Nuclear Functions of the Tyrosine Kinase Src
Abstract
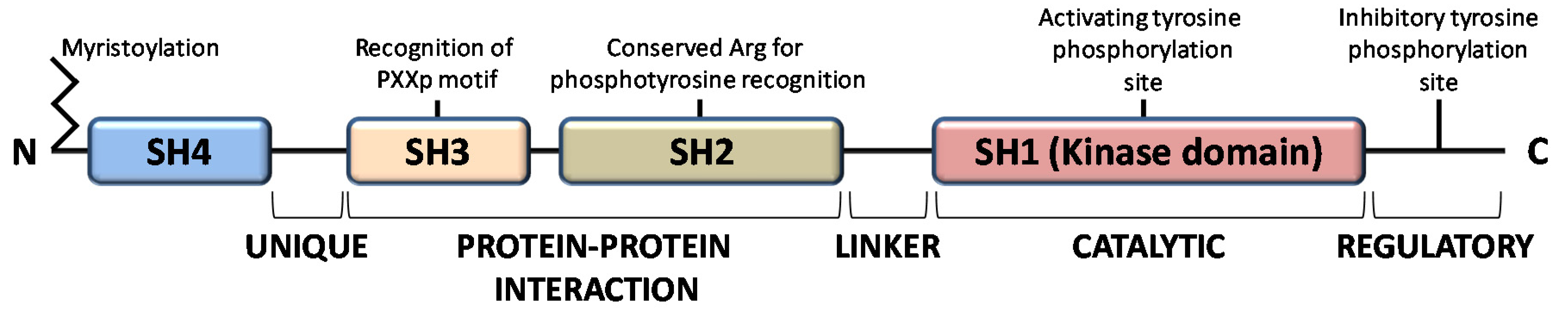
:1. Introduction
2. Nuclear Functions of SFKs other than Src
3. Src Translocation into the Nucleus
4. Physiopathological Roles of Nuclear Src
4.1. Regulation of Gene Transcription and Chromatin Architecture
4.2. Src-Dependent Regulation of Tumor Suppressors
4.3. Src and Estrogen Receptor
4.4. Interaction with the Nuclear Envelope Protein Emerin
4.5. Src and the Mechanotransduction
5. Prognostic Roles of Nuclear Src
6. Conclusions
Funding
Conflicts of Interest
Abbreviations
| Blk | B Lymphoid tyrosine Kinase |
| Brk | BReast tumor Kinase |
| CAFs | Cancer Related Fibroblasts |
| Csk | C-terminal Src Kinase |
| ERα | Estrogen Receptor alpha |
| FAK | Focal Adhesion Kinase |
| Fgr | Gardner-Rasheed Feline Sarcoma Viral Oncogene Homolog |
| Frk | Fyn Related Src family tyrosine Kinase |
| Hck | Hematopoietic Cell Kinase |
| Lck | Lymphocyte-specific protein tyrosine kinase |
| Lyn | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| NADPH | Nicotinamide Adenine Dinucleotide PHosphate |
| NLS | Nuclear Localization Signal/Sequence |
| NMTs | N-myristoyltransferase |
| PRG | Progesterone |
| PTP | Protein Tyrosine Phosphatase |
| RTKs | Receptor Tyrosine Kinases |
| ROS | Reactive Oxygen Species |
| SAPK | Stress-Activated Protein Kinase |
| SFKs | Src Family Kinases |
| Shc | Src Homology 2 domain Containing |
| SHP2 | Src Homology 2 containing protein tyrosine Phosphatase 2 |
| Srm | Src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites |
| TAZ | Tafazzin |
| YAP | Yes-Associated Protein |
| Yes | Yamaguchi sarcoma viral oncogene homolog |
References
- Espada, J.; Martin-Perez, J. An Update on Src Family of Nonreceptor Tyrosine Kinases Biology. Int. Rev. Cell Mol. Biol. 2017, 331, 83–122. [Google Scholar]
- Parsons, S.J.; Parsons, J.T. Src family kinases, key regulators of signal transduction. Oncogene 2004, 23, 7906–7909. [Google Scholar] [CrossRef] [Green Version]
- Shah, N.H.; Amacher, J.F.; Nocka, L.M.; Kuriyan, J. The Src module: An ancient scaffold in the evolution of cytoplasmic tyrosine kinases. Crit. Rev. Biochem. Mol. Boil. 2018, 53, 535–563. [Google Scholar] [CrossRef] [Green Version]
- Owen, D.M.; Rentero, C.; Rossy, J.; Magenau, A.; Williamson, D.J.; Rodríguez, M.; Gaus, K. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J. Biophotonics 2010, 3, 446–454. [Google Scholar] [CrossRef]
- Smith, A.W.; Huang, H.H.; Endres, N.F.; Rhodes, C.; Groves, J.T. Dynamic Organization of Myristoylated Src in the Live Cell Plasma Membrane. J. Phys. Chem. B 2016, 120, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Spassov, D.S.; Ruiz-Saenz, A.; Piple, A.; Moasser, M.M. A Dimerization Function in the Intrinsically Disordered N-Terminal Region of Src. Cell Rep. 2018, 25, 449–463. [Google Scholar] [CrossRef] [Green Version]
- Le Roux, A.-L.; Busquets, M.A.; Sagués, F.; Pons, M. Kinetics characterization of c-Src binding to lipid membranes: Switching from labile to persistent binding. Colloids Surf. B Biointerfaces 2016, 138, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Le Roux, A.-L.; Castro, B.; Garbacik, E.T.; Parajo, M.F.G.; Pons, M. Single molecule fluorescence reveals dimerization of myristoylated Src N-terminal region on supported lipid bilayers. ChemistrySelect 2016, 1, 642–647. [Google Scholar] [CrossRef] [Green Version]
- Teyra, J.; Huang, H.; Jain, S.; Guan, X.; Dong, A.; Liu, Y.; Tempel, W.; Min, J.; Tong, Y.; Kim, P.M.; et al. Comprehensive Analysis of the Human SH3 Domain Family Reveals a Wide Variety of Non-canonical Specificities. Structure 2017, 25, 1598–1610.e3. [Google Scholar] [CrossRef] [Green Version]
- Boggon, T.J.; Eck, M.J. Structure and regulation of Src family kinases. Oncogene 2004, 23, 7918–7927. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Doshi, A.; Lei, M.; Eck, M.J.; Harrison, S.C. Crystal Structures of c-Src Reveal Features of Its Autoinhibitory Mechanism. Mol. Cell 1999, 3, 629–638. [Google Scholar] [CrossRef]
- Sato, K.-I.; Nagao, T.; Kakumoto, M.; Kimoto, M.; Otsuki, T.; Iwasaki, T.; Tokmakov, A.A.; Owada, K.; Fukami, Y. Adaptor Protein Shc Is an Isoform-specific Direct Activator of the Tyrosine Kinase c-Src. J. Boil. Chem. 2002, 277, 29568–29576. [Google Scholar] [CrossRef] [Green Version]
- Boczek, E.E.; Luo, Q.; Dehling, M.; Röpke, M.; Mader, S.L.; Seidl, A.; Kaila, V.R.I.; Buchner, J. Autophosphorylation activates c-Src kinase through global structural rearrangements. J. Boil. Chem. 2019, 294, 13186–13197. [Google Scholar] [CrossRef]
- Meng, Y.; Roux, B. Locking the active conformation of c-Src kinase through the phosphorylation of the activation loop. J. Mol. Biol. 2014, 426, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Pond, M.P.; Roux, B. Tyrosine Kinase Activation and Conformational Flexibility: Lessons from Src-Family Tyrosine Kinases. Acc. Chem. Res. 2017, 50, 1193–1201. [Google Scholar] [CrossRef]
- Roskoski, R. Src kinase regulation by phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun. 2005, 331, 1–14. [Google Scholar] [CrossRef]
- Fan, G.; Aleem, S.; Yang, M.; Miller, W.T.; Tonks, N.K. Protein-tyrosine Phosphatase and Kinase Specificity in Regulation of SRC and Breast Tumor Kinase* ♦. J. Boil. Chem. 2015, 290, 15934–15947. [Google Scholar] [CrossRef] [Green Version]
- Bjorge, J.D.; Pang, A.; Fujita, D.J. Identification of Protein-tyrosine Phosphatase 1B as the Major Tyrosine Phosphatase Activity Capable of Dephosphorylating and Activating c-Src in Several Human Breast Cancer Cell Lines. J. Boil. Chem. 2000, 275, 41439–41446. [Google Scholar] [CrossRef] [Green Version]
- Okada, M. Regulation of the Src Family Kinases by Csk. Int. J. Boil. Sci. 2012, 8, 1385–1397. [Google Scholar] [CrossRef] [Green Version]
- Soriano, P.; Montgomery, C.; Geske, R.; Bradley, A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 1991, 64, 693–702. [Google Scholar] [CrossRef]
- Miyazaki, T.; Sanjay, A.; Neff, L.; Tanaka, S.; Horne, W.C.; Baron, R. Src Kinase Activity Is Essential for Osteoclast Function. J. Boil. Chem. 2004, 279, 17660–17666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peruzzi, B.; Teti, A.M. The Physiology and Pathophysiology of the Osteoclast. Clin. Rev. Bone Miner. Metab. 2011, 10, 71–97. [Google Scholar] [CrossRef]
- Marzia, M.; Sims, N.A.; Voit, S.; Migliaccio, S.; Taranta, A.; Bernardini, S.; Faraggiana, T.; Yoneda, T.; Mundy, G.R.; Boyce, B.F.; et al. Decreased C-Src Expression Enhances Osteoblast Differentiation and Bone Formation. J. Cell Boil. 2000, 151, 311–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djeungoue-Petga, M.-A.; Lurette, O.; Jean, S.; Hamel-Côté, G.; Martín-Jiménez, R.; Bou, M.; Cannich, A.; Roy, P.; Hebert-Chatelain, E. Intramitochondrial Src kinase links mitochondrial dysfunctions and aggressiveness of breast cancer cells. Cell Death Dis. 2019, 10, 9401–9415. [Google Scholar] [CrossRef]
- Hikita, T.; Kuwahara, A.; Watanabe, R.; Miyata, M.; Oneyama, C. Src in endosomal membranes promotes exosome secretion and tumor progression. Sci. Rep. 2019, 9, 3265. [Google Scholar] [CrossRef] [Green Version]
- Kostenko, S.; Heu, C.C.; Yaron, J.R.; Singh, G.; De Oliveira, C.; Muller, W.J.; Singh, V.P. c-Src regulates cargo transit via the Golgi in pancreatic acinar cells. Sci. Rep. 2018, 8, 11903. [Google Scholar] [CrossRef]
- Miyazaki, T.; Neff, L.; Tanaka, S.; Horne, W.C.; Baron, R. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J. Cell Boil. 2003, 160, 709–718. [Google Scholar] [CrossRef] [Green Version]
- Reinecke, J.; Caplan, S. Endocytosis and the Src family of non-receptor tyrosine kinases. Biomol. Concepts 2014, 5, 143–155. [Google Scholar] [CrossRef] [Green Version]
- Dubois, F.; Leroy, C.; Simon, V.; Benistant, C.; Roche, S. YES oncogenic activity is specified by its SH4 domain and regulates RAS/MAPK signaling in colon carcinoma cells. Am. J. Cancer Res. 2015, 5, 1972–1987. [Google Scholar]
- Saito, Y.D.; Jensen, A.R.; Salgia, R.; Posadas, E.M. Fyn: A novel molecular target in cancer. Cancer 2010, 116, 1629–1637. [Google Scholar] [CrossRef]
- Matsushima, S.; Kuroda, J.; Zhai, P.; Liu, T.; Ikeda, S.; Nagarajan, N.; Oka, S.-I.; Yokota, T.; Kinugawa, S.; Hsu, C.-P.; et al. Tyrosine kinase FYN negatively regulates NOX4 in cardiac remodeling. J. Clin. Investig. 2016, 126, 3403–3416. [Google Scholar] [CrossRef]
- Dwyer, A.; Mouchemore, K.; Steer, J.H.; Sunderland, A.J.; Sampaio, N.; Greenland, E.L.; A Joyce, D.; Pixley, F.J. Src family kinase expression and subcellular localization in macrophages: Implications for their role in CSF-1-induced macrophage migration. J. Leukoc. Boil. 2016, 100, 163–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephen, L.; Elmaghloob, Y.; McIlwraith, M.J.; Yelland, T.; Sanchez, P.C.; Roda-Navarro, P.; Ismail, S. The Ciliary Machinery Is Repurposed for T Cell Immune Synapse Trafficking of LCK. Dev. Cell 2018, 47, 122–132.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poh, A.; O’Donoghue, R.J.; Ernst, M. Hematopoietic cell kinase (HCK) as a therapeutic target in immune and cancer cells. Oncotarget 2015, 6, 15752–15771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, D.L.; Berthelsen, J.; Willerslew-Olsen, A.; Fredholm, S.; Dabelsteen, S.; Bonefeld, C.M.; Geisler, C.; Woetmann, A. A novel BLK-induced tumor model. Tumor Boil. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serfas, M.S.; Tyner, A.L. Brk, Srm, Frk, and Src42A Form a Distinct Family of Intracellular Src-Like Tyrosine Kinases. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2003, 13, 409–419. [Google Scholar]
- Derry, J.J.; Richard, S.; Carvajal, H.V.; Ye, X.; Vasioukhin, V.; Cochrane, A.W.; Chen, T.; Tyner, A.L. Sik (BRK) Phosphorylates Sam68 in the Nucleus and Negatively Regulates Its RNA Binding Ability. Mol. Cell. Boil. 2000, 20, 6114–6126. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Weichselbaum, R.; Kharbanda, S.; Kufe, D. Role for Lyn Tyrosine Kinase as a Regulator of Stress-Activated Protein Kinase Activity in Response to DNA Damage. Mol. Cell. Boil. 2000, 20, 5370–5380. [Google Scholar] [CrossRef] [Green Version]
- Ogunbolude, Y.; Dai, C.; Bagu, E.T.; Goel, R.K.; Miah, S.; MacAusland-Berg, J.; Ng, C.Y.; Chibbar, R.; Napper, S.; Raptis, L.; et al. FRK inhibits breast cancer cell migration and invasion by suppressing epithelial-mesenchymal transition. Oncotarget 2017, 8, 113034–113065. [Google Scholar] [CrossRef]
- Kim, J.-L.; Ha, G.-H.; Campo, L.; Denning, M.F.; Patel, T.B.; Osipo, C.; Lin, S.-Y.; Breuer, E.-K. The role of Rak in the regulation of stability and function of BRCA1. Oncotarget 2015, 8, 86799–86815. [Google Scholar] [CrossRef] [Green Version]
- Maejima, Y.; Kuroda, J.; Matsushima, S.; Ago, T.; Sadoshima, J. Regulation of myocardial growth and death by NADPH oxidase. J. Mol. Cell. Cardiol. 2011, 50, 408–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, S.J.; Shalloway, D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature 1994, 368, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Dallari, S.; Macal, M.; Loureiro, M.E.; Jo, Y.; Swanson, L.; Hesser, C.; Ghosh, P.; Zuniga, E.I. Src family kinases Fyn and Lyn are constitutively activated and mediate plasmacytoid dendritic cell responses. Nat. Commun. 2017, 8, 14830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E. A short amino acid sequence able to specify nuclear location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef]
- David-Pfeuty, T.; Bagrodia, S.; Shalloway, D. Differential localization patterns of myristoylated and nonmyristoylated c-Src proteins in interphase and mitotic c-Src overexpresser cells. J. Cell Sci. 1993, 105, 105. [Google Scholar] [CrossRef] [Green Version]
- Urciuoli, E.; Coletta, I.; Rizzuto, E.; De Vito, R.; Petrini, S.; D’Oria, V.; Pezzullo, M.; Milano, G.; Cozza, R.; Locatelli, F.; et al. Src nuclear localization and its prognostic relevance in human osteosarcoma. J. Cell. Physiol. 2017, 233, 1658–1670. [Google Scholar] [CrossRef]
- Le Roux, A.-L.; Mohammad, I.-L.; Mateos, B.; Arbesú, M.; Gairí, M.; Khan, F.A.; Teixeira, J.M.C.; Pons, M. A Myristoyl-Binding Site in the SH3 Domain Modulates c-Src Membrane Anchoring. iScience 2019, 12, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, K.; Nakayama, Y.; Togashi, Y.; Obata, Y.; Kuga, T.; Kasahara, K.; Fukumoto, Y.; Yamaguchi, N. Nuclear localization of Lyn tyrosine kinase mediated by inhibition of its kinase activity. Exp. Cell Res. 2008, 314, 3392–3404. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, T.; Dietzel, S.; Müller, S.; Solovei, I.; Fakan, S. Chromosome territories—A functional nuclear landscape. Curr. Opin. Cell Boil. 2006, 18, 307–316. [Google Scholar] [CrossRef]
- Leonetti, E.; Gesualdi, L.; Scheri, K.C.; DiNicola, S.; Fattore, L.; Masiello, M.G.; Cucina, A.; Mancini, R.; Bizzarri, M.; Ricci, G.; et al. c-Src Recruitment is Involved in c-MET-Mediated Malignant Behaviour of NT2D1 Non-Seminoma Cells. Int. J. Mol. Sci. 2019, 20, 320. [Google Scholar] [CrossRef] [Green Version]
- Scheri, K.C.; Leonetti, E.; Laino, L.; Gigantino, V.; Gesualdi, L.; Grammatico, P.; Bizzari, M.; Franco, R.; Oosterhuis, J.W.; Stoop, H.; et al. c-MET receptor as potential biomarker and target molecule for malignant testicular germ cell tumors. Oncotarget 2018, 9, 31842–31860. [Google Scholar] [PubMed]
- Takahashi, A.; Obata, Y.; Fukumoto, Y.; Nakayama, Y.; Kasahara, K.; Kuga, T.; Higashiyama, Y.; Saito, T.; Yokoyama, K.K.; Yamaguchi, N. Nuclear localization of Src-family tyrosine kinases is required for growth factor-induced euchromatinization. Exp. Cell Res. 2009, 315, 1117–1141. [Google Scholar] [CrossRef] [PubMed]
- Giotopoulos, G.; Chan, W.I.; Horton, S.J.; Ruau, D.; Gallipoli, P.; Fowler, A.; Crawley, C.; Papaemmanuil, E.; Campbell, P.J.; Göttgens, B.; et al. The epigenetic regulators CBP and p300 facilitate leukemogenesis and represent therapeutic targets in acute myeloid leukemia. Oncogene 2016, 35, 279–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paladino, D.; Yue, P.; Furuya, H.; Acoba, J.; Rosser, C.J.; Turkson, J. A novel nuclear Src and p300 signaling axis controls migratory and invasive behavior in pancreatic cancer. Oncotarget 2016, 7, 7253–7267. [Google Scholar] [CrossRef] [Green Version]
- George, T.J.; Trevino, J.G.; Liu, C. Src Inhibition Is Still a Relevant Target in Pancreatic Cancer. Oncologist 2014, 19, 211. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Thakur, S.; Leong-Quong, R.Y.; Suzuki, K.; Pang, A.; Bjorge, J.D.; Riabowol, K.; Fujita, N.J. Src Regulates the Activity of the ING1 Tumor Suppressor. PLoS ONE 2013, 8, 60943. [Google Scholar] [CrossRef] [Green Version]
- Greer, C.B.; Tanaka, Y.; Kim, Y.J.; Xie, P.; Zhang, M.Q.; Park, I.-H.; Kim, T.H. Histone Deacetylases Positively Regulate Transcription through the Elongation Machinery. Cell Rep. 2015, 13, 1444–1455. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.J.; Greer, C.B.; Cecchini, K.R.; Harris, L.N.; Tuck, D.P.; Kim, T.H. HDAC inhibitors induce transcriptional repression of high copy number genes in breast cancer through elongation blockade. Oncogene 2013, 32, 2828–2835. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.A.; Piao, M.J.; Ryu, Y.S.; Maeng, Y.H.; Hyun, J.W. Cytoplasmic Localization of RUNX3 via Histone Deacetylase-Mediated SRC Expression in Oxidative-Stressed Colon Cancer Cells. J. Cell. Physiol. 2017, 232, 1914–1921. [Google Scholar] [CrossRef]
- Streicher, C.; Heyny, A.; Andrukhova, O.; Haigl, B.; Slavic, S.; Schüler, C.; Kollmann, K.; Kantner, I.; Sexl, V.; Kleiter, M.; et al. Estrogen Regulates Bone Turnover by Targeting RANKL Expression in Bone Lining Cells. Sci. Rep. 2017, 7, 6460. [Google Scholar] [CrossRef] [Green Version]
- Manolagas, S. Sex Steroids and Bone. Recent Prog. Horm. Res. 2002, 57, 385–409. [Google Scholar] [CrossRef] [PubMed]
- Kousteni, S.; Han, L.; Chen, J.R.; Almeida, M.; Plotkin, L.I.; Bellido, T.; Manolagas, S.C. Kinase-mediated regulation of common transcription factors accounts for the bone-protective effects of sex steroids. J. Clin. Investig. 2003, 111, 1651–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, H.; Kong, S.; Zhang, S.; Cheng, J.; Zhou, C.; He, B.; Xin, Q.; Lydon, J.P.; DeMayo, F.J.; Feng, G.-S.; et al. Nuclear Shp2 directs normal embryo implantation via facilitating the ERα tyrosine phosphorylation by the Src kinase. Proc. Natl. Acad. Sci. USA 2017, 114, 4816–4821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.; Yu, W.M.; Xu, M.; Qu, C.K. SHP-2 phosphatase regulates DNA damage-induced apoptosis and G2/M arrest in catalytically dependent and independent manners, respectively. J. Biol. Chem. 2005, 280, 42701–42706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castoria, G.; Giovannelli, P.; Lombardi, M.; De Rosa, C.; Giraldi, T.; De Falco, A.; Barone, M.V.; Abbondanza, C.; Migliaccio, A.; Auricchio, F. Tyrosine phosphorylation of estradiol receptor by Src regulates its hormone-dependent nuclear export and cell cycle progression in breast cancer cells. Oncogene 2012, 31, 4868–4877. [Google Scholar] [CrossRef] [Green Version]
- Arnold, S.F.; Vorojeikina, D.P.; Notides, A.C. Phosphorylation of tyrosine 537 on the human estrogen receptor is required for binding to an estrogen response element. J. Biol. Chem. 1995, 270, 30205–30212. [Google Scholar] [PubMed] [Green Version]
- Viggiano, E.; Pilarczyk, M.; Carboni, N.; Picillo, E.; Ergoli, M.; Del Gaudio, S.; Marchel, M.; Nigro, G.; Palladino, A.; Politano, L. X-Linked Emery–Dreifuss Muscular Dystrophy: Study Of X-Chromosome Inactivation and Its Relation with Clinical Phenotypes in Female Carriers. Genes 2019, 10, 919. [Google Scholar] [CrossRef] [Green Version]
- Tifft, K.E.; Bradbury, K.A.; Wilson, K.L. Tyrosine phosphorylation of nuclear-membrane protein emerin by Src, Abl and other kinases. J. Cell Sci. 2009, 122, 3780–3790. [Google Scholar] [CrossRef] [Green Version]
- Prasad, T.S.K.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2008, 37, D767–D772. [Google Scholar] [CrossRef] [Green Version]
- Berk, J.M.; Simon, D.N.; Jenkins-Houk, C.R.; Westerbeck, J.W.; Grønning-Wang, L.M.; Carlson, C.R.; Wilson, K.L. The molecular basis of emerin-emerin and emerin-BAF interactions. J. Cell Sci. 2014, 127, 3956–3969. [Google Scholar] [CrossRef] [Green Version]
- Guilluy, C.; Osborne, L.D.; Van Landeghem, L.; Sharek, L.; Superfine, R.; Garcia-Mata, R.; Burridge, K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nature 2014, 16, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, R.; Ranade, D.; Sengupta, K. Emerin modulates spatial organization of chromosome territories in cells on softer matrices. Nucleic Acids Res. 2018, 46, 5561–5586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, K.A.; Atherton, P.; Ballestrem, C. Mechanotransduction at the cell-matrix interface. Semin. Cell Dev. Boil. 2017, 71, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Watt, K.I.; Harvey, K.F.; Gregorevic, P. Regulation of Tissue Growth by the Mammalian Hippo Signaling Pathway. Front. Physiol. 2017, 8, 942. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.-X.; Zhao, B.; Guan, K.-L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yu, A.; Yu, F.-X. The Hippo pathway in tissue homeostasis and regeneration. Protein Cell 2017, 8, 349–359. [Google Scholar] [CrossRef] [Green Version]
- Muslin, A.J.; Xing, H. 14-3-3 proteins: Regulation of subcellular localization by molecular interference. Cell. Signal 2000, 12, 703–709. [Google Scholar] [CrossRef]
- Dobrokhotov, O.; Samsonov, M.; Sokabe, M.; Hirata, H. Mechanoregulation and pathology of YAP/TAZ via Hippo and non-Hippo mechanisms. Clin. Transl. Med. 2018, 7, 23. [Google Scholar] [CrossRef]
- Ege, N.; Dowbaj, A.; Jiang, M.; Howell, M.; Hooper, S.; Foster, C.; Jenkins, R.P.; Sahai, E. Quantitative Analysis Reveals that Actin and Src-Family Kinases Regulate Nuclear YAP1 and Its Export. Cell Syst. 2018, 6, 692–708.e13. [Google Scholar] [CrossRef] [Green Version]
- Low, B.C.; Pan, C.Q.; Shivashankar, G.; Bershadsky, A.D.; Sudol, M.; Sheetz, M. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 2014, 588, 2663–2670. [Google Scholar] [CrossRef] [Green Version]
- Haelterman, N.; Lim, J. Sensing the load. eLife 2019, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Yavropoulou, M.; Yovos, J. The molecular basis of bone mechanotransduction. J. Musculoskelet. Neuronal Interact. 2016, 16, 221–236. [Google Scholar]
- Uda, Y.; Azab, E.; Sun, N.; Shi, C.; Pajevic, P.D.; Sun, N. Osteocyte Mechanobiology. Curr. Osteoporos. Rep. 2017, 15, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Hum, J.M.; Day, R.N.; Bidwell, J.P.; Wang, Y.; Pavalko, F.M. Mechanical loading in osteocytes induces formation of a Src/Pyk2/MBD2 complex that suppresses anabolic gene expression. PLoS ONE 2014, 9, e97942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, U.; Kumar, S.; Dimmock, J.R.; Sharma, R.K. Inhibition of protein N-myristoylation: A therapeutic protocol in developing anticancer agents. Curr. Cancer Drug Targets 2012, 12, 667–692. [Google Scholar] [CrossRef] [PubMed]
- Sulejmani, E.; Cai, H. Targeting protein myristoylation for the treatment of prostate cancer. Oncoscience 2018, 5, 3–5. [Google Scholar]
- Campbell, E.J.; McDuff, E.; Tatarov, O.; Tovey, S.; Brunton, V.; Cooke, T.G.; Edwards, J. Phosphorylated c-Src in the nucleus is associated with improved patient outcome in ER-positive breast cancer. Br. J. Cancer 2008, 99, 1769–1774. [Google Scholar] [CrossRef] [Green Version]
- Yogi, V.; Pareek, A.; Singh, O.P.; Ghori, H.U.; Tiwari, V.; Redhu, P. Bone metastases incidence and its correlation with hormonal and human epidermal growth factor receptor 2 neu receptors in breast cancer. J. Cancer Res. Ther. 2019, 15, 971–975. [Google Scholar] [CrossRef]

| SFKs | Subcellular Localization | References | ||
|---|---|---|---|---|
| CM | C | N | ||
| Yes | X | X | Dubois et al. [29] | |
| Fyn | X | X | X | Saito et al. [30] - Matsushima et al. [31] |
| Fgr | X | X | Dwyer et al. [32] | |
| Lck | X | Stephen et al. [33] | ||
| Hck | X | X | Poh et al. [34] | |
| Blk | X | Petersen et al. [35] | ||
| Srm | X | Serfas and Tyner [36] | ||
| Brk | X | X | X | Derry et al. [37] |
| Lyn | X | X | X | Yoshida et al. [38] |
| Frk (Rak) | X | X | Ogunbolude et al. [39] - Kim et al. [40] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bagnato, G.; Leopizzi, M.; Urciuoli, E.; Peruzzi, B. Nuclear Functions of the Tyrosine Kinase Src. Int. J. Mol. Sci. 2020, 21, 2675. https://doi.org/10.3390/ijms21082675
Bagnato G, Leopizzi M, Urciuoli E, Peruzzi B. Nuclear Functions of the Tyrosine Kinase Src. International Journal of Molecular Sciences. 2020; 21(8):2675. https://doi.org/10.3390/ijms21082675
Chicago/Turabian StyleBagnato, Giulia, Martina Leopizzi, Enrica Urciuoli, and Barbara Peruzzi. 2020. "Nuclear Functions of the Tyrosine Kinase Src" International Journal of Molecular Sciences 21, no. 8: 2675. https://doi.org/10.3390/ijms21082675





