Identification of the Repressive Domain of the Negative Circadian Clock Component CHRONO
Abstract
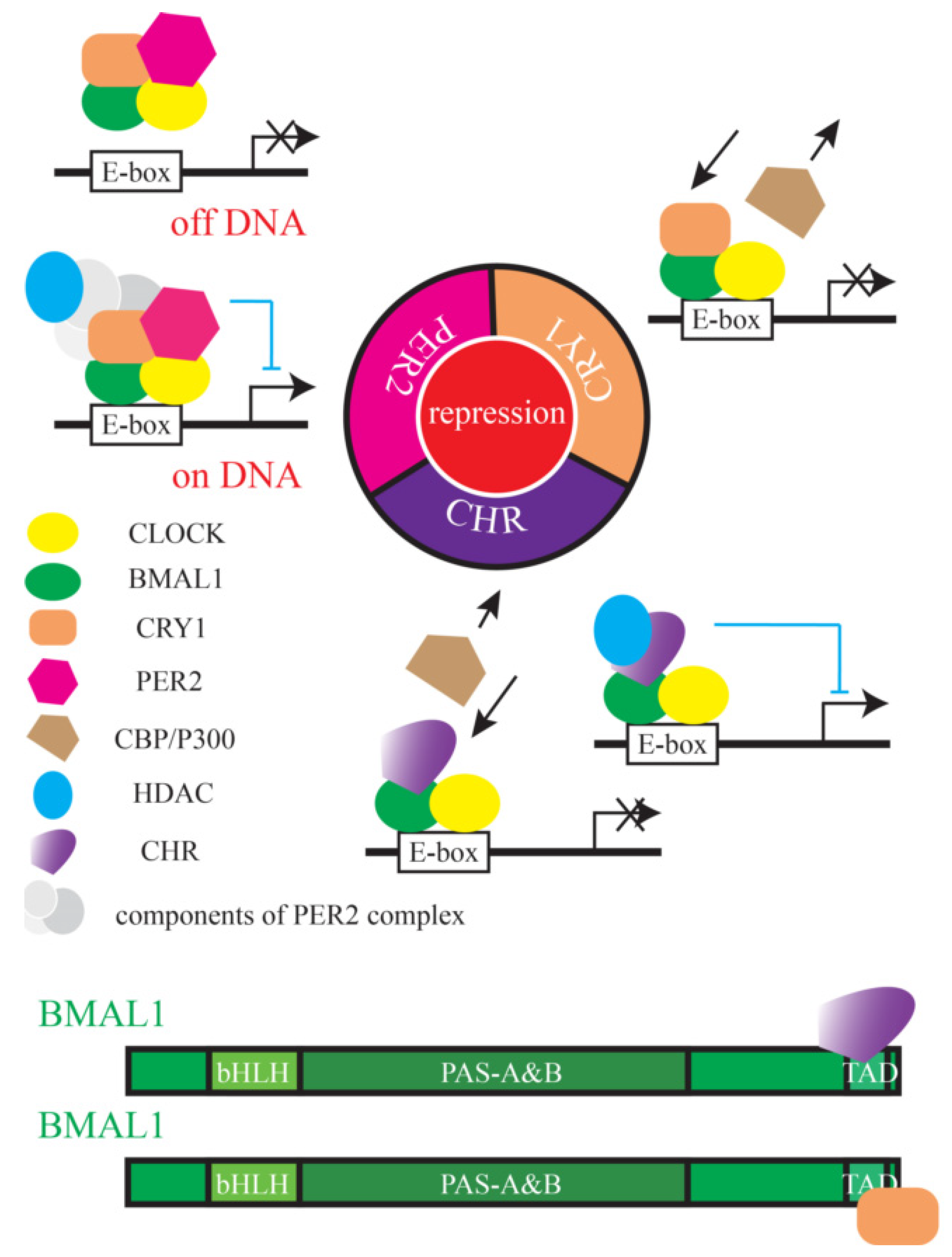
:1. Introduction
2. Materials and Methods
2.1. Strains and Constructs
2.2. Site-Directed Mutagenesis
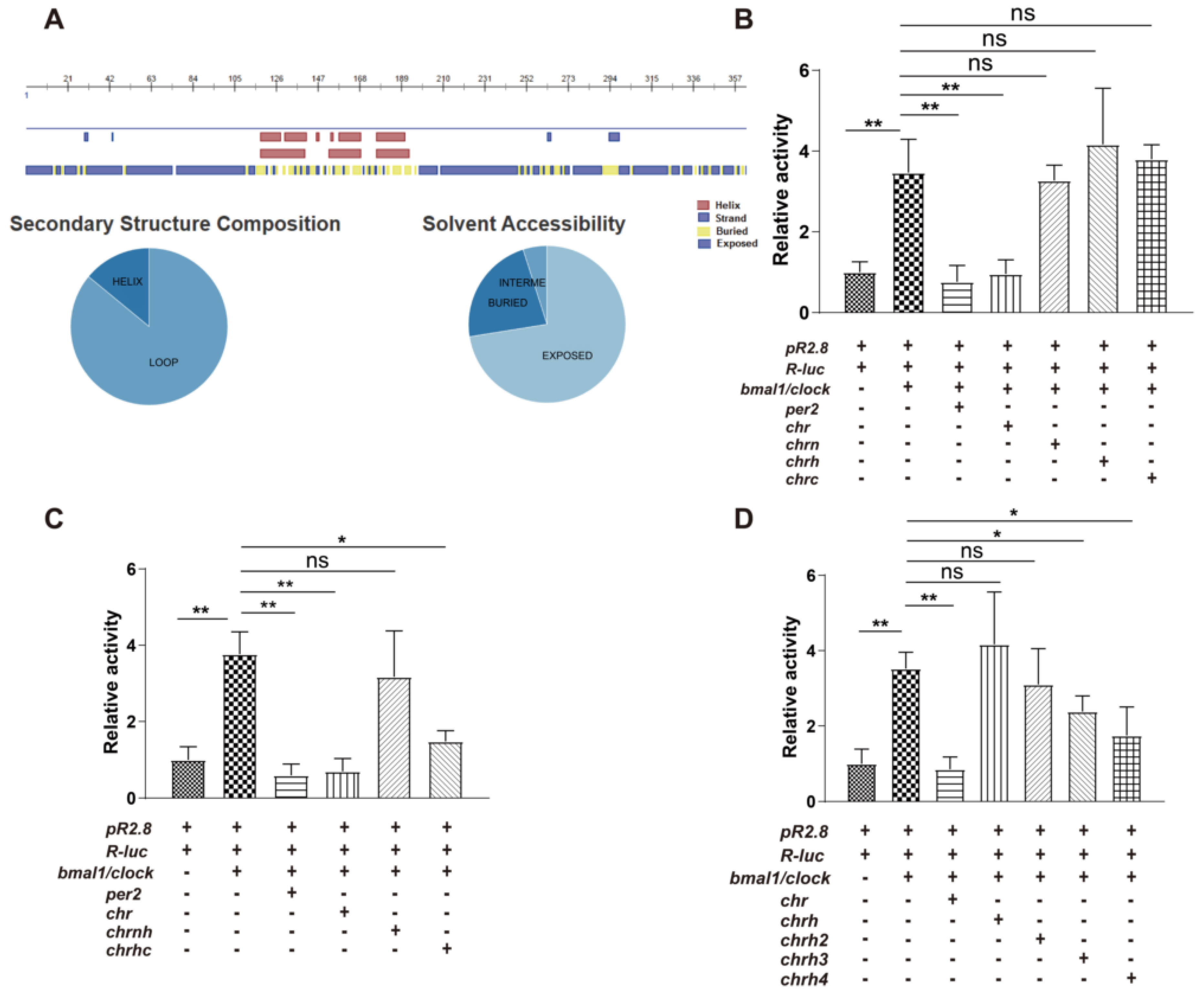
2.3. Secondary Structure Prediction
2.4. Nuclear Location Signal Prediction
2.5. Codon Optimization
2.6. Protein Expression and Purification
2.7. Circular Dichroism Spectroscopy
2.8. Luciferase Reporter Assay
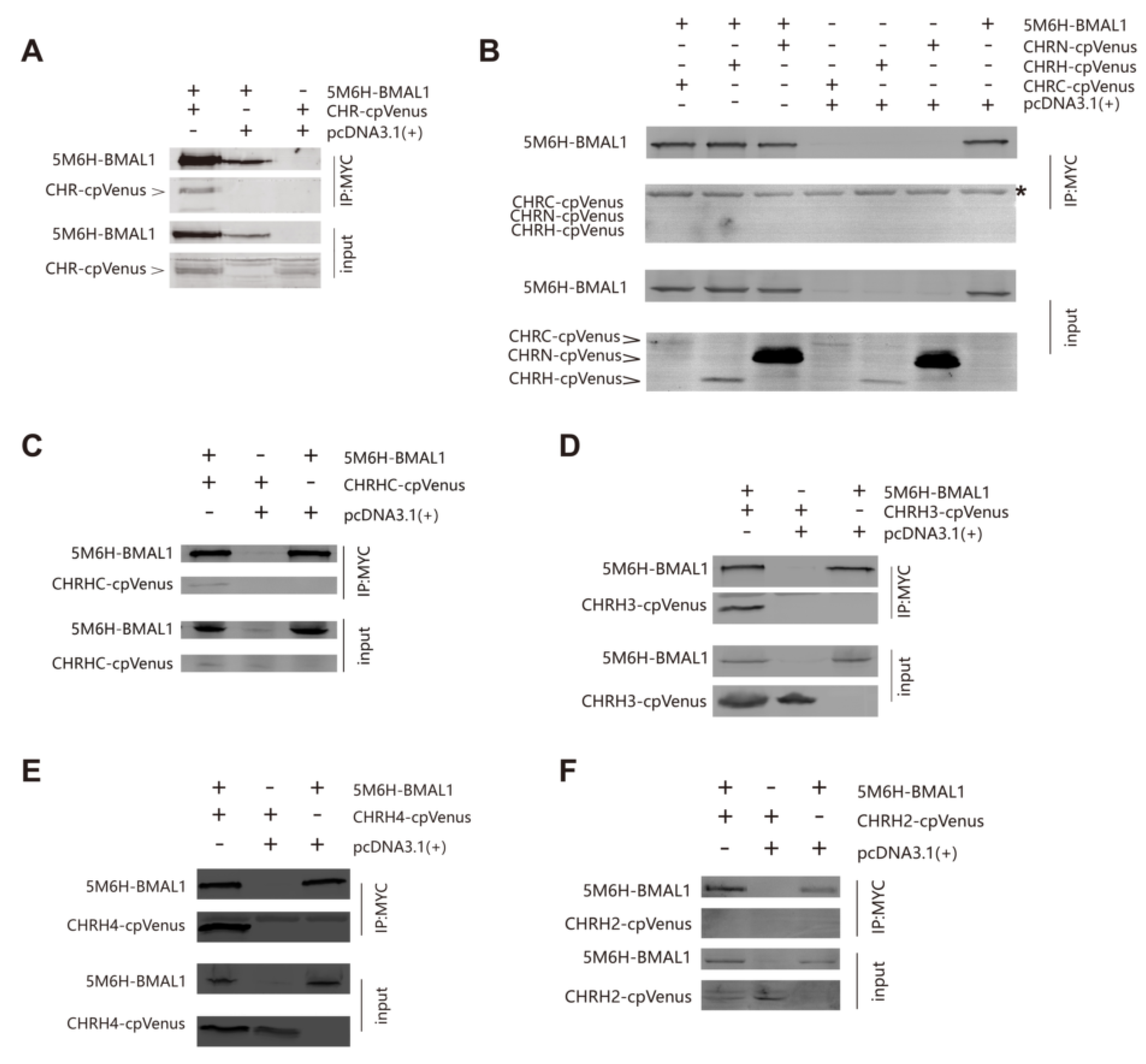
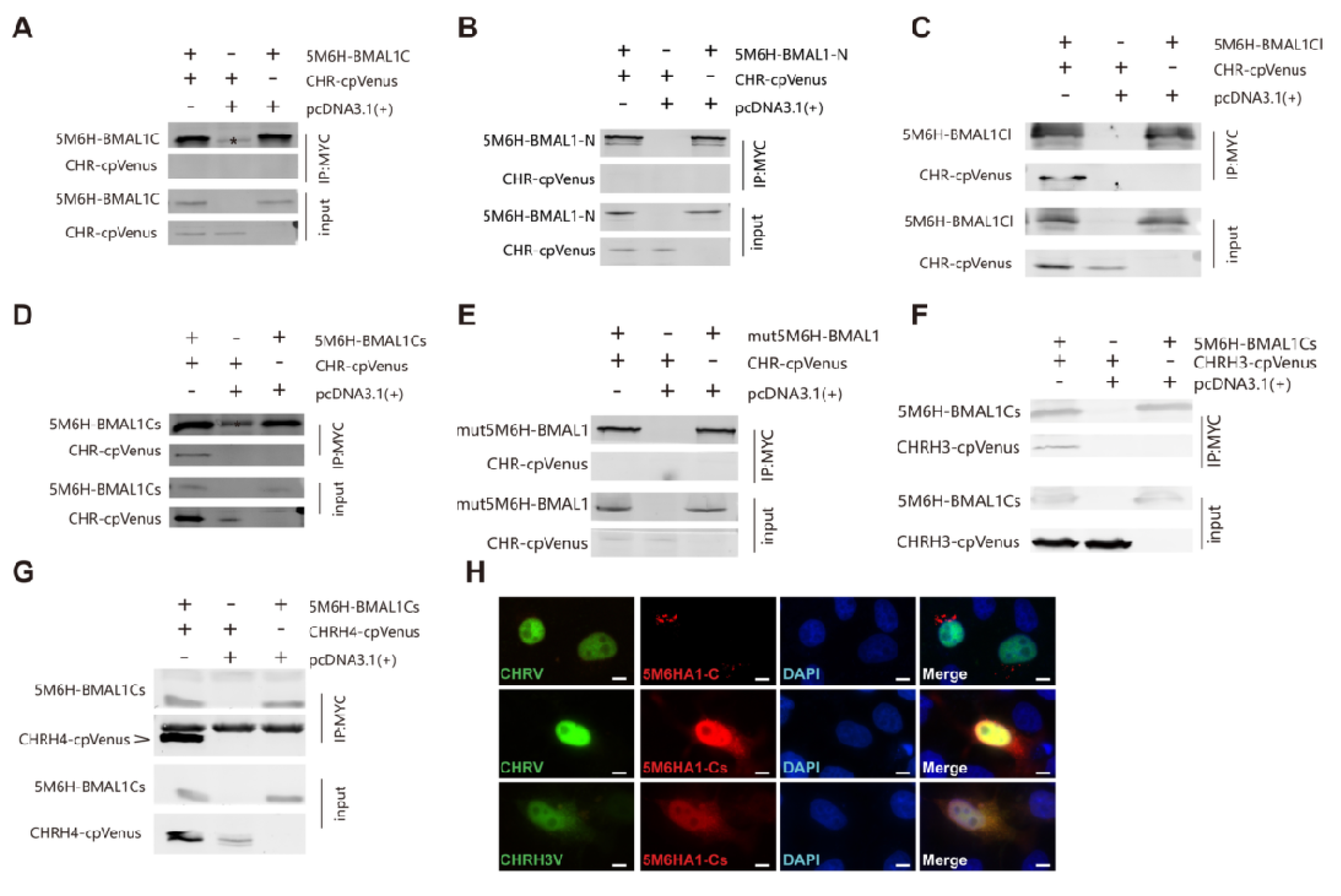
2.9. Western Blotting and Immunoprecipitation
2.10. Immunofluorescence and Localization
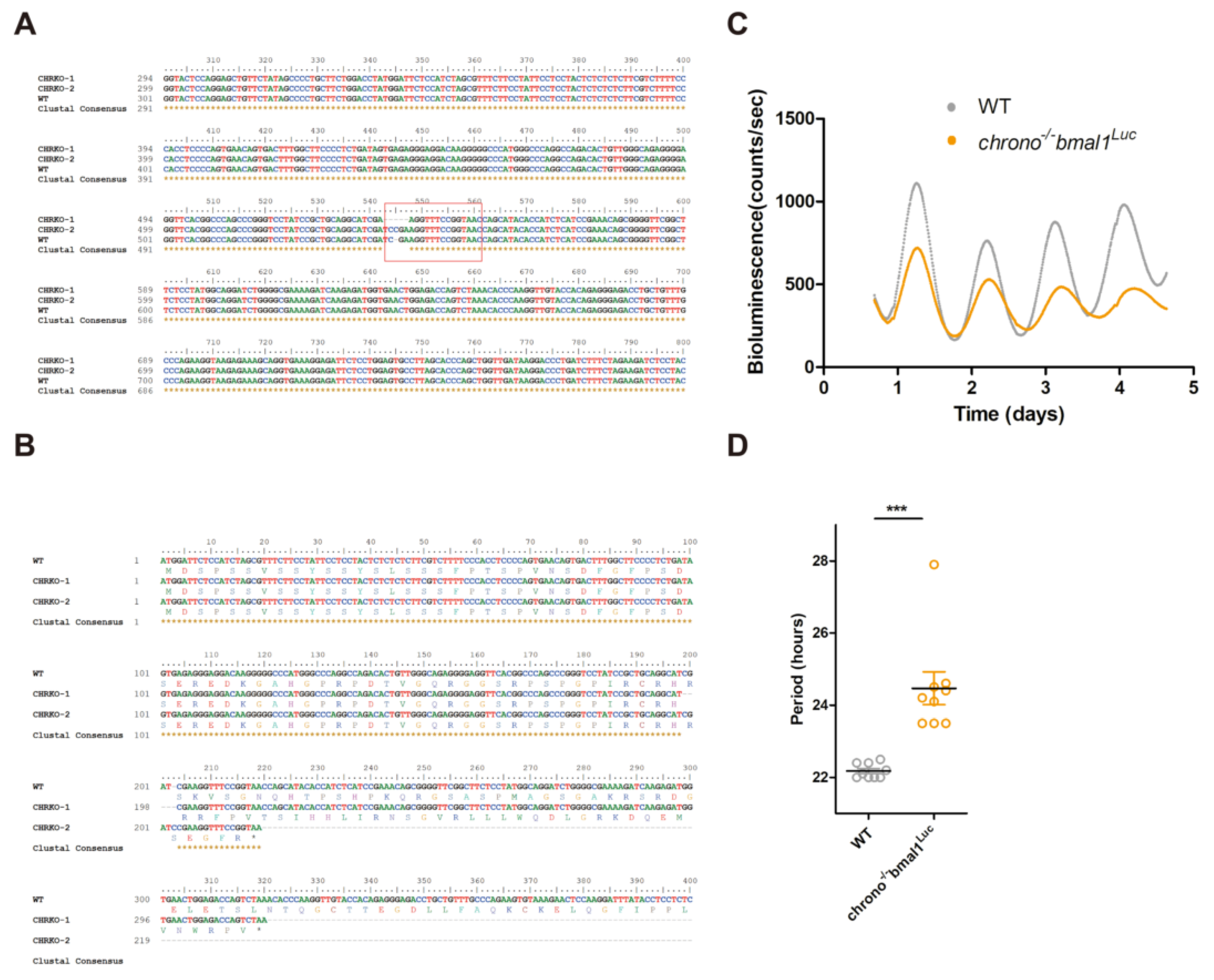
2.11. Chrono Knock Out in U2-OS Cells
2.12. Bioluminescence Recoding and Data Analysis
3. Results
3.1. The Helical Domain of CHR Represses the Transcriptional Activity of B/C Complexes
3.2. Secondary Structure of MBP-CHRH/CHRH3 (CHRH or CHRH3 fused with MBP) Measured by CD Spectroscopy
3.3. The Helix-Rich Domain is Necessary for CHR/BMAL1 Interaction
3.4. The C-Terminus of BMAL1 is Responsible for CHR Binding
3.5. CRISPR Knocking-Out of CHR Disturbs the Circadian Timekeeping
3.6. cpVenus Fused to the N-Terminus of CHR Interferes with Its Nuclear Localization
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TTFL | transcriptional and translational feedback loop |
| BMAL1 | brain and Muscle Arnt-like protein-1 |
| CLOCK | circadian locomotor output cycles kaput |
| CBP | CREB-binding protein |
| ccg | clock controlled genes |
| ciart | circadian associated repressor of transcription |
| CD | circular dichroism |
| ChIP | chromatin Immunoprecipitation |
| co-IP | co-Immunoprecipitation |
| HDAC | histone deacetylase |
| IF | immunofluorescence |
| CRISPR | clustered Regularly Interspaced Short Palindromic Repeat |
References
- Huang, N.; Chelliah, Y.; Shan, Y.; Taylor, C.A.; Yoo, S.H.; Partch, C.; Green, C.B.; Zhang, H.; Takahashi, J.S. Crystal structure of the heterodimericCLOCK:BMAL1 transcriptional activator complex. Science 2012, 337, 189–194. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Sehgal, A. Speed control: Cogs and gears that drive the circadian clock. Trends Neurosci. 2012, 35, 574–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, J.S. Molecular components of the circadian clock in mammals. Diabetes Obes. Metab. 2015, 17, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Kume, K.; Zylka, M.J.; Sriram, S.; Shearman, L.P.; Weaver, D.R.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999, 98, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Hardin, P.E.; Panda, S. Circadian timekeeping and output mechanisms in animals. Curr. Opin. Neurobiol. 2013, 23, 724–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef] [Green Version]
- Eide, E.J.; Woolf, M.F.; Kang, H.; Woolf, P.; Hurst, W.; Camacho, F.; Vielhaber, E.L.; Giovanni, A.; Virshup, D.M. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 2005, 25, 2795–2807. [Google Scholar] [CrossRef] [Green Version]
- Maywood, E.S.; Chesham, J.E.; Smyllie, N.J.; Hastings, M.H. The Tau mutation of casein kinase 1ε sets the period of the mammalian pacemaker via regulation of Period1 or Period2 clock proteins. J. Biol. Rhythms 2014, 29, 110–118. [Google Scholar] [CrossRef] [Green Version]
- Hirano, A.; Yumimoto, K.; Tsunematsu, R.; Matsumoto, M.; Oyama, M.; Kozuka-Hata, H.; Nakagawa, T.; Lanjakornsiripan, D.; Nakayama, K.I.; Fukada, Y. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 2013, 152, 1106–1118. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.H.; Mohawk, J.A.; Siepka, S.M.; Shan, Y.; Huh, S.K.; Hong, H.K.; Kornblum, I.; Kumar, V.; Koike, N.; Xu, M.; et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 2013, 152, 1091–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Q.J.; Maywood, E.S.; Bechtold, D.A.; Lu, W.Q.; Li, J.; Gibbs, J.E.; Dupré, S.M.; Chesham, J.E.; Rajamohan, F.; Knafels, J.; et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 15240–15245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goriki, A.; Hatanaka, F.; Myung, J.; Kim, J.K.; Yoritaka, T.; Tanoue, S.; Abe, T.; Kiyonari, H.; Fujimoto, K.; Kato, Y.; et al. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 2014, 12, e1001839. [Google Scholar] [CrossRef] [PubMed]
- Anafi, R.C.; Lee, Y.; Sato, T.K.; Venkataraman, A.; Ramanathan, C.; Kavakli, I.H.; Hughes, M.E.; Baggs, J.E.; Growe, J.; Liu, A.C.; et al. Machine learning helps identify CHRONO as a circadian clock component. PLoS Biol. 2014, 12, e1001840. [Google Scholar] [CrossRef]
- Annayev, Y.; Adar, S.; Chiou, Y.Y.; Lieb, J.D.; Sancar, A.; Ye, R. Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression. J. Biol. Chem. 2014, 289, 5013–5024. [Google Scholar] [CrossRef] [Green Version]
- Ye, R.; Selby, C.P.; Chiou, Y.Y.; Ozkan-Dagliyan, I.; Gaddameedhi, S.; Sancar, A. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 2014, 28, 1989–1998. [Google Scholar] [CrossRef] [Green Version]
- Chiou, Y.Y.; Yang, Y.; Rashid, N.; Ye, R.; Selby, C.P.; Sancar, A. Mammalian Period represses and de-represses transcription by displacing CLOCK-BMAL1 from promoters in a Cryptochrome-dependent manner. Proc. Natl. Acad. Sci. USA 2016, 113, E6072–E6079. [Google Scholar] [CrossRef] [Green Version]
- Koike, K.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef] [Green Version]
- Duong, H.A.; Weitz, C.J. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat. Struct. Mol. Biol. 2014, 21, 126–132. [Google Scholar] [CrossRef]
- Langmesser, S.; Tallone, T.; Bordon, A.; Rusconi, S.; Albrecht, U. Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol. Biol. 2008, 9, 41. [Google Scholar] [CrossRef] [Green Version]
- Ye, R.; Selby, C.P.; Ozturk, N.; Annayev, Y.; Sancar, A. Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 2011, 286, 25891–25902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.K.; Xu, H.; Ukai-Tadenuma, M.; Burton, B.; Wang, Y.; Ueda, H.R.; Liu, A.C. Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J. Biol. Chem. 2012, 287, 25917–25926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Gustafson, C.L.; Sammons, P.J.; Khan, S.K.; Parsley, N.C.; Ramanathan, C.; Lee, H.W.; Liu, A.C.; Partch, C.L. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat. Struct. Mol. Biol. 2015, 22, 476–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, A.K.; Fribourgh, J.L.; Chelliah, Y.; Sandate, C.R.; Hura, G.L.; Schneidman-Duhovny, D.; Tripathi, S.M.; Takahashi, J.S.; Partch, C.L. Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc. Natl. Acad. Sci. USA 2017, 114, 1560–1565. [Google Scholar] [CrossRef] [Green Version]
- Kiyohara, Y.B.; Tagao, S.; Tamanini, F.; Morita, A.; Sugisawa, Y.; Yasuda, M.; Yamanaka, I.; Ueda, H.R.; van der Horst, G.T.; Kondo, T.; et al. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci. USA 2006, 103, 10074–10079. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, S.; Takahashi, J.S. Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 2005, 393, 288–301. [Google Scholar]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Hönigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. PredictProtein-an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, W337–W343. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.Y.; Bullock, C.M.; Li, C.; Lee, A.G.; Bermak, J.C.; Belluzzi, J.; Weaver, D.R.; Leslie, F.M.; Zhou, Q.Y. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 2002, 417, 405–410. [Google Scholar] [CrossRef]
- Andrade, M.A.; Chacón, P.; Merelo, J.J.; Morán, F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993, 6, 383–390. [Google Scholar] [CrossRef]
- Czarna, A.; Breitkreuz, H.; Mahrenholz, C.C.; Arens, J.; Strauss, H.M.; Wolf, E. Quantitative analyses of cryptochrome-mBMAL1 interactions: Mechanistic insights into the transcriptional regulation of the mammalian circadian clock. J. Biol. Chem. 2011, 286, 22414–22425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahata, S.; Ozaki, T.; Mimura, J.; Kikuchi, Y.; Sogawa, K.; Fujii-Kuriyama, Y. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes Cells 2000, 5, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Duong, H.A.; Robles, M.S.; Knutti, D.; Weitz, C.J. A molecular mechanism for circadian clock negative feedback. Science 2011, 332, 1436–1439. [Google Scholar] [CrossRef] [Green Version]
- van der Horst, G.T.; Muijtjens, M.; Kobayashi, K.; Takano, R.; Kanno, S.; Takao, M.; de Wit, J.; Verkerk, A.; Eker, A.P.; van Leenen, D.; et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 1999, 398, 627–630. [Google Scholar] [CrossRef]
- Vitaterna, M.H.; Selby, C.P.; Todo, T.; Niwa, H.; Thompson, C.; Fruechte, E.M.; Hitomi, K.; Thresher, R.J.; Ishikawa, T.; Miyazaki, J.; et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA 1999, 96, 12114–12119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, D.; Honma, S.; Honma, K. Differential roles of AVP and VIP signaling in the postnatal changes of neural networks for coherent circadian rhythms in the SCN. Sci. Adv. 2016, 2, e1600960. [Google Scholar] [CrossRef] [Green Version]
- Ono, D.; Honma, S.; Honma, K. Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat. Commun. 2013, 4, 1666. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Li, N.; Qiu, J.; Ge, H.; Qin, X. Identification of the Repressive Domain of the Negative Circadian Clock Component CHRONO. Int. J. Mol. Sci. 2020, 21, 2469. https://doi.org/10.3390/ijms21072469
Yang Y, Li N, Qiu J, Ge H, Qin X. Identification of the Repressive Domain of the Negative Circadian Clock Component CHRONO. International Journal of Molecular Sciences. 2020; 21(7):2469. https://doi.org/10.3390/ijms21072469
Chicago/Turabian StyleYang, Yu, Ning Li, Jiameng Qiu, Honghua Ge, and Ximing Qin. 2020. "Identification of the Repressive Domain of the Negative Circadian Clock Component CHRONO" International Journal of Molecular Sciences 21, no. 7: 2469. https://doi.org/10.3390/ijms21072469
APA StyleYang, Y., Li, N., Qiu, J., Ge, H., & Qin, X. (2020). Identification of the Repressive Domain of the Negative Circadian Clock Component CHRONO. International Journal of Molecular Sciences, 21(7), 2469. https://doi.org/10.3390/ijms21072469




