Chromosomal Signatures Corroborate the Phylogenetic Relationships within Akodontini (Rodentia, Sigmodontinae)
Abstract
:1. Introduction
2. Results
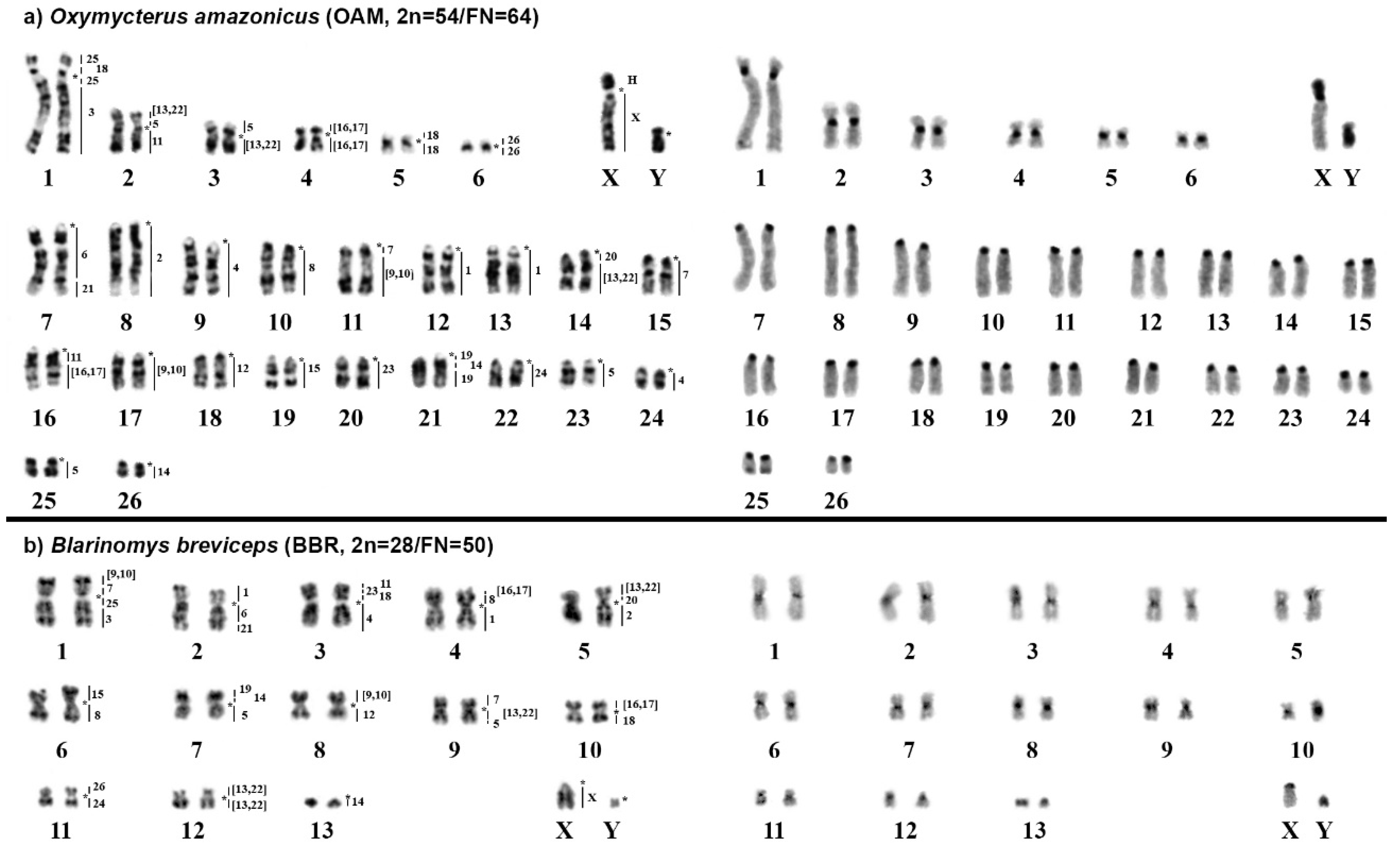
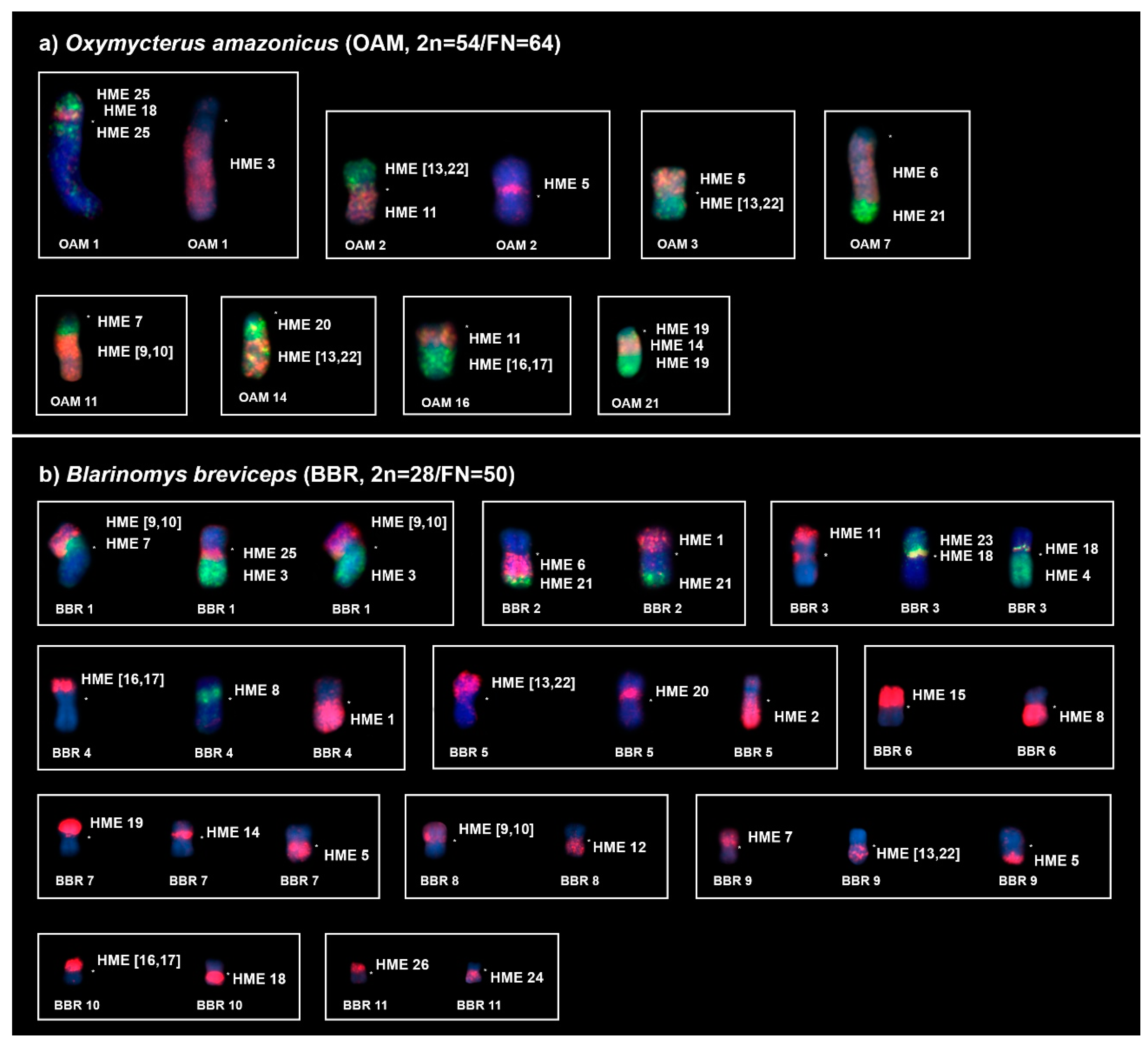
2.1. Oxymycterus Amazonicus (OAM; 2n = 54, FN = 64)
2.2. Blarinomys Breviceps (BBR; 2n = 28, FN = 50)
3. Discussion
3.1. New Cytogenetic Data for Oxymycterus and Blarinomys
3.2. Speciation Hypothesis in Oxymycterus and Blarinomys
3.3. Chromosomal Signatures in Akodontini Reinforce Clade A Monophyly
4. Materials and Methods
4.1. Samples
4.2. Cytogenetics
4.3. Image Capture and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Neill, R.J.W.; Eldridge, M.D.B.; Graves, J.A.M. Chromosome heterozygosity and de novo chromosome rearrangements in mammalian interspecies hybrids. Mamm. Genome 2001, 12, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.; Wang, J.; Su, W.; Wang, W.; Tanomtong, A.; Perelman, P.L.; Graphodatsky, A.S.; Yang, F. Chromosomal rearrangements and karyotype evolution in carnivores revealed by chromosome painting. Heredity 2012, 108, 17–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, N.F.; Svartman, M.; Manchester, A.; Moraes-Barros, N.; Stanyon, R.; Vianna-Morgante, A.M. Chromosome painting in three-toed sloths: A cytogenetic signature and ancestral karyotype for Xenarthra. BMC Evol. Biol. 2012, 12, 36. [Google Scholar] [CrossRef] [Green Version]
- Scherthan, H.; Cremer, T.; Arnason, U.; Weier, H.; Lima-de-Faria, A.; Frönicke, L. Comparative chromosome painting discloses homologous segments in distantly related mammals. Nat. Genet. 1994, 6, 342–347. [Google Scholar] [CrossRef] [Green Version]
- Wienberg, J. The evolution of eutherian chromosomes. Curr. Opin. Genet. Dev. 2004, 14, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.J.; Ruiz-Herrera, A. Defining the ancestral eutherian karyotype: A cladistic interpretation of chromosome painting and genome sequence assembly data. Chromosome Res. 2008, 16, 1133–1141. [Google Scholar] [CrossRef]
- Hass, I.; Sbalqueiro, I.J.; Müller, S. Chromosomal phylogeny of four Akodontini species (Rodentia, Cricetidae) from Southern Brazil established by ZOO-FISH using Mus musculus (Muridae) painting probes. Chromosome Res. 2008, 16, 75–88. [Google Scholar] [CrossRef]
- Oliveira da Silva, W.; Pieczarka, J.C.; Ferguson-Smith, M.A.; O’Brien, P.C.M.; Mendes-Oliveira, A.C.; Sampaio, I.; Carneiro, J.; Nagamachi, C.Y. Chromosomal diversity and molecular divergence among three undescribed species of Neacomys (Rodentia, Sigmodontinae) separated by Amazonian rivers. PLoS ONE 2017, 12, e0182218. [Google Scholar] [CrossRef] [Green Version]
- Oliveira da Silva, W.; Pieczarka, J.C.; Rodrigues da Costa, M.J.; Ferguson-Smith, M.A.; O’Brien, P.C.; Mendes-Oliveira, A.C.; Rossi, R.V.; Nagamachi, C.Y. Chromosomal phylogeny and comparative chromosome painting among Neacomys species (Rodentia, Sigmodontinae) from eastern Amazonia. BMC Evol. Biol. 2019, 19, 184. [Google Scholar] [CrossRef] [Green Version]
- Gomes, A.J.B.; Nagamachi, C.Y.; Rodrigues, L.R.R.; Ferguson-Smith, M.A.; O’Brien, P.C.; Pieczarka, J.C. Chromosomal evolution and phylogeny in the Nullicauda group (Chiroptera, Phyllostomidae): Evidence from multidirectional chromosome painting. BMC Evol. Biol. 2018, 18, 62. [Google Scholar] [CrossRef]
- da Silva, W.O.; Rodrigues da Costa, M.J.; Pieczarka, J.C.; Rissino, J.; Pereira, J.C.; Ferguson-Smith, M.A.; Nagamachi, C.Y. Identification of two independent X-autosome translocations in closely related mammalian (Proechimys) species. Sci. Rep. 2019, 9, 4047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; O’Brien, P.C.M.; Wienberg, J.; Neitzel, H.; Lin, C.C.; Ferguson-Smith, M.A. Chromosomal evolution of the Chinese muntjac (Muntiacus reevesi). Chromosoma 1997, 106, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Perelman, P.L.; Beklemisheva, V.R.; Yudkin, D.V.; Petrina, T.N.; Rozhnov, V.V.; Nie, W.; Graphodatsky, A.S. Comparative chromosome painting in Carnivora and Pholidota. Cytogenet. Genome Res. 2012, 137, 174–193. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Holland, P.W.H. Rare genomic changes as a tool for phylogenetics. TREE 2000, 15, 454–459. [Google Scholar] [CrossRef]
- Romanenko, S.A.; Perelman, P.L.; Trifonov, V.A.; Graphodatsky, A.S. Chromosomal evolution in Rodentia. Heredity 2012, 108, 4–16. [Google Scholar] [CrossRef] [Green Version]
- Ribas, T.F.A.; Rodrigues, L.R.R.; Nagamachi, C.Y.; Gomes, A.J.B.; Rissino, J.D.; O’Brien, P.C.M.; Fengtang, Y.; Ferguson-Smith, M.A.; Pieczarka, J.C. Phylogenetic reconstruction by cross-species chromosome painting and G-banding in four species of Phyllostomini tribe (Chiroptera, Phyllostomidae) in the Brazilian Amazon: An independent evidence for monophyly. PLoS ONE 2015, 10, e0122845. [Google Scholar] [CrossRef]
- Ye, J.; Biltueva, L.; Huang, L.; Nie, W.; Wang, J.; Jing, M.; Su, W.; Vorobieva, N.V.; Jiang, X.; Graphodatsky, A.S.; et al. Cross-species chromosome painting unveils cytogenetic signatures for the Eulipotyphla and evidence for the polyphyly of Insectivora. Chromosome Res. 2006, 14, 151–159. [Google Scholar] [CrossRef]
- Deakin, J.E.; Potter, S.; O’Neill, R.; Ruiz-Herrera, A.; Cioffi, M.B.; Eldridge, M.D.B.; Fukui, K.; Graves, J.A.M.; Griffin, D.; Grutzner, F.; et al. Chromosomics: Bridging the gap between genomes and chromosomes. Genes 2019, 10, 627. [Google Scholar] [CrossRef] [Green Version]
- Musser, G.G.; Carleton, M.D. Superfamily Muroidea. In Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Wilson, D.E., Reeder, D.M., Eds.; John Hopkins University Press: Baltimore, MD, USA, 2005; Volume 2, pp. 894–1531. [Google Scholar]
- Patton, J.L.; Pardiñas, U.F.J.; D’Elía, G. Mammals of South America. Volume 2, Rodents; The University of Chicago Press: Chicago, MI, USA, 2015; pp. 140–277. [Google Scholar]
- Ventura, K.; O’Brien, P.C.M.; Yonenaga-Yassuda, Y.; Ferguson-Smith, M.A. Chromosome homologies of the highly rearranged karyotypes of four Akodon species (Rodentia, Cricetidae) resolved by reciprocal chromosome painting: The evolution of the lowest diploid number in rodents. Chromosome Res. 2009, 17, 1063–1078. [Google Scholar] [CrossRef]
- Hass, I.; Muller, S.; Artoni, R.F.; Sbalqueiro, I.J. Comparative chromosome maps of neotropical rodents Necromys lasiurus and Thaptomys nigrita (Cricetidae) established by ZOO-FISH. Cytogenet. Genome Res. 2011, 135, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Nagamachi, C.Y.; Pieczarka, J.C.; O’Brien, P.C.M.; Pinto, J.A.; Malcher, S.M.; Pereira, A.L.; Rissino, J.; Mendes-Oliveira, A.C.; Rossi, R.V.; Ferguson-Smith, M.A. FISH with whole chromosome and telomeric probes demonstrates huge karyotypic reorganization with ITS between two species of Oryzomyini (Sigmodontinae, Rodentia): Hylaeamys megacephalus probes on Cerradomys langguthi karyotype. Chromosome Res. 2013, 21, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Suárez, P.; Nagamachi, C.Y.; Lanzone, C.; Malleret, M.M.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Pieczarka, J.C. Clues on syntenic relationship among some species of Oryzomyini and Akodontini Tribes (Rodentia: Sigmodontinae). PLoS ONE 2015, 10, e0143482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, A.L.; Malcher, S.M.; Nagamachi, C.Y.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Mendes-Oliveira, A.C.; Pieczarka, J.C. Extensive chromosomal reorganization among species of New World muroid rodents (Cricetidae, Sigmodontinae): Searching for phylogenetic ancestral traits. PLoS ONE 2016, 11, e0146179. [Google Scholar] [CrossRef] [Green Version]
- Malcher, S.M.; Pieczarka, J.C.; Geise, L.; Rossi, R.V.; Pereira, A.L.; O’Brien, P.C.M.; Asfora, P.H.; Silva, V.F.; Sampaio, I.; Ferguson-Smith, M.A.; et al. Oecomys catherinae (Sigmodontinae, Cricetidae): Evidence for chromosomal speciation? PLoS ONE 2017, 12, e0181434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Elía, G. Phylogenetics of Sigmodontinae (Rodentia, Muroidea, Cricetidae), with special reference to the akodont group, and with additional comments on historical biogeography. Cladistics 2003, 19, 307–323. [Google Scholar] [CrossRef] [Green Version]
- Pardiñas, U.; Geise, L.; Ventura, K.; Lessa, G. A new genus for Habrothrix angustidens and Akodon serrensis (Rodentia: Cricetidae): Again paleontology meets neontology in the legacy of Lund. Mastozool. Neotrop. 2016, 23, 93–115. [Google Scholar]
- Svartman, M.; Almeida, C.J.E. The karyotype of Oxymycterus sp. (Cricetidae, Rodentia) from Central Brazil. Experientia 1993, 49, 718–720. [Google Scholar] [CrossRef]
- Silva, M.J.J.; Patton, J.L.; Yonenaga-Yassuda, Y. Phylogenetic relationships and karyotype evolution in the Sigmodontinae rodent Akodon (2n = 10 and 2n = 16) from Brazil. Genet. Mol. Biol. 2006, 3, 469–474. [Google Scholar] [CrossRef]
- Vitullo, A.D.; Merani, M.S.; Reig, O.A.; Kajon, A.E.; Scaglia, O.; Espinosa, M.B.; Perez-Zapata, A. Cytogenetics of South American Akodont rodents (Cricetidae): New karyotypes and chromosomal banding patterns of Argentinian and Uruguayan forms. J. Mamm. 1986, 67, 69–80. [Google Scholar] [CrossRef]
- Reig, O.A. An assessment of the systematics and evolution of the Akodontini, with the description of new fossil species of Akodon (Cricetidae: Sigmodontinae). Fieldiana Zool. 1987, 39, 347–399. [Google Scholar]
- Geise, L.; Bergallo, H.G.; Esbérard, C.E.L.; Rocha, C.F.D.; Sluys, M.V. The karyotype of Blarinomys breviceps (Mammalia: Rodentia: Cricetidae) with comments on its morphology and some ecological notes. Zootaxa 2008, 1907, 47–60. [Google Scholar] [CrossRef]
- Barros, M.C.; Sampaio, I.; Schneider, H.; Langguth, A. Molecular phylogenies, chromosomes and dispersion in Brazilian akodontines (Rodentia, Sigmodontinae). Ser. Zool. 2009, 99, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Peç, W.T.; Quintela, F.M.; Ribas, L.E.J.; Althoff, S.L.; Maestri, R.; Gonçalves, G.L.; de Freitas, T.R. A new species of Oxymycterus (Rodentia: Cricetidae: Sigmodontinae) from a transitional area of Cerrado—Atlantic Forest in southeastern Brazil. J. Mamm. 2019, 100, 578–598. [Google Scholar] [CrossRef]
- Bonvicino, C.R.; Penna-Firme, V.; Seuánez, H.N. The karyotype of Brucepattersonius griserufescens Hershkovitz, 1998 (Rodentia, Sigmodontinae) with comments on distribution and taxonomy. Z. Saügetierkd. 1998, 63, 329–335. [Google Scholar]
- Geise, L.; Pereira, L.G.; Bossi, D.E.P.; Bergallo, H.G. Pattern of elevational distribution and richness of non volant mammals in Itatiaia National Park and its surroundings, in Southeastern Brazil. Braz. J. Biol. 2004, 64, 599–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, J.C.; Manduca, E.G.; Gonçalves, P.R.; Morais, M.M., Jr.; Pereira, R.F.; Lessa, G.; Dergam, J.A. Small mammals from Serra do Brigadeiro State Park, Minas Gerais, Southeastern Brazil: Species Composition and Elevational Distribution. Arq. Mus. Nac. 2009, 67, 103–118. [Google Scholar]
- Bonvicino, C.R.; Lemos, B.; Weskler, M. Small mammals of Chapada dos Veadeiros National Park (Cerrado of Central Brazil): Ecologic, karyologic and taxonomic considerations. Braz. J. Biol. 2005, 65, 359–406. [Google Scholar] [CrossRef] [Green Version]
- Bonvicino, C.R. Diversidade cariotípica em roedores Akodontini do Brasil. Bol. Soc. Bras. Mastozool. 2011, 62, 7–13. [Google Scholar]
- Quintela, F.M.; Santos, M.B.; Christoff, A.U.; Gava, A. Pequenos mamíferos não-voadores (Didelphimorphia, Rodentia) em dois fragmentos de mata de restinga de Rio Grande, Planície Costeira do Rio Grande do Sul. Biota Neotrop. 2012, 12, 261–266. [Google Scholar] [CrossRef] [Green Version]
- Ventura, K.; Sato-Kuwabara, Y.; Fagundes, V.; Geise, L.; Leite, Y.L.R.; Costa, L.P.; Silva, M.J.J.; Yonenaga-Yassuda, Y.; Rodrigues, M.T. Phylogeographic structure and karyotypic diversity of the Brazilian shrew mouse (Blarinomys breviceps, Sigmodontinae) in the Atlantic Forest. Cytogenet. Genome Res. 2012, 138, 19–30. [Google Scholar] [CrossRef]
- Di-Nizo, C.B.; Banci, K.R.S.; Sato-Kuwabara, Y.; Silva, M.J.J. Advances in cytogenetics of Brazilian rodents: Cytotaxonomy, chromosome evolution and new karyotypic data. Comp. Cytogenet. 2017, 11, 833–892. [Google Scholar] [CrossRef] [Green Version]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Graphodatsky, A.; Ferguson-Smith, M.A.; Stanyon, R. A short introduction to cytogenetic studies in mammals with reference to the present volume. Cytogenet. Genome Res. 2012, 137, 83–96. [Google Scholar] [CrossRef] [PubMed]
- da Silva, W.O.; Pieczarka, J.C.; Rossi, R.V.; Schneider, H.; Sampaio, I.; Miranda, C.L.; da Silva, C.R.; Cardoso, E.M.; Nagamachi, C.Y. Diversity and Karyotypic Evolution in the Genus Neacomys (Rodentia, Sigmodontinae). Cytogenet. Genome Res. 2015, 146, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Bradley, R.D.; Baker, R.J. A test of the genetic species concept: Cytochrome-B sequences and mammals. J. Mamm. 2001, 82, 960–973. [Google Scholar] [CrossRef]
- Baker, R.J.; Bradley, R.D. Speciation in mammals and the genetic species concept. J. Mamm. 2006, 87, 643–662. [Google Scholar] [CrossRef] [PubMed]
- Peçanha, W.T.; Althoff, S.L.; Galiano, D.; Quintela, F.M.; Maestri, R.; Gonçalves, G.L.; Freitas, T.L.O. Pleistocene climatic oscillations in Neotropical open areas: Refuge isolation in the rodent Oxymycterus nasutus endemic to grasslands. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Hoorn, C.; Wesselingh, F.P.; Steege, H.; Bermudez, M.A.; Mora, A.; Sevink, J.; Sanmartín, I.; Sanchez-Meseguer, A.; Anderson, C.L.; Figueiredo, J.P.; et al. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 2010, 330, 927–931. [Google Scholar] [CrossRef] [Green Version]
- Vargas, R. Abundance and population structure of Oxymycterus paramensis (Rodentia) in forest fragments in the Andes of Bolivia. Rev. Cien. Technol. Innov. 2019, 17, 23–44. [Google Scholar]
- Gonçalves, P.R.; Oliveira, J.A. Morphological and genetic variation between two sympatric forms of Oxymycterus (Rodentia: Sigmodontinae): An evaluation of hypotheses of differentiation within the genus. J. Mamm. 2004, 85, 148–161. [Google Scholar] [CrossRef] [Green Version]
- Costa, L.P.; Leite, Y.L. Historical fragmentation shaping vertebrate diversification in the Atlantic forest biodiversity hotspot. In Bones, Clones and Biomes: The History and Geography of Recent Neotropical Mammals; Patterson, B., Costa, L.P., Eds.; The University of Chicago Press: Chicago, IL, USA, 2012. [Google Scholar]
- Pellegrino, K.C.; Rodrigues, M.T.; Waite, N.A.; Morando, M.; Yonenaga-Yassuda, Y.; Sites, J.W., Jr. Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): Genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biol. J. Linn. Soc. 2005, 85, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Carnaval, A.C.; Hickerson, M.J.; Haddad, C.F.; Rodrigues, M.T.; Moritz, C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 2009, 323, 785–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabanne, G.S.; Santos, F.R.; Miyaki, C.Y. Phylogeography of Xiphorhynchus fuscus (Passeriformes, Dendrocolaptidae): Vicariance and recent demographic expansion in southern Atlantic forest. Biol. J. Linn. Soc. 2007, 91, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Costa, L.P.; Leite, Y.L.; da Fonseca, G.A.; Fonseca, M.T. Biogeography of South American forest mammals: Endemism and diversity in the Atlantic Forest. Biotropica 2000, 32, 872–881. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Lagos, A.R.; Muller, B.L.A. Hotspot Brasileiro—Mata Atlântica. Saúde Amb. Rev. 2007, 2, 35–45. [Google Scholar]
- Joly, C.A.; Metzger, J.P.; Tabarelli, M. Experiences from the Brazilian Atlantic Forest: Ecological findings and conservation initiatives. New Phytol. 2014. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, M.C.; Metzger, J.P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How much is left, and how is the remaining forest distributed? Implications for conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- Haag, T.; Santos, A.S.; Sana, D.A.; Morato, R.G.; Cullen, L., Jr.; Crawshaw, P.G., Jr.; De Angelo, C.; Di Bitetti, M.S.; Salzano, F.M.; Eizirik, E. The effect of habitat fragmentation on the genetic structure of a top predator: Loss of diversity and high differentiation among remnant populations of Atlantic Forest jaguars (Panthera onca). Mol. Ecol. 2010, 19, 4906–4921. [Google Scholar] [CrossRef]
- Hillman, S.S.; Drewes, R.C.; Hedrick, M.S.; Hancock, T.V. Physiological vagility and its relationship to dispersal and neutral genetic heterogeneity in vertebrates. J. Exp. Biol. 2014, 217, 3356–3364. [Google Scholar] [CrossRef] [Green Version]
- Lande, R. Effective deme sizes during long-term evolution estimated from rates of chromosomal rearrangement. Evolution 1979, 33, 234–251. [Google Scholar] [CrossRef] [PubMed]
- Lande, R. The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 1985, 54, 323–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corn, P.S. Straight-line drift fences and pitfall traps. In Measuring and Monitoring Biological Standard Methods for Amphibians, 1st ed.; Heyer, W.R., Donnelly, M.A., McDiarmid, R.W., Hayek, L.C., Foster, M.S., Eds.; Smithsonian Institution Press: Washington, DC, USA, 1994; pp. 109–117. [Google Scholar]
- Ford, C.E.; Hamerton, J.L. A colchicine, hypotonic—Citrate, squash sequence for mammalian chromosomes. Stain. Technol. 1956, 31, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Sumner, A.T.; Evans, H.J.; Buckland, R.A. New technique for distinguishing between human chromosomes. Nat. New Biol. 1971, 31, 282. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]

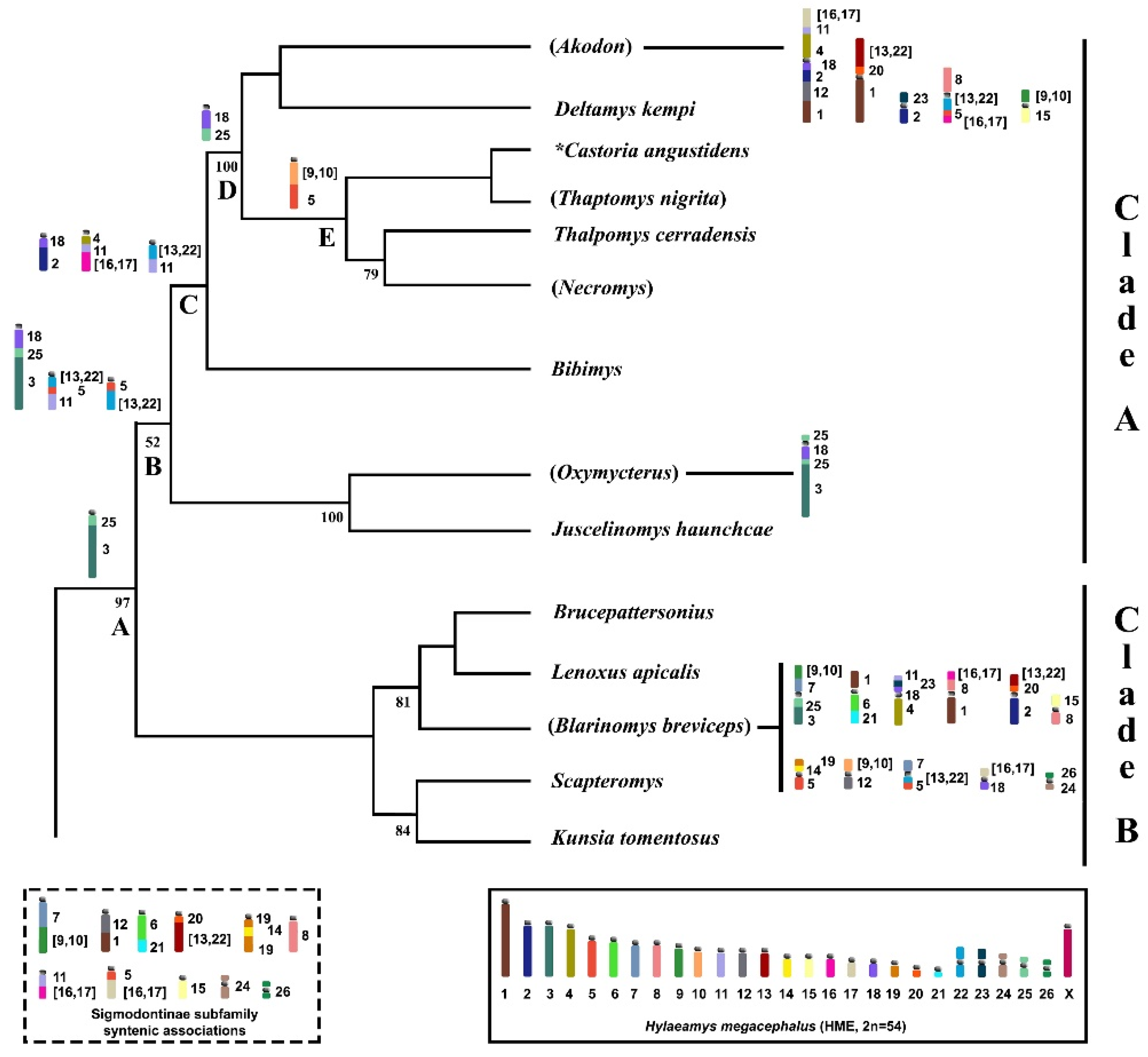
| CLADE A | CLADE B | ||||
|---|---|---|---|---|---|
| Division | Akodon | Oxymycterus | Bibimys | Blarinomys | Scapteromys |
| Genus | Akodon | Oxymycterus | Bibimys | Blarinomys | Scapteromys |
| Deltamys | Juscelinomys | Brucepattersonius | Kunsia | ||
| Necromys | Lenoxus | ||||
| Thalpomys | |||||
| Thaptomys | |||||
| * Castoria angustidens | |||||
| Species * | Karyotype | Reference |
|---|---|---|
| Oxymycterus amazonicus | 2n = 54, FN = 64 | Present study |
| O. caparaoe | 2n = 54, FN = 64 | [36] |
| O. dasytrichus | 2n = 54, FN = 62 | [37,38] |
| O. delator | 2n = 54, FN = 62, 64 | [36,39,40] |
| O. nasutus | 2n = 54, FN = 64 | [41] |
| O. paramensis | 2n = 54, FN = 60, 64 | [31,36] |
| O. quaestor | 2n = 54, FN = 64 | [36] |
| O. rufus | 2n = 54, FN = 60, 64 | [31,36] |
| Oxymycterus sp. | 2n = 54, FN = 64 | [29] |
| Blarinomys breviceps | 2n = 52 (+2Bs), FN = 50; 2n = 52, FN = 50; 2n = 45 (+1B), FN = 50; 2n = 43 (+4Bs), FN = 50; 2n = 37 (+1B), FN = 50; 2n = 34, FN = 50; 2n = 31 (+2Bs), FN = 50; 2n = 28, FN = 50 | [33,42] |
| B. breviceps | 2n = 28, FN = 50 | Present study |
| HME | OAM | BBR |
|---|---|---|
| 1 | 12, 13 | 2p, 4q |
| 2 | 8 | 5q |
| 3 | 1q dist. | 1q dist. |
| 4 | 9, 24 | 3q |
| 5 | 2p prox., 3p, 23, 25 | 7q, 9q dist. |
| 6 | 7q prox. | 2q prox. |
| 7 | 11q prox., 15 | 1p prox., 9p |
| 8 | 10 | 4p prox., 6q |
| (9,10) | 11q dist., 17 | 1p dist., 8p |
| 11 | 2q dist., 16q prox. | 3p dist. |
| 12 | 18 | 8q |
| (13,22) | 2p dist., 3q, 14q dist. | 5p dist., 9q prox., 12 |
| 14 | 21q int., 26 | 7p proximal, 13 |
| 15 | 19 | 6p |
| (16,17) | 4, 16q dist. | 4p dist., 10p |
| 18 | 1p prox., 5 | 3p prox., 10q |
| 19 | 21q (prox. and dist.) | 7p dist. |
| 20 | 14q prox. | 5p prox. |
| 21 | 7q dist. | 2q dist. |
| 23 | 20 | 3p int. |
| 24 | 22 | 11q |
| 25 | 1p dist., 1q int. | 1q prox. |
| 26 | 6 | 11p |
| X | Xq | X |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira da Silva, W.; Malcher, S.M.; Pereira, A.L.; Pieczarka, J.C.; Ferguson-Smith, M.A.; O’Brien, P.C.M.; Mendes-Oliveira, A.C.; Geise, L.; Nagamachi, C.Y. Chromosomal Signatures Corroborate the Phylogenetic Relationships within Akodontini (Rodentia, Sigmodontinae). Int. J. Mol. Sci. 2020, 21, 2415. https://doi.org/10.3390/ijms21072415
Oliveira da Silva W, Malcher SM, Pereira AL, Pieczarka JC, Ferguson-Smith MA, O’Brien PCM, Mendes-Oliveira AC, Geise L, Nagamachi CY. Chromosomal Signatures Corroborate the Phylogenetic Relationships within Akodontini (Rodentia, Sigmodontinae). International Journal of Molecular Sciences. 2020; 21(7):2415. https://doi.org/10.3390/ijms21072415
Chicago/Turabian StyleOliveira da Silva, Willam, Stella Miranda Malcher, Adenilson Leão Pereira, Julio Cesar Pieczarka, Malcolm Andrew Ferguson-Smith, Patricia Caroline Mary O’Brien, Ana Cristina Mendes-Oliveira, Lena Geise, and Cleusa Yoshiko Nagamachi. 2020. "Chromosomal Signatures Corroborate the Phylogenetic Relationships within Akodontini (Rodentia, Sigmodontinae)" International Journal of Molecular Sciences 21, no. 7: 2415. https://doi.org/10.3390/ijms21072415
APA StyleOliveira da Silva, W., Malcher, S. M., Pereira, A. L., Pieczarka, J. C., Ferguson-Smith, M. A., O’Brien, P. C. M., Mendes-Oliveira, A. C., Geise, L., & Nagamachi, C. Y. (2020). Chromosomal Signatures Corroborate the Phylogenetic Relationships within Akodontini (Rodentia, Sigmodontinae). International Journal of Molecular Sciences, 21(7), 2415. https://doi.org/10.3390/ijms21072415






