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Mod. 1999, 17, 57–61. [Google Scholar]
- Delano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Fährrolfes, R.; Bietz, S.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Otto, T.; Volkamer, A.; Rarey, M. ProteinsPlus: A web portal for structure analysis of macromolecules. Nucleic Acids Res. 2017, 45, 337–343. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; Van Gunsteren, W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces; Pullman, B., Ed.; The Jerusalem Symposia on Quantum Chemistry and Biochemistry; Springer: Dordrecht, The Netherlands, 1981; pp. 331–342. [Google Scholar]
- Malde, A.K.; Zuo, L.; Breeze, M.; Stroet, M.; Poger, D.; Nair, P.C.; Oostenbrink, C.; Mark, A.E. An Automated force field Topology Builder (ATB) and repository: Version 1.0. J. Chem. Theory Comput. 2011, 7, 4026–4037. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Sondergard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Lemkul, A.J. Scripts and Programs, OSF. 2018. Available online: osf.io/bafn4 (accessed on 27 January 2019).

| Flavonoids | T (K) | Kb a (× 104 M−1) | na | ΔH (kcal.mol−1) | ΔG (kcal.mol−1) | T·ΔS (kcal.mol−1) |
|---|---|---|---|---|---|---|
| HST | 288 | 8.40 | 1.05 | −29.0 | −6.5 | −22.5 |
| 298 | 1.53 | 1.04 | −5.7 | −23.3 | ||
| HSD | 288 | 0.96 | 0.89 | −1.7 | −5.2 | 3.5 |
| 298 | 0.87 | 0.93 | −5.4 | 3.7 |
| Flavonoids | First MD Simulation ΔGbinding (kcal.mol−1) | Second MD Simulation ΔGbinding (kcal.mol−1) | Average (kcal.mol−1) |
|---|---|---|---|
| HST | −40.4 ± 0.8 | −30.0 ± 1.0 | −35.2 |
| HSD | −27.1 ± 0.4 | −29.3 ± 0.5 | −28.2 |
| First MD Simulation | Second MD Simulation | >10% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor a | Acceptor a | %Acc.b | Donor a | Acceptor a | %Acc.b | Res.c | Atm.d | %Ave. | ||||
| K169 | NZ | HST | O2,3 | 0.7 | K169 | NZ | HST | O2–4 | 27.036 | K8 | N | 70.315 |
| N174 | N | HST | O3,4 | 11.794 | N174 | N | HST | O4 | 2.499 | N174 | O2 | 69.815 |
| N174 | ND2 | HST | O4 | 0.2 | R4 | NH1 | HST | O3 | 0.2 | N17 | ND2 | 26.762 |
| N5 | ND2 | HST | O5 | 7.596 | R4 | NH2 | HST | O3 | 0.15 | K8 | NZ | 25.812 |
| K8 | N | HST | O3–6 | 44.428 | K8 | N | HST | O5 | 96.202 | R20 | O | 14.643 |
| K8 | NZ | HST | O3–5 | 51.174 | K8 | NZ | HST | O4 | 0.45 | K169 | NZ | 13.868 |
| F9 | N | HST | O3,4 | 6.447 | F9 | N | HST | O5 | 0.1 | |||
| N17 | ND2 | HST | O1,2,6 | 30.735 | N17 | ND2 | HST | O1,2 | 22.789 | |||
| R20 | NH2 | HST | O1 | 0.05 | R20 | NE | HST | O1,2 | 25.737 | |||
| H22 | N | HST | O1,2 | 1.05 | R20 | NH1 | HST | O1,2 | 6.947 | |||
| H22 | NE2 | HST | O2 | 0.15 | R20 | NH2 | HST | O1,2 | 9.845 | |||
| F23 | N | HST | O2 | 0.5 | H22 | N | HST | O1,2 | 3.199 | |||
| HST | O5 | I173 | O | 0.05 | H22 | NE2 | HST | O3 | 0.05 | |||
| HST | O5 | N5 | OD1 | 0.5 | HST | O4 | N174 | O1 | 1.0 | |||
| HST | O5 | N5 | O | 5.547 | HST | O4 | N174 | O2 | 89.455 | |||
| HST | O5 | P6 | O | 0.55 | HST | O2 | E81 | OE1 | 30.385 | |||
| HST | O4 | N174 | O1 | 11.894 | HST | O2 | E81 | OE2 | 9.895 | |||
| HST | O4 | N174 | O2 | 50.175 | HST | O2 | N17 | OD1 | 0.5 | |||
| HST | O2 | N17 | OD1 | 6.797 | HST | O2 | R20 | O | 1.949 | |||
| HST | O2 | N17 | O | 0.05 | HST | O2 | H22 | ND1 | 9.345 | |||
| HST | O2 | G18 | O | 0.05 | ||||||||
| HST | O2 | R20 | O | 27.336 | ||||||||
| HST | O2 | H22 | ND1 | 3.348 | ||||||||
| First MD Simulation | Second MD Simulation | >10% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor a | Acceptor a | %Acc.b | Donor a | Acceptor a | %Acc.b | Res.c | Atm.d | %Ave. | ||||
| K169 | NZ | HSD | O4 | 0.05 | K169 | NZ | HSD | O6,8 O14,15 | 1.75 | R20 | O | 22.564 |
| N174 | N | HSD | O4 | 4.298 | N174 | N | HSD | O3,4 | 2.898 | H22 | ND1 | 17.043 |
| R4 | NE | HSD | O13–15 | 0.2 | N174 | ND2 | HSD | O3,4 | 4.098 | K8 | N | 14.793 |
| R4 | NH1 | HSD | O13–15 | 1.35 | K8 | N | HSD | O2 | 3.998 | F9 | N | 10.495 |
| R4 | NH2 | HSD | O13–15 | 0.6 | K8 | NZ | HSD | O3,4 | 2.649 | |||
| N5 | N | HSD | O15 | 0.1 | F9 | N | HSD | O2 | 11.294 | |||
| N5 | ND2 | HSD | O3,4 O13–15 | 2.699 | N17 | ND2 | HSD | O6,8,9 | 10.245 | |||
| K8 | N | HSD | O2,4 | 25.587 | G18 | N | HSD | O8,9 | 0.80 | |||
| K8 | NZ | HSD | O3,4 | 1.2 | R20 | N | HSD | O8 | 0.05 | |||
| F9 | N | HSD | O2 | 9.695 | R20 | NE | HSD | O8–12 | 2.1 | |||
| N17 | ND2 | HSD | O5,6,8 | 1.699 | R20 | NH1 | HSD | O7–12,15 | 8.946 | |||
| R20 | NE | HSD | O6,8–10 | 3.299 | R20 | NH2 | HSD | O7,9 O10,12 | 2.149 | |||
| R20 | NH1 | HSD | O6,8,9 | 4.998 | H22 | N | HSD | O7,11–15 | 11.094 | |||
| R20 | NH2 | HSD | O6,8–10 | 0.85 | H22 | NE2 | HSD | O7,11 O12,14,15 | 4.848 | |||
| H22 | N | HSD | O7,13 | 1.1 | F23 | N | HSD | O5,13,14 | 0.4 | |||
| H22 | NE2 | HSD | O8,10 | 0.95 | R4 | NH2 | HSD | O15 | 0.15 | |||
| F23 | N | HSD | O5,13,15 | 2.799 | HSD | O5,10 O13–15 | H22 | ND1 | 16.193 | |||
| HSD | O8,10 O13–15 | H22 | ND1 | 17.892 | HSD | O13 | H22 | NE2 | 0.05 | |||
| HSD | O13–15 | H22 | O | 2.899 | HSD | O5,8 O10,13,15 | R20 | O | 41.83 | |||
| HSD | O15 | H22 | NE2 | 0.05 | HSD | O8 | R20 | NH2 | 0.05 | |||
| HSD | O4,14,15 | N5 | OD1 | 5.347 | HSD | O15 | C21 | N | 0.05 | |||
| HSD | O8,13 | R20 | O | 3.248 | HSD | O13–15 | H22 | O | 1.5 | |||
| HSD | O8 | N17 | OD1 | 0.8 | HSD | O8–10 | E81 | OE1 | 35.382 | |||
| HSD | O4 | I173 | O | 0.05 | HSD | O8,9 | E81 | OE2 | 40.329 | |||
| HSD | O4 | N174 | O1 | 1.549 | HSD | O8,9 | N17 | OD1 | 1.649 | |||
| HSD | O4 | N174 | O2 | 2.849 | HSD | O8 | G18 | O | 1.399 | |||
| HSD | O4 | N5 | O | 2.299 | HSD | O4 | K169 | O | 0.1 | |||
| HSD | O4 | P6 | O | 1.949 | HSD | O4 | T172 | OG1 | 0.05 | |||
| HSD | O4 | T172 | O | 4.598 | ||||||||
| HSD | O4 | N174 | O1 | 0.1 | ||||||||
| HSD | O4 | N174 | O2 | 3.698 | ||||||||
| HSD | O4 | N5 | OD1 | 0.15 | ||||||||
| HSD | O4 | P6 | O | 0.05 | ||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piva, H.M.R.; Sá, J.M.; Miranda, A.S.; Tasic, L.; Fossey, M.A.; Souza, F.P.; Caruso, Í.P. Insights into Interactions of Flavanones with Target Human Respiratory Syncytial Virus M2-1 Protein from STD-NMR, Fluorescence Spectroscopy, and Computational Simulations. Int. J. Mol. Sci. 2020, 21, 2241. https://doi.org/10.3390/ijms21062241
Piva HMR, Sá JM, Miranda AS, Tasic L, Fossey MA, Souza FP, Caruso ÍP. Insights into Interactions of Flavanones with Target Human Respiratory Syncytial Virus M2-1 Protein from STD-NMR, Fluorescence Spectroscopy, and Computational Simulations. International Journal of Molecular Sciences. 2020; 21(6):2241. https://doi.org/10.3390/ijms21062241
Chicago/Turabian StylePiva, Hêmily M. R., Jéssica M. Sá, Artemiza S. Miranda, Ljubica Tasic, Marcelo A. Fossey, Fátima P. Souza, and Ícaro P. Caruso. 2020. "Insights into Interactions of Flavanones with Target Human Respiratory Syncytial Virus M2-1 Protein from STD-NMR, Fluorescence Spectroscopy, and Computational Simulations" International Journal of Molecular Sciences 21, no. 6: 2241. https://doi.org/10.3390/ijms21062241
APA StylePiva, H. M. R., Sá, J. M., Miranda, A. S., Tasic, L., Fossey, M. A., Souza, F. P., & Caruso, Í. P. (2020). Insights into Interactions of Flavanones with Target Human Respiratory Syncytial Virus M2-1 Protein from STD-NMR, Fluorescence Spectroscopy, and Computational Simulations. International Journal of Molecular Sciences, 21(6), 2241. https://doi.org/10.3390/ijms21062241