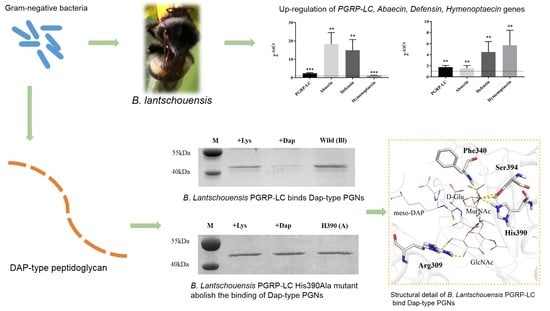
Structural and Functional Analysis of PGRP-LC Indicates Exclusive Dap-Type PGN Binding in Bumblebees
Abstract
:1. Introduction
2. Results
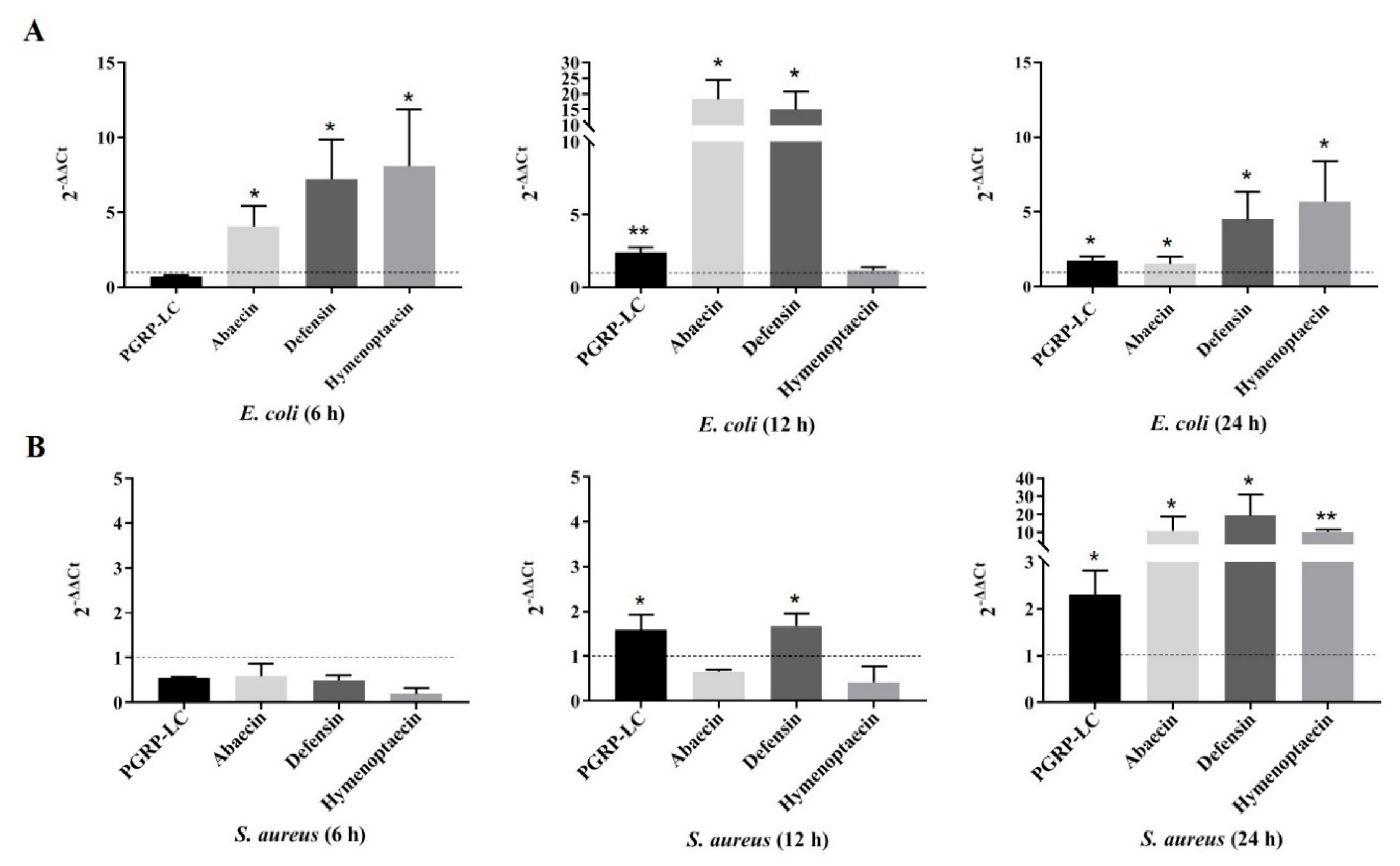
2.1. Response of Bl-PGPR-LC to Infection with Gram-Negative and Gram-Positive Bacteria
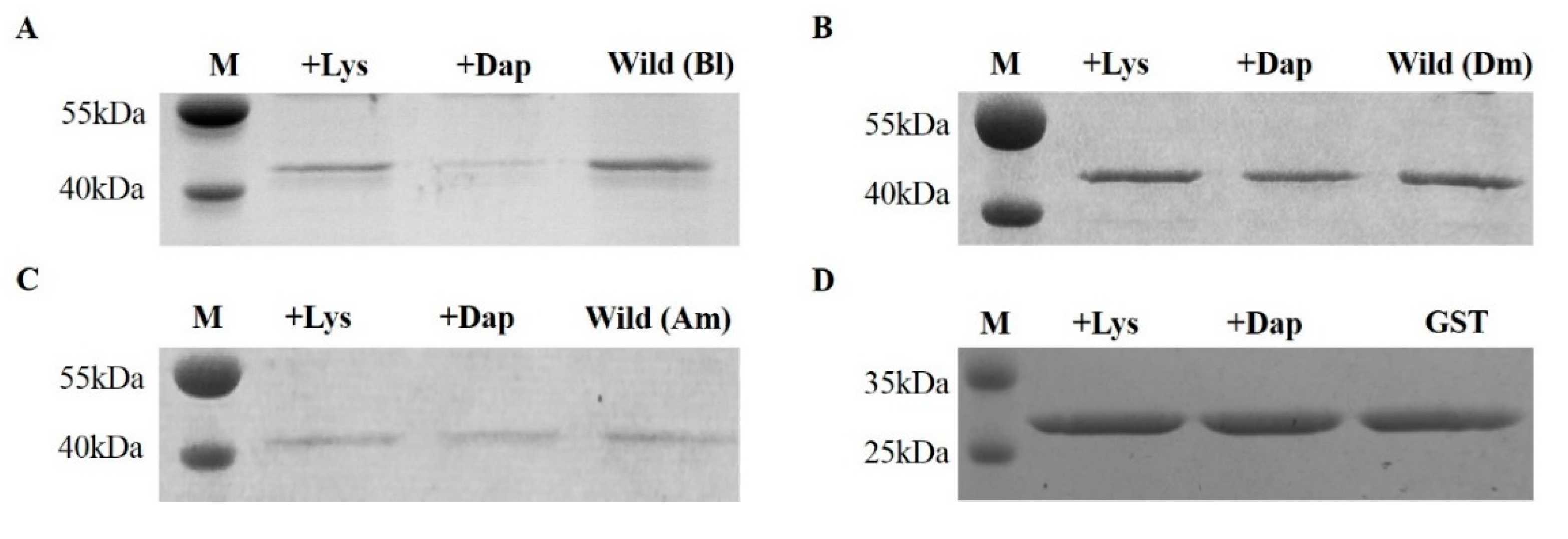
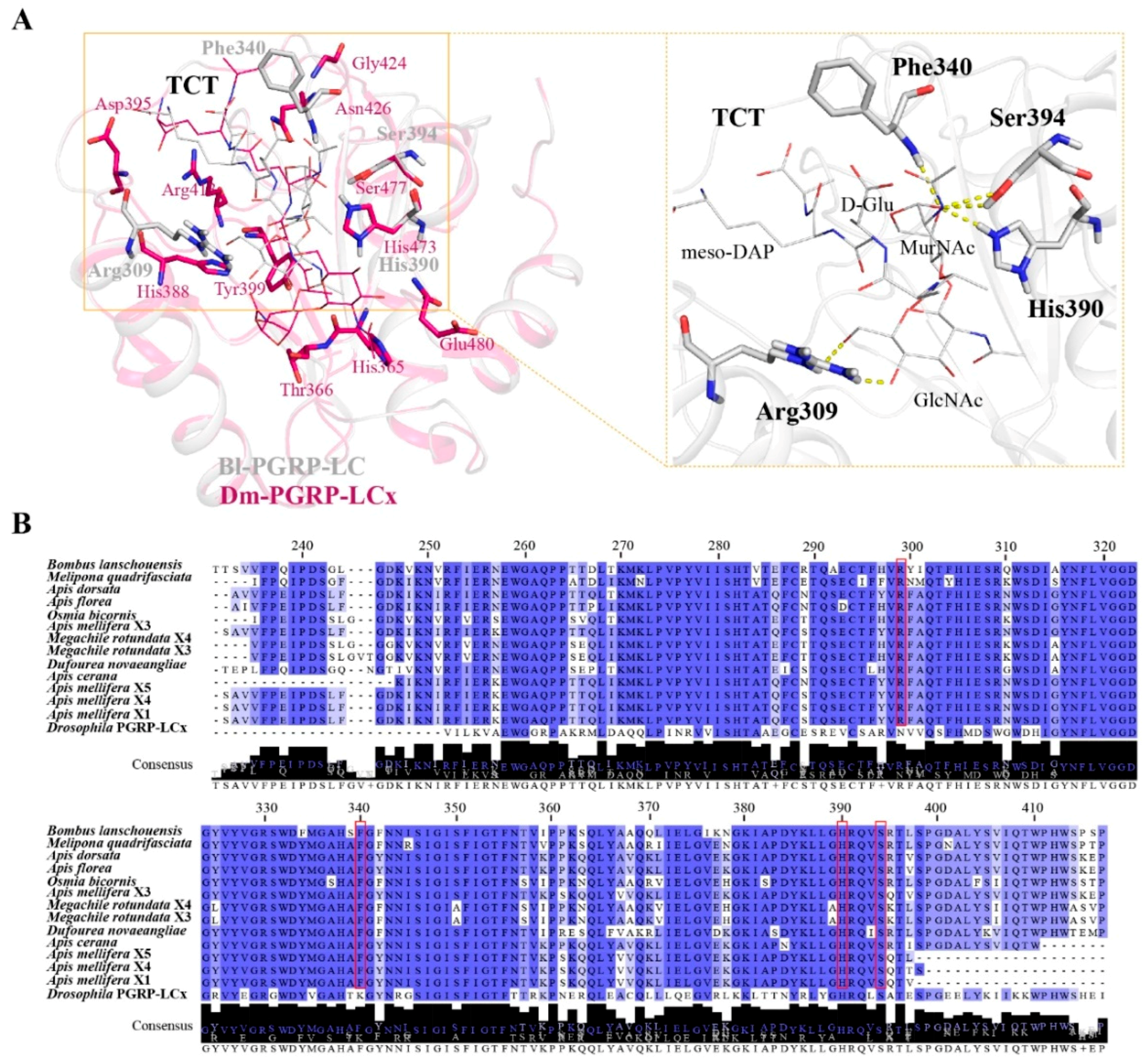
2.2. Bombus PGRP-LC Strongly Bound to Dap-Type PGN and Exhibited Unique Anchor Residues in Comparison with Drosophila PGRP-LCx
2.3. Structural Insight into PGRP-LC from Different Bee Species
3. Discussion
4. Materials and Methods
4.1. Bacterial Infection and Real-Time Quantitative PCR Verification
4.2. Preparation of Proteins
4.3. Lys-Type and Dap-Type PGN-Binding Assays
4.4. Amino Acid Sequence Alignment and Phylogenetic and Structural Analyses
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PGRPs | peptidoglycan recognition proteins |
| PGN | peptidoglycan |
References
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medzhitov, R.; Janeway, C.A., Jr. Decoding the patterns of self and nonself by the innate immune system. Science 2002, 296, 298–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, J.A. The immune response of Drosoph. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Kinoshita, K.; Ashida, M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 1996, 271, 13854–13860. [Google Scholar] [CrossRef] [Green Version]
- Dziarski, R.; Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome biol. 2006, 7, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choe, K.M.; Werner, T.; Stoven, S.; Hultmark, D.; Anderson, K.V. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 2002, 296, 359–362. [Google Scholar] [CrossRef] [Green Version]
- Michel, T.; Reichhart, J.M.; Hoffmann, J.A.; Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001, 414, 756–759. [Google Scholar] [CrossRef]
- Schleifer, K.H.; Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.S.; Byun, M.; Oh, B.H. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol. 2003, 4, 787–793. [Google Scholar] [CrossRef]
- Reiser, J.B.; Teyton, L.; Wilson, I.A. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution. J. Mol. Biol. 2004, 340, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.I.; Pili-Floury, S.; Herve, M.; Parquet, C.; Chelliah, Y.; Lemaitre, B.; Mengin-Lecreulx, D.; Deisenhofer, J. A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual L,D-carboxypeptidase activity. PLoS Biol. 2004, 2, E277. [Google Scholar] [CrossRef] [PubMed]
- Leone, P.; Bischoff, V.; Kellenberger, C.; Hetru, C.; Royet, J.; Roussel, A. Crystal structure of Drosophila PGRP-SD suggests binding to DAP-type but not lysine-type peptidoglycan. Mol. Immunol. 2008, 45, 2521–2530. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.I.; Ihara, K.; Chelliah, Y.; Mengin-Lecreulx, D.; Wakatsuki, S.; Deisenhofer, J. Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition. Proc. Natl. Acad. Sci. USA 2005, 102, 10279–10284. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.H.; Kim, M.S.; Kim, H.E.; Yano, T.; Oshima, Y.; Aggarwal, K.; Goldman, W.E.; Silverman, N.; Kurata, S.; Oh, B.H. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 2006, 281, 8286–8295. [Google Scholar] [CrossRef] [Green Version]
- Basbous, N.; Coste, F.; Leone, P.; Vincentelli, R.; Royet, J.; Kellenberger, C.; Roussel, A. The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway. EMBO Rep. 2011, 12, 327–333. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.I.; Chelliah, Y.; Borek, D.; Mengin-Lecreulx, D.; Deisenhofer, J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 2006, 311, 1761–1764. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, T.; Yano, T.; Aggarwal, K.; Lim, J.H.; Ueda, K.; Oshima, Y.; Peach, C.; Erturk-Hasdemir, D.; Goldman, W.E.; Oh, B.H.; et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006, 7, 715–723. [Google Scholar] [CrossRef]
- Senapathi, D.; Carvalheiro, L.G.; Biesmeijer, J.C.; Dodson, C.A.; Evans, R.L.; McKerchar, M.; Morton, R.D.; Moss, E.D.; Roberts, S.P.; Kunin, W.E.; et al. The impact of over 80 years of land cover changes on bee and wasp pollinator communities in England. Proc. Roy. Soc. B Biol. Sci. 2015, 282, 20150294. [Google Scholar] [CrossRef] [Green Version]
- Parmentier, L.; Meeus, I.; Cheroutre, L.; Mommaerts, V.; Louwye, S.; Smagghe, G. Commercial bumblebee hives to assess an anthropogenic environment for pollinator support: A case study in the region of Ghent (Belgium). Environ. Monit. Assess. 2014, 186, 2357–2367. [Google Scholar] [CrossRef]
- Klatt, B.K.; Holzschuh, A.; Westphal, C.; Clough, Y.; Smit, I.; Pawelzik, E.; Tscharntke, T. Bee pollination improves crop quality, shelf life and commercial value. Proc. Roy. Soc. B Biol. Sci. 2014, 281, 20132440. [Google Scholar] [CrossRef] [PubMed]
- Eilers, E.J.; Kremen, C.; Smith Greenleaf, S.; Garber, A.K.; Klein, A.M. Contribution of pollinator-mediated crops to nutrients in the human food supply. PLoS ONE 2011, 6, e21363. [Google Scholar] [CrossRef] [PubMed]
- Velthuis, H.H.W.; van Doorn, A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 2006, 37, 421–451. [Google Scholar] [CrossRef] [Green Version]
- Morandin, L.A.; Laverty, T.M.; Kevan, P.G. Bumble bee (Hymenoptera: Apidae) activity and pollination levels in commercial tomato greenhouses. J. Econ. Entomol. 2001, 94, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Kraus, B.; Page, R.E., Jr. Parasites, pathogens, and polyandry in social insects. Am. Nat. 1998, 151, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meeus, I.; Smagghe, G. Israeli acute paralysis virus associated paralysis symptoms, viral tissue distribution and Dicer-2 induction in bumblebee workers (Bombus terrestris). J. Gen. Virol. 2016, 97, 1981–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parmentier, L.; Smagghe, G.; de Graaf, D.C.; Meeus, I. Varroa destructor Macula-like virus, Lake Sinai virus and other new RNA viruses in wild bumblebee hosts (Bombus pascuorum, Bombus lapidarius and Bombus pratorum). J. Invertebr. Pathol. 2016, 134, 6–11. [Google Scholar] [CrossRef]
- Cappelle, K.; Smagghe, G.; Dhaenens, M.; Meeus, I. Israeli Acute Paralysis Virus Infection Leads to an Enhanced RNA Interference Response and Not Its Suppression in the Bumblebee Bombus terrestris. Viruses 2016, 8, 334. [Google Scholar] [CrossRef] [Green Version]
- Meeus, I.; de Miranda, J.R.; de Graaf, D.C.; Wackers, F.; Smagghe, G. Effect of oral infection with Kashmir bee virus and Israeli acute paralysis virus on bumblebee (Bombus terrestris) reproductive success. J. Invertebr. Pathol. 2014, 121, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Fürst, M.; McMahon, D.P.; Osborne, J.; Paxton, R.; Brown, M. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 2014, 506, 364. [Google Scholar] [CrossRef]
- Honeybee Genome Sequencing, C. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931–949. [Google Scholar] [CrossRef] [PubMed]
- Sadd, B.M.; Barribeau, S.M.; Bloch, G.; De Graaf, D.C.; Dearden, P.K.; Elsik, C.G.; Gadau, J.; Grimmelikhuijzen, C.J.P.; Hasselmann, M.; Lozier, J.D. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015, 16, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, H.; Wan, H.; Li, J.; Jin, B.R. Molecular cloning and characterization of a short peptidoglycan recognition protein (PGRP-S) with antibacterial activity from the bumblebee Bombus ignitus. Dev. Comp. Immunol. 2010, 34, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect. Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.J.; Zhao, X.M.; Huang, J.X.; Chen, M.M.; An, J.D. Structural Insights into the Preferential Binding of PGRP-SAs from Bumblebees and Honeybees to Dap-Type Peptidoglycans Rather than Lys-Type Peptidoglycans. J. Immunol. 2019, 202, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhao, X.; Naeem, M.; An, J. Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae). Protein. Sci. 2018, 27, 893–897. [Google Scholar] [CrossRef]
- Chen, M.; Ye, N.; Liu, Y.; An, J. Preliminary analysis of PGRP-LC gene and structure characteristics in bumblebees. Sociobiology 2019, 66, 348–357. [Google Scholar] [CrossRef] [Green Version]
- Cardinal, S.; Danforth, B.N. The antiquity and evolutionary history of social behavior in bees. PLoS ONE 2011, 6, e21086. [Google Scholar] [CrossRef] [Green Version]
- Simone-Finstrom, M. Social immunity and the superorganism: Behavioral defenses protecting honey bee colonies from pathogens and parasites. Bee World 2017, 94, 21–29. [Google Scholar] [CrossRef]
- Cremer, S.; Armitage, S.A.; Schmid-Hempel, P. Social immunity. Curr. Biol. 2007, 17, R693–R702. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30 (Suppl. S1), S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Bordoli, L.; Kiefer, F.; Arnold, K.; Benkert, P.; Battey, J.; Schwede, T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009, 4, 1. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]

| Genes | Primers Names | Primers Sequences |
|---|---|---|
| Actin | B-actin F4 | ACCTCCCTTGAGAAGAGCTACG |
| B-actin R4 | TACCCAGGAAGGAAGGTTGG | |
| PGRP-LC | LC F1 | GCAGTCTTGCCAGTTCCTCTAC |
| LC R1 | GAAGCACCACATTCCCATC | |
| Abaecin | Abaecin F1 | ATATAATCCGCCACGACCG |
| Abaecin R1 | GGTTTGGTAATGGGTATGGC | |
| Defensin | Defensin F2 | TCTTGTCGCTCTTCTCTTTGTG |
| Defensin R2 | TCTTCTTTGTCTGTCAGCACG | |
| Hymenoptaecin | Hymenoptaecin F1 | CCCGTTCTTCGGTAACTGTG |
| Hymenoptaecin R1 | TCACTCCGTTTCTGTCGTAGAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Ye, N.; Chen, M.; Zhao, H.; An, J. Structural and Functional Analysis of PGRP-LC Indicates Exclusive Dap-Type PGN Binding in Bumblebees. Int. J. Mol. Sci. 2020, 21, 2441. https://doi.org/10.3390/ijms21072441
Liu Y, Ye N, Chen M, Zhao H, An J. Structural and Functional Analysis of PGRP-LC Indicates Exclusive Dap-Type PGN Binding in Bumblebees. International Journal of Molecular Sciences. 2020; 21(7):2441. https://doi.org/10.3390/ijms21072441
Chicago/Turabian StyleLiu, Yanjie, Nanhui Ye, Minming Chen, Huiyue Zhao, and Jiandong An. 2020. "Structural and Functional Analysis of PGRP-LC Indicates Exclusive Dap-Type PGN Binding in Bumblebees" International Journal of Molecular Sciences 21, no. 7: 2441. https://doi.org/10.3390/ijms21072441