Effective MSTN Gene Knockout by AdV-Delivered CRISPR/Cas9 in Postnatal Chick Leg Muscle
Abstract
:1. Introduction
2. Results
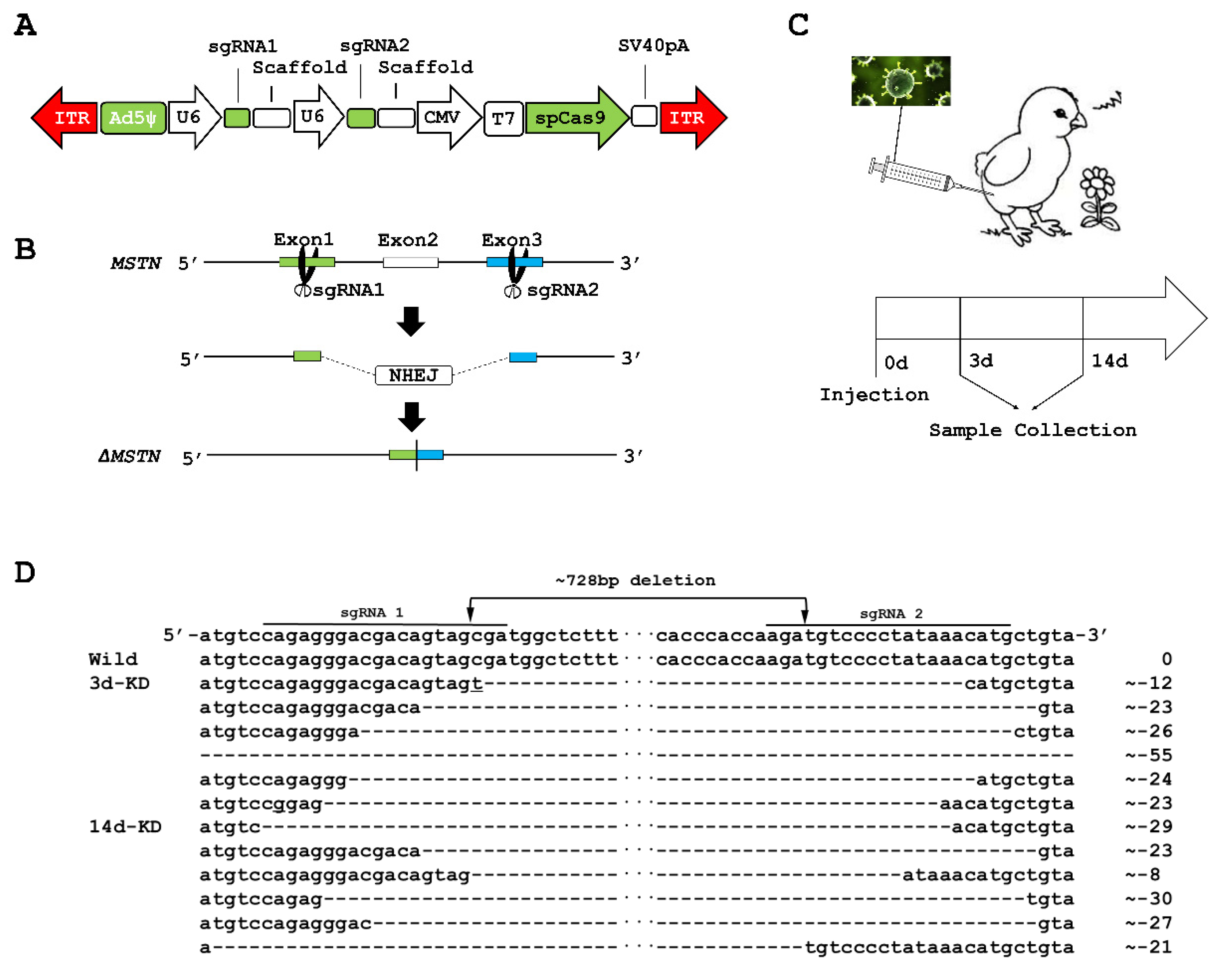
2.1. MSTN Knockout (KO) in the Muscles of Chicks
2.2. Differentially Expressed Genes (DEGs) in KO Muscles
2.3. Gene Ontology (GO) and Pathway Analyses of DEGs
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Feasibility Assessment of the AdV-CRISPR System in DF-1 Cells
4.3. Fertilized Eggs, Incubation, and Chick Breeding
4.4. Intramuscular Injection of the AdV-CRISPR System
4.5. Detection of Mutations in the MSTN Gene
4.6. Transcriptome Sequencing and Differentially Expressed Gene (DEG) Analysis
4.7. Gene Ontology (GO) and Pathway Analysis of Differentially Expressed Genes (DEGs)
4.8. Ethics Statement
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MSTN | Myostatin |
| AdV | adenovirus |
| KO | knockout |
| WT | wild-type |
| 3d | 3-day-old |
| 14d | 14-day-old |
| DEGs | differentially expressed genes |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| sgRNA | single guide RNA |
| NHEJ | non-homologous end joining |
| RT-PCR | reverse transcription polymerase chain reaction |
| PCA | Principal components analysis |
| IPA | Ingenuity Pathway Analysis |
| GO | Gene ontology |
| DAVID | Database for Annotation, Visualization and Integrated Discovery |
References
- Chen, F.; Wu, P.; Shen, M.; He, M.; Chen, L.; Qiu, C.; Shi, H.; Zhang, T.; Wang, J.; Xie, K.; et al. Transcriptome Analysis of Differentially Expressed Genes Related to the Growth and Development of the Jinghai Yellow Chicken. Genes 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksbjerg, N.; Therkildsen, M. Myogenesis and muscle growth and meat quality. In New Aspects of Meat Quality; Elsevier: Amsterdam, The Netherlands, 2017; pp. 33–62. [Google Scholar]
- Xue, Q.; Zhang, G.; Li, T.; Ling, J.; Zhang, X.; Wang, J. Transcriptomic profile of leg muscle during early growth in chicken. PLoS ONE 2017, 12, e0173824. [Google Scholar] [CrossRef]
- Davis, R.V.N.; Lamont, S.J.; Rothschild, M.F.; Persia, M.E.; Ashwell, C.M.; Schmidt, C.J. Transcriptome Analysis of Post-Hatch Breast Muscle in Legacy and Modern Broiler Chickens Reveals Enrichment of Several Regulators of Myogenic Growth. PLoS ONE 2015, 10, e0122525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef]
- Lee, S.J. Regulation of muscle mass by myostatin. Annu. Rev. Cell Dev. Biol. 2004, 20, 61–86. [Google Scholar] [CrossRef] [PubMed]
- Parsons, S.A.; Millay, D.P.; Sargent, M.A.; McNally, E.M.; Molkentin, J.D. Age-dependent effect of myostatin blockade on disease severity in a murine model of limb-girdle muscular dystrophy. Am. J. Pathol. 2006, 168, 1975–1985. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Pang, D.; Wang, K.; Xu, A.; Yao, C.; Li, M.; You, W.; Wang, Q.; Yu, H. The Possible Role of Complete Loss of Myostatin in Limiting Excessive Proliferation of Muscle Cells (C2C12) via Activation of MicroRNAs. Int. J. Mol. Sci. 2019, 20, 643. [Google Scholar] [CrossRef] [Green Version]
- Kambadur, R.; Sharma, M.; Smith, T.P.; Bass, J.J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res 1997, 7, 910–916. [Google Scholar] [CrossRef] [Green Version]
- Cyranoski, D. Super-muscly pigs created by small genetic tweak. Nature 2015, 523, 13–14. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.S.; Bobbili, N.K.; Paek, K.S.; Jin, H.J. Production of a monoclonal anti-myostatin antibody and the effects of in ovo administration of the antibody on posthatch broiler growth and muscle mass. Poult Sci. 2006, 85, 1062–1071. [Google Scholar] [CrossRef]
- Lee, S.J. Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS ONE 2007, 2, e789. [Google Scholar] [CrossRef]
- Mosher, D.S.; Quignon, P.; Bustamante, C.D.; Sutter, N.B.; Mellersh, C.S.; Parker, H.G.; Ostrander, E.A. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet 2007, 3, e79. [Google Scholar] [CrossRef]
- Schuelke, M.; Wagner, K.R.; Stolz, L.E.; Hubner, C.; Riebel, T.; Komen, W.; Braun, T.; Tobin, J.F.; Lee, S.J. Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 2004, 350, 2682–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yu, H.; Lei, A.; Zhou, J.; Zeng, W.; Zhu, H.; Dong, Z.; Niu, Y.; Shi, B.; Cai, B.; et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci. Rep. 2015, 5, 13878. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Cai, B.; Zhou, S.; Zhu, H.; Qu, L.; Wang, X.; Chen, Y. RNA-seq reveals transcriptome changes in goats following myostatin gene knockout. PLoS ONE 2017, 12, e0187966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.X.; Zhao, X.H.; Wang, J.Y.; Ding, F.X.; Zhang, L. Effect of an exon 1 mutation in the myostatin gene on the growth traits of the Bian chicken. Anim. Genet. 2012, 43, 458–459. [Google Scholar] [CrossRef] [PubMed]
- Paswan, C.; Bhattacharya, T.K.; Nagaraj, C.S.; Chaterjee, R.N.; Jayashankar, M.R. SNPs in minimal promoter of myostatin (GDF-8) gene and its association with body weight in broiler chicken. J. Appl. Anim. Res. 2014, 42, 304–309. [Google Scholar] [CrossRef] [Green Version]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kim, S.W.; Park, T.S. Myostatin gene knockout mediated by Cas9-D10A nickase in chicken DF1 cells without off-target effect. Asian Australas. J. Anim. Sci. 2017, 30, 743–748. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Q.S.; Wang, Y.J.; Cheng, S.Z.; Lian, C.; Tang, B.B.; Wang, F.; Lu, Z.Y.; Ji, Y.Q.; Zhao, R.F.; Zhang, W.H.; et al. Site-Directed Genome Knockout in Chicken Cell Line and Embryos Can Use CRISPR/Cas Gene Editing Technology. G3 Genes Genom. Genet. 2016, 6, 1787–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, Y.; Zuo, Q.; Li, D.; Zhang, W.; Wang, F.; Ji, Y.; Jin, J.; Lu, Z.; Wang, M.; et al. CRISPR/Cas9 mediated chicken Stra8 gene knockout and inhibition of male germ cell differentiation. PLoS ONE 2017, 12, e0172207. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.H.; Ferguson, I.; Li, M.; Kim, A.; Onishi, A.; Li, L.; Su, B.; Gong, X. Essential function of NHE8 in mouse retina demonstrated by AAV-mediated CRISPR/Cas9 knockdown. Exp. Eye Res. 2018, 176, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [Green Version]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.R.; Doumit, M.E.; Merkel, R.A. Transforming growth factor-beta, basic fibroblast growth factor, and platelet-derived growth factor-BB interact to affect proliferation of clonally derived porcine satellite cells. J. Cell. Physiol. 1993, 157, 307–312. [Google Scholar] [CrossRef]
- Doumit, M.E.; Cook, D.R.; Merkel, R.A. Fibroblast growth factor, epidermal growth factor, insulin-like growth factors, and platelet-derived growth factor-BB stimulate proliferation of clonally derived porcine myogenic satellite cells. J. Cell. Physiol. 1993, 157, 326–332. [Google Scholar] [CrossRef]
- Zhang, C.C.; Li, Y.L.; Wu, Y.N.; Wang, L.Y.; Wang, X.N.; Du, J. Interleukin-6/Signal Transducer and Activator of Transcription 3 (STAT3) Pathway Is Essential for Macrophage Infiltration and Myoblast Proliferation during Muscle Regeneration. J. Biol. Chem. 2013, 288, 1489–1499. [Google Scholar] [CrossRef] [Green Version]
- Doran, T.J.; Cooper, C.A.; Jenkins, K.A.; Tizard, M.L. Advances in genetic engineering of the avian genome: “Realising the promise”. Transgenic Res. 2016, 25, 307–319. [Google Scholar] [CrossRef]
- Wang, D.; Mou, H.; Li, S.; Li, Y.; Hough, S.; Tran, K.; Li, J.; Yin, H.; Anderson, D.G.; Sontheimer, E.J.; et al. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum. Gene Ther. 2015, 26, 432–442. [Google Scholar] [CrossRef] [Green Version]
- Bi, Y.Z.; Hua, Z.D.; Liu, X.M.; Hua, W.J.; Ren, H.Y.; Xiao, H.W.; Zhang, L.P.; Li, L.; Wang, Z.R.; Laible, G.; et al. Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- DENG, S.; Kongpan, L.; WANG, F.; Ning, L.; LIU, G.; ZHAO, Y.; LIAN, Z. One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front. Agr. Sci. Eng. 2014, 1, 2–5. [Google Scholar]
- Lv, Q.; Yuan, L.; Deng, J.; Chen, M.; Wang, Y.; Zeng, J.; Li, Z.; Lai, L. Efficient Generation of Myostatin Gene Mutated Rabbit by CRISPR/Cas9. Sci. Rep. 2016, 6, 25029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayuso, M.; Fernandez, A.; Nunez, Y.; Benitez, R.; Isabel, B.; Barragan, C.; Fernandez, A.I.; Rey, A.I.; Medrano, J.F.; Canovas, A.; et al. Comparative Analysis of Muscle Transcriptome between Pig Genotypes Identifies Genes and Regulatory Mechanisms Associated to Growth, Fatness and Metabolism. PLoS ONE 2015, 10, e0145162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.F.; Dai, G.J.; Chen, F.X.; Chen, L.; Zhang, T.; Xie, K.Z.; Wang, J.Y.; Zhan, G.X. Transcriptome profile analysis of leg muscle tissues between slow- and fast-growing chickens. PLoS ONE 2018, 13, e0206131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welle, S.; Cardillo, A.; Zanche, M.; Tawil, R. Skeletal muscle gene expression after myostatin knockout in mature mice. Physiol. Genom. 2009, 38, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Cassar-Malek, I.; Passelaigue, F.; Bernard, C.; Leger, J.; Hocquette, J.F. Target genes of myostatin loss-of-function in muscles of late bovine fetuses. BMC Genom. 2007, 8, 63. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.J.; Jan, A.T.; Baig, M.H.; Ashraf, J.M.; Nahm, S.S.; Kim, Y.W.; Park, S.Y.; Choi, I. Fibromodulin: A master regulator of myostatin controlling progression of satellite cells through a myogenic program. FASEB J. 2016, 30, 2708–2719. [Google Scholar] [CrossRef]
- Verbrugge, S.A.J.; Schonfelder, M.; Becker, L.; Yaghoob Nezhad, F.; Hrabe de Angelis, M.; Wackerhage, H. Genes Whose Gain or Loss-Of-Function Increases Skeletal Muscle Mass in Mice: A Systematic Literature Review. Front. Physiol. 2018, 9, 553. [Google Scholar] [CrossRef] [Green Version]
- Biga, P.R.; Froehlich, J.M.; Greenlee, K.J.; Galt, N.J.; Meyer, B.M.; Christensen, D.J. Gelatinases impart susceptibility to high-fat diet-induced obesity in mice. J. Nutr. Biochem. 2013, 24, 1462–1468. [Google Scholar] [CrossRef] [Green Version]
- Blitz, E.; Viukov, S.; Sharir, A.; Shwartz, Y.; Galloway, J.L.; Pryce, B.A.; Johnson, R.L.; Tabin, C.J.; Schweitzer, R.; Zelzer, E. Bone Ridge Patterning during Musculoskeletal Assembly Is Mediated through SCX Regulation of Bmp4 at the Tendon-Skeleton Junction. Dev. Cell 2009, 17, 861–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutet, S.C.; Biressi, S.; Iori, K.; Natu, V.; Rando, T.A. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol. Cell 2010, 40, 749–761. [Google Scholar] [CrossRef] [Green Version]
- Srikanchai, T.; Murani, E.; Wimmers, K.; Ponsuksili, S. Four loci differentially expressed in muscle tissue depending on water-holding capacity are associated with meat quality in commercial pig herds. Mol. Biol. Rep. 2010, 37, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Descamps, S.; Arzouk, H.; Bacou, F.; Bernardi, H.; Fedon, Y.; Gay, S.; Reyne, Y.; Rossano, B.; Levin, J. Inhibition of myoblast differentiation by Sfrp1 and Sfrp2. Cell Tissue Res. 2008, 332, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.X.; Banks, A.; Liu, T.; Kazak, L.; Rao, R.R.; Cohen, P.; Wang, X.; Yu, S.; Lo, J.C.; Tseng, Y.H.; et al. IRF4 Is a Key Thermogenic Transcriptional Partner of PGC-1 alpha. Cell 2014, 158, 69–83. [Google Scholar] [CrossRef] [Green Version]
- Bruschetta, D.G.; Esposito, S.D.E. The Role of Central PRCP in the Metabolism Regulation. Ph.D. Thesis, UNIVERSITÀ DEGLI STUDI DI MESSINA, Messina, Italy, 2016. [Google Scholar]
- Wang, L.M.; Ma, S.; Ding, Q.; Wang, X.L.; Chen, Y.L. CRISPR/Cas9-mediated MSTN gene editing induced mitochondrial alterations in C2C12 myoblast cells. Electron. J. Biotechnol. 2019, 40, 30–39. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, Y.; Ma, G.; Zhao, X.; Chen, Y.; Zhu, D. Identification of gene expression modifications in myostatin-stimulated myoblasts. Biochem. Biophys. Res. Commun. 2005, 326, 660–666. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, K.; Han, C.X.; Zhou, H.; Ding, J.M.; Xu, Z.; Yang, L.Y.; He, C.; Akinyemi, F.; Zheng, Y.M.; Qin, C.; et al. Effective MSTN Gene Knockout by AdV-Delivered CRISPR/Cas9 in Postnatal Chick Leg Muscle. Int. J. Mol. Sci. 2020, 21, 2584. https://doi.org/10.3390/ijms21072584
Xu K, Han CX, Zhou H, Ding JM, Xu Z, Yang LY, He C, Akinyemi F, Zheng YM, Qin C, et al. Effective MSTN Gene Knockout by AdV-Delivered CRISPR/Cas9 in Postnatal Chick Leg Muscle. International Journal of Molecular Sciences. 2020; 21(7):2584. https://doi.org/10.3390/ijms21072584
Chicago/Turabian StyleXu, Ke, Cheng Xiao Han, Hao Zhou, Jin Mei Ding, Zhong Xu, Ling Yu Yang, Chuan He, Fisayo Akinyemi, Yu Ming Zheng, Chao Qin, and et al. 2020. "Effective MSTN Gene Knockout by AdV-Delivered CRISPR/Cas9 in Postnatal Chick Leg Muscle" International Journal of Molecular Sciences 21, no. 7: 2584. https://doi.org/10.3390/ijms21072584




