The Potential Role of Complement System in the Progression of Ovarian Clear Cell Carcinoma Inferred from the Gene Ontology-Based Immunofunctionome Analysis
Abstract
1. Introduction
2. Results
2.1. DNA Microarray Gene Expression Datasets for OCCC and Gene Sets Definition
2.2. Comparison of Functionomes between the Four OCCC Stage Groups and Normal Controls
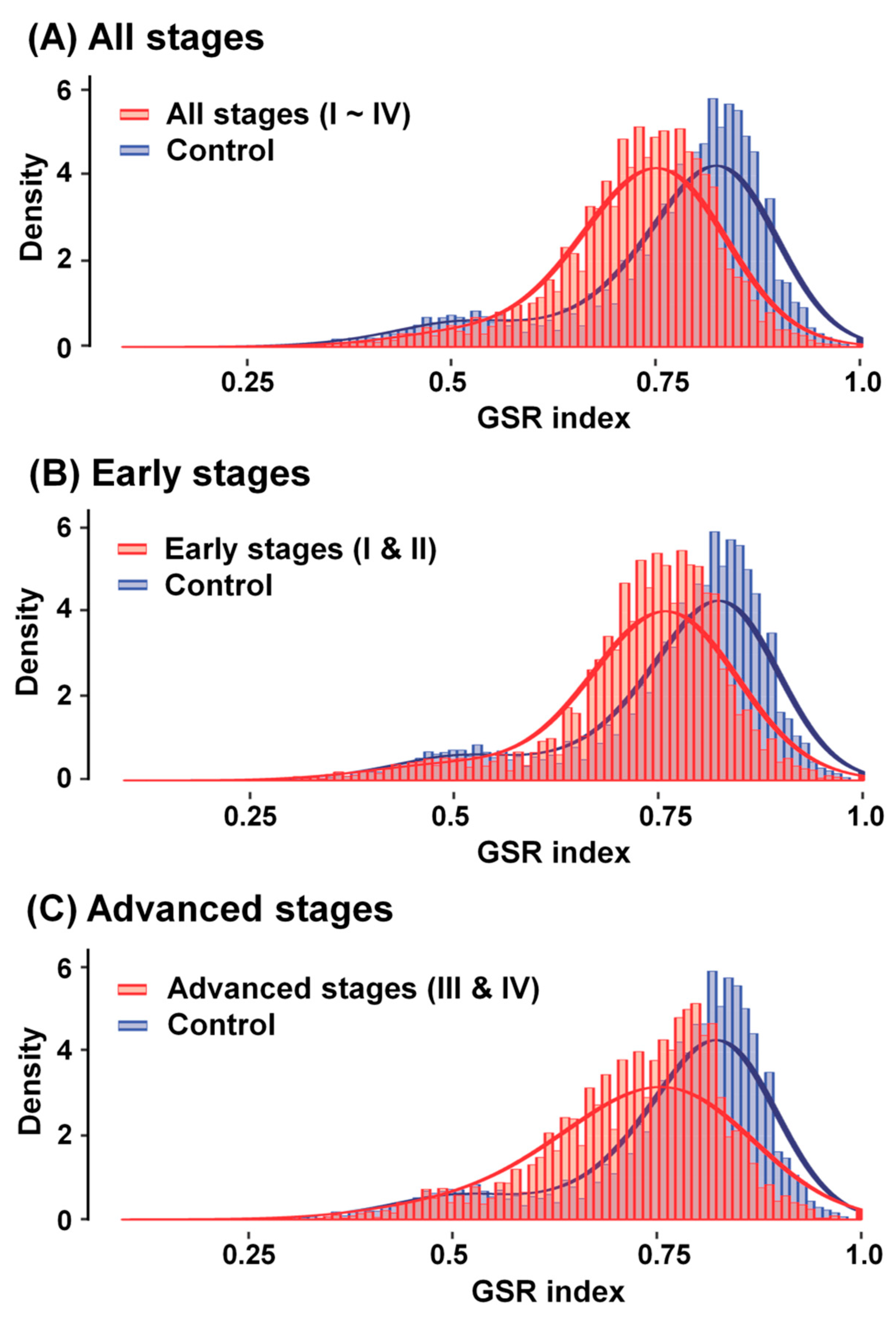
2.3. Reestablishment of Means and Histograms of GSR Indices for Immunofunctionomes and Comparison of the Functionomes to Determine the Relationship between OCCC Stages
2.4. Calculating and Rearranging the Accurate Functional Regulation Patterns of the Early and Advanced OCCC Stages Using Machine Learning
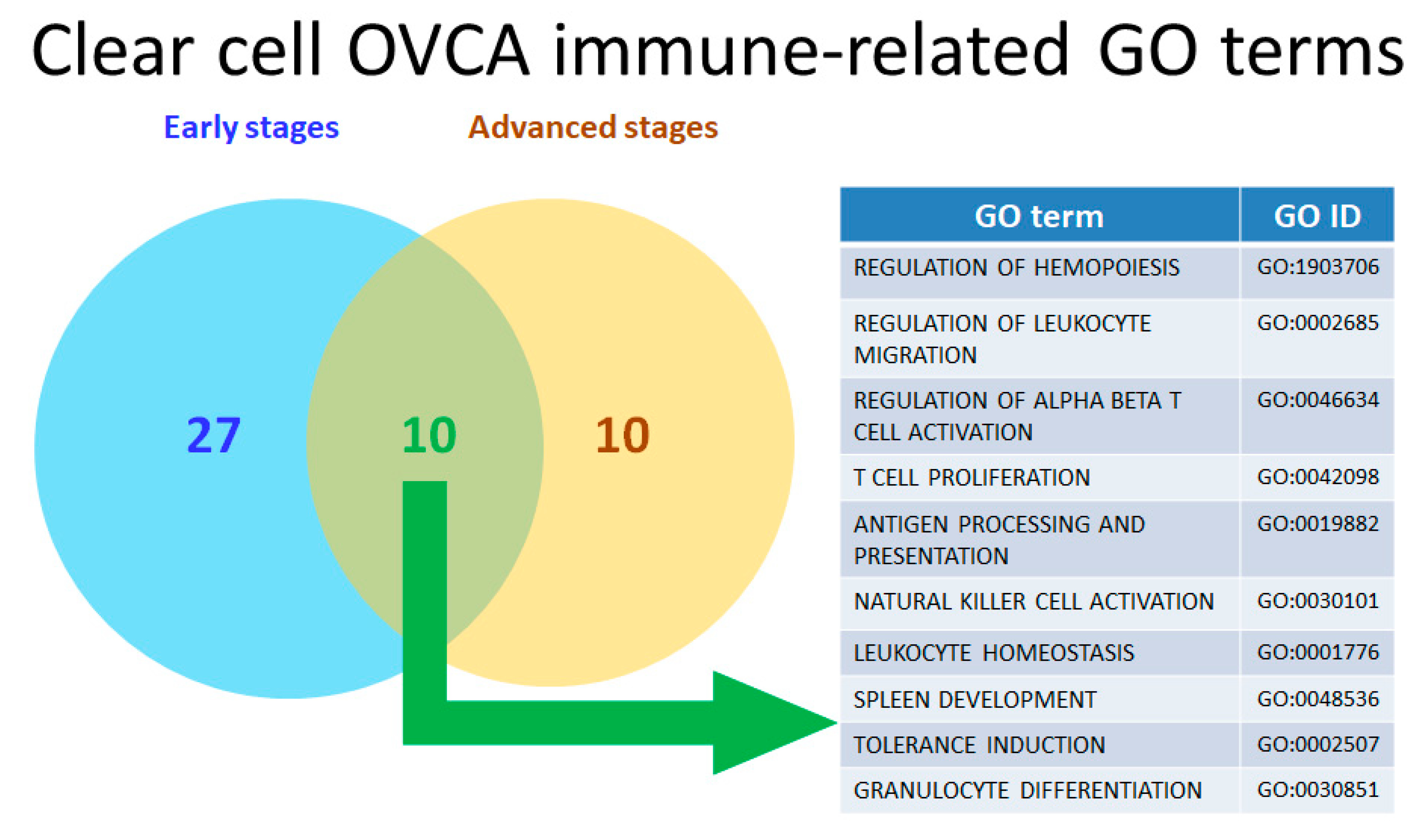
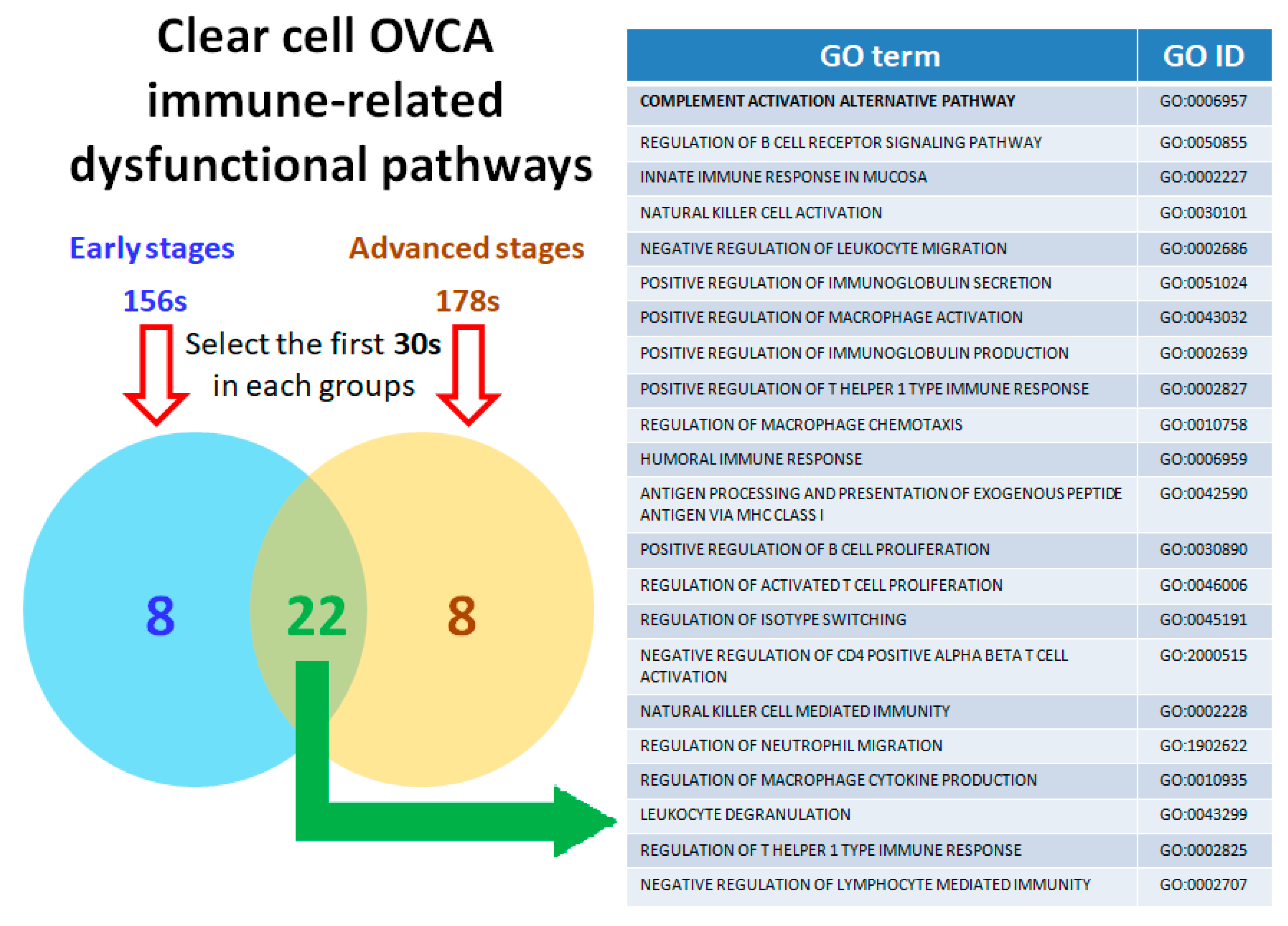
2.5. Twenty-Two Commonly Dysregulated GO Terms Are the Most Meaningful Dysfunctional Immunological Pathways in OCCC Progression
2.6. Distinct Genes Involved in the Key Components of the Dysregulated Immunological Functions Expressed during OCCC Progression
2.7. The Immune-related Genes of the Complement System Have Influence on Progression of OCCC
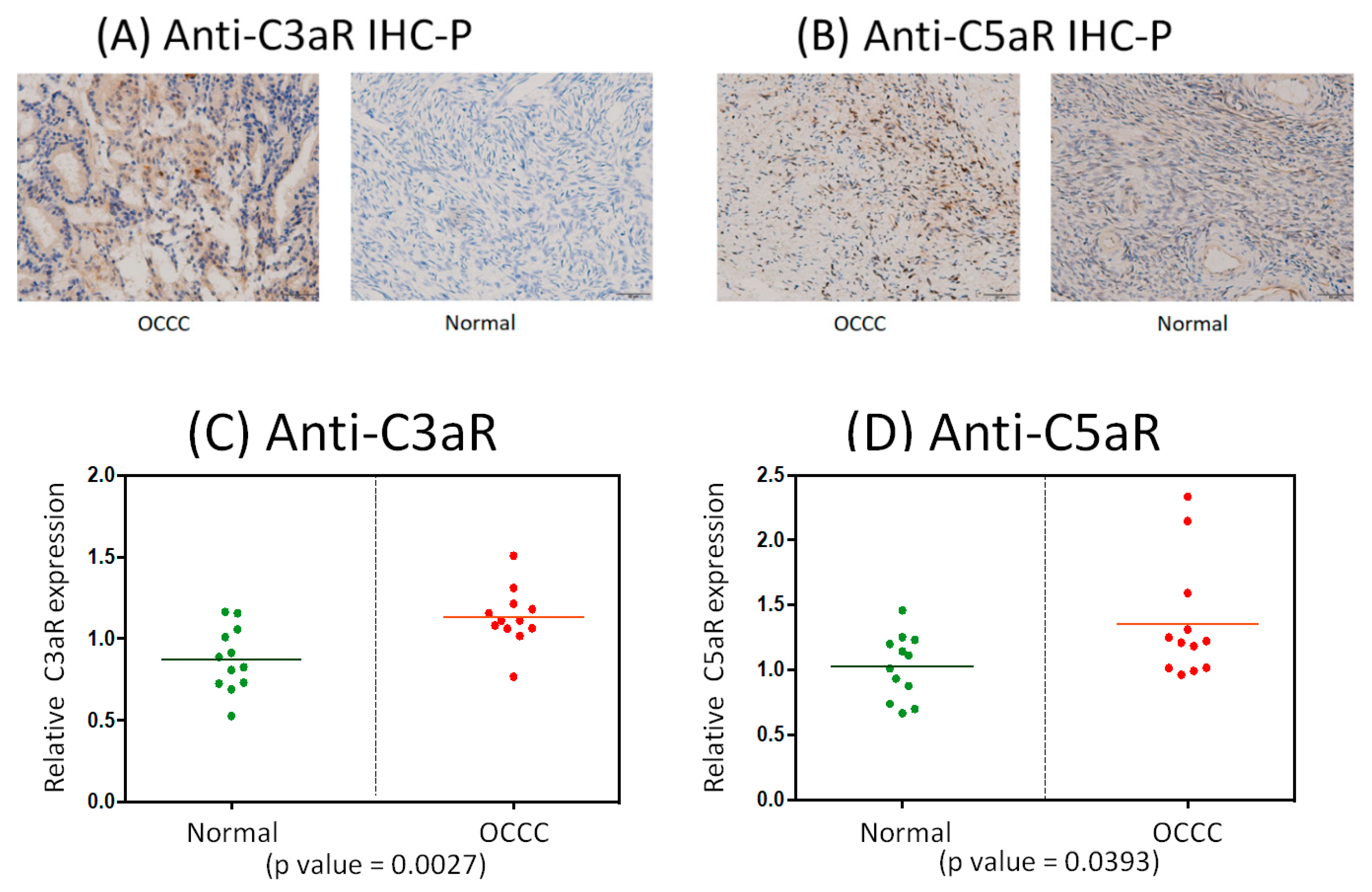
2.8. Immunohistochemistrical Analysis of Anti-C3aR and Anti-C5aR Expression between OCCC and Normal Ovarian Tissues
3. Discussion
3.1. CFP/Complement Factor Properdin
3.2. C9/Complement C9
3.3. C5/Complement C5
3.4. VSIG4/V-set and Immunoglobulin Domain Containing 4
3.5. C8B/Complement C8 Beta Chain
3.6. C7/Complement C7
3.7. C3/Complement C3
4. Materials and Methods
4.1. Computing the GSR Indices and Reconstructing the Functionome and Immunofunctionome
4.2. Microarray Dataset Collection
4.3. Statistical Analysis
4.4. Classification and Prediction by Machine Learning
4.5. Cluster Weight Index
4.6. Set Analysis
4.7. Clinical Samples
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OCCC | Ovarian clear cell carcinoma |
| EOC | Epithelial ovarian carcinoma |
| HGSOC | High grade serous ovarian carcinoma |
| EAOC | Endometriosis-associated ovarian cancer/carcinoma |
| EC | Endometrioid ovarian carcinoma |
| GO | Gene Ontology |
| GEO | Gene Expression Omnibus |
| DEGs | Differentially expressed genes |
| GSR | Gene set regularity |
| FIGO | The International Federation of Gynecology and Obstetrics |
| NCBI | National Center for Biotechnology Information |
| MSigDB | Molecular signatures database |
| PFS | Progression-free survival |
| OS | Overall survival |
| AUC | Area under curve |
| SD | Standard deviation |
| N/A | Unconfirmed stages |
| SVM | Support vector machine |
| EFA | Exploratory factor analysis |
| NK | Natural killer |
| DCs | Dendritic cells |
| TMB | Tumor mutational burden |
| PD-L1 | Programmed death-ligand 1 |
| PD-1 | Programmed death 1 |
| ARID1A | AT-rich interactive domain-containing protein 1A |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| MET | Met proto-oncogene |
| BRCA1 | Breast cancer type 1 susceptibility protein |
| BRCA2 | Breast cancer type 2 susceptibility protein |
| PTEN | Phosphatase and tensin homolog |
| mTOR | Mammalian target of rapamycin |
| PI3K | Phosphoinositide 3-kinase |
| CD4 | CD4 Molecule |
| CD8 | CD8 Molecule |
| MBLs | Mannose-binding lectins |
| MAC | Membrane attack complex |
| MDSC | Myeloid-derived suppressor cells |
| CDC/MAC/PF | Cholesterol-dependent cytolysin/membrane attack complex/perforin-like domain |
| CDC | Complement-dependent cytotoxicity |
| CFP | Complement factor properdin |
| C9 | Complement C9 |
| C5 | Complement C5 |
| VSIG4 | V-set and immunoglobulin domain containing 4 |
| C8B | Complement C8 beta chain |
| C7 | Complement C7 |
| C3 | Complement C3 |
| C5aR | C5a receptor |
| C3aR | C3a receptor |
| OVCA | Ovarian cancer |
| DIRAC | Differential rank conservation |
References
- Zhang, Y.; Cao, L.; Nguyen, D.; Lu, H. TP53 mutations in epithelial ovarian cancer. Transl. Cancer Res. 2016, 5, 650. [Google Scholar] [CrossRef]
- Pearce, C.L.; Templeman, C.; Rossing, M.A.; Lee, A.; Near, A.M.; Webb, P.M.; Nagle, C.M.; Doherty, J.A.; Cushing-Haugen, K.L.; Wicklund, K.G. Association between endometriosis and risk of histological subtypes of ovarian cancer: A pooled analysis of case–control studies. Lancet Oncol. 2012, 13, 385–394. [Google Scholar] [CrossRef]
- Okamoto, A.; Glasspool, R.M.; Mabuchi, S.; Matsumura, N.; Nomura, H.; Itamochi, H.; Takano, M.; Takano, T.; Susumu, N.; Aoki, D. Gynecologic Cancer InterGroup (GCIG) consensus review for clear cell carcinoma of the ovary. Int. J. Gynecol. Cancer 2014, 24 (Suppl. 3), S20–S25. [Google Scholar] [CrossRef] [PubMed]
- Anglesio, M.S.; Carey, M.S.; Köbel, M.; MacKay, H.; Huntsman, D.G. Clear cell carcinoma of the ovary: A report from the first Ovarian Clear Cell Symposium, June 24th, 2010. Gynecol. Oncol. 2011, 121, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Benedet, J. FIGO staging classifications and clinical practice guidelines in the management of gynecologic cancers. FIGO Committee on Gynecologic Oncology. Int. J. Gynaecol. Obstet. 2000, 70, 209–262. [Google Scholar] [PubMed]
- Sugiyama, T.; Kamura, T.; Kigawa, J.; Terakawa, N.; Kikuchi, Y.; Kita, T.; Suzuki, M.; Sato, I.; Taguchi, K. Clinical characteristics of clear cell carcinoma of the ovary: A distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer: Interdiscip. Int. J. Am. Cancer Soc. 2000, 88, 2584–2589. [Google Scholar] [CrossRef]
- Okamoto, A.; Sugiyama, T.; Hamano, T.; Kim, J.W.; Kim, B.G.; Enomoto, T.; Aoki, D.; Terao, Y.; Suzuki, N.; Mikami, M. Randomized phase III trial of paclitaxel/carboplatin (PC) versus cisplatin/irinotecan (CPT-P) as first-line chemotherapy in patients with clear cell carcinoma (CCC) of the ovary: A Japanese Gynecologic Oncology Group (JGOG)/GCIG study. Am. Soc. Clin. Oncol. 2014, 32, 5507. [Google Scholar] [CrossRef]
- Winter, W.E.; Maxwell, G.L.; Tian, C.; Carlson, J.W.; Ozols, R.F.; Rose, P.G.; Markman, M.; Armstrong, D.K.; Muggia, F.; McGuire, W.P. Prognostic factors for stage III epithelial ovarian cancer: A Gynecologic Oncology Group Study. J. Clin. Oncol. 2007, 25, 3621–3627. [Google Scholar] [CrossRef]
- Kok, V.C.; Tsai, H.-J.; Su, C.-F.; Lee, C.-K. The risks for ovarian, endometrial, breast, colorectal, and other cancers in women with newly diagnosed endometriosis or adenomyosis: A population-based study. Int. J. Gynecol. Cancer 2015, 25, 968–976. [Google Scholar] [CrossRef]
- Kobayashi, H.; Sumimoto, K.; Moniwa, N.; Imai, M.; Takakura, K.; Kuromaki, T.; Morioka, E.; Arisawa, K.; Terao, T. Risk of developing ovarian cancer among women with ovarian endometrioma: A cohort study in Shizuoka, Japan. Int. J. Gynecol. Cancer 2007, 17, 37–43. [Google Scholar] [CrossRef]
- Sampson, J.A. Endometrial carcinoma of the ovary, arising in endometrial tissue in that organ. Arch. Surg. 1925, 10, 1–72. [Google Scholar] [CrossRef]
- Sekizawa, A.; Amemiya, S.; Otsuka, J.; Saito, H.; Farina, A.; Okai, T.; Tachikawa, T. Malignant transformation of endometriosis: Application of laser microdissection for analysis of genetic alterations according to pathological changes. Med. Electron Microsc. 2004, 37, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Parazzini, F.; Esposito, G.; Tozzi, L.; Noli, S.; Bianchi, S. Epidemiology of endometriosis and its comorbidities. Eur. J. Obstet. & Gynecol. Reprod. Biol. 2017, 209, 3–7. [Google Scholar]
- Symons, L.K.; Miller, J.E.; Kay, V.R.; Marks, R.M.; Liblik, K.; Koti, M.; Tayade, C. The immunopathophysiology of endometriosis. Trends Mol. Med. 2018, 24, 748–762. [Google Scholar] [CrossRef] [PubMed]
- Samstein, R.M.; Lee, C.-H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Oda, K.; Hamanishi, J.; Matsuo, K.; Hasegawa, K. Genomics to immunotherapy of ovarian clear cell carcinoma: Unique opportunities for management. Gynecol. Oncol 2018, 151, 381–389. [Google Scholar] [CrossRef]
- Infante, J.; Braiteh, F.; Emens, L.; Balmanoukian, A.; Oaknin, A.; Wang, Y.; Liu, B.; Molinero, L.; Fasso, M.; O’Hear, C. Safety, clinical activity and biomarkers of atezolizumab (atezo) in advanced ovarian cancer (OC). Eur. Soc. Med. Oncol. 2016, 27, vi300. [Google Scholar] [CrossRef]
- Disis, M.L.; Patel, M.R.; Pant, S.; Infante, J.R.; Lockhart, A.C.; Kelly, K.; Beck, J.T.; Gordon, M.S.; Weiss, G.J.; Ejadi, S. Avelumab (MSB0010718C), an anti-PD-L1 antibody, in patients with previously treated, recurrent or refractory ovarian cancer: A phase Ib, open-label expansion trial. Am. Soc. Clin. Oncol. 2015, 33, 5509. [Google Scholar] [CrossRef]
- Hamanishi, J.; Mandai, M.; Ikeda, T.; Minami, M.; Kawaguchi, A.; Matsumura, N.; Abiko, K.; Baba, T.; Yamaguchi, K.; Ueda, A. Durable tumor remission in patients with platinum-resistant ovarian cancer receiving nivolumab. Am. Soc. Clin. Oncol. 2015, 33, 5570. [Google Scholar] [CrossRef]
- Varga, A.; Piha-Paul, S.A.; Ott, P.A.; Mehnert, J.M.; Berton-Rigaud, D.; Johnson, E.A.; Cheng, J.D.; Yuan, S.; Rubin, E.H.; Matei, D.E. Antitumor activity and safety of pembrolizumab in patients (pts) with PD-L1 positive advanced ovarian cancer: Interim results from a phase Ib study. Am. Soc. Clin. Oncol. 2015, 33, 5510. [Google Scholar] [CrossRef]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. InnateDB: Systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acid. Res. 2012, 41, D1228–D1233. [Google Scholar] [CrossRef]
- Győrff Fekete, J.T.; Ősz, Á.; Pete, I.; Nagy, G.R.; Vereczkey, I.; Győrffy, B. Predictive biomarkers of platinum and taxane resistance using the transcriptomic data of 1816 ovarian cancer patients. Gynecol. Oncol. 2020, 156, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Fisher, A.J.; Mickler, E.A.; Duerson III, F.; Cummings, O.W.; Peters-Golden, M.; Twigg III, H.L.; Woodruff, T.M.; Wilkes, D.S.; Vittal, R. Contribution of the anaphylatoxin receptors, C3aR and C5aR, to the pathogenesis of pulmonary fibrosis. FASEB J. 2016, 30, 2336–2350. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Ortiz-Espinosa, S.; Pio, R. In complement anaphylatoxins C3a and C5a: Emerging roles in cancer progression and treatment. Semin. Cell. & Dev. Biol. 2019, 85, 153–163. [Google Scholar]
- Sato, N.; Tsunoda, H.; Nishida, M.; Morishita, Y.; Takimoto, Y.; Kubo, T.; Noguchi, M. Loss of heterozygosity on 10q23. 3 and mutation of the tumor suppressor gene PTEN in benign endometrial cyst of the ovary: Possible sequence progression from benign endometrial cyst to endometrioid carcinoma and clear cell carcinoma of the ovary. Cancer Res. 2000, 60, 7052–7056. [Google Scholar]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Prim. 2016, 2, 16061. [Google Scholar] [CrossRef]
- Abbas, A.K.; Lichtman, A.H.; Pillai, S. Basic Immunology: Functions and Disorders of the Immune System; Elsevier Health Sciences: Amsterdam, The Netherlands, 2019; pp. 164–173. [Google Scholar]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860. [Google Scholar] [CrossRef]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. New England J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef]
- Chang, C.-M.; Chuang, C.-M.; Wang, M.-L.; Yang, M.-J.; Chang, C.-C.; Yen, M.-S.; Chiou, S.-H. Gene set-based functionome analysis of pathogenesis in epithelial ovarian serous carcinoma and the molecular features in different FIGO stages. Int. J. Mol. Sci. 2016, 17, 886. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-M.; Chuang, C.-M.; Wang, M.-L.; Yang, Y.-P.; Chuang, J.-H.; Yang, M.-J.; Yen, M.-S.; Chiou, S.-H.; Chang, C.-C. Gene Set− Based Integrative Analysis Revealing Two Distinct Functional Regulation Patterns in Four Common Subtypes of Epithelial Ovarian Cancer. Int. J. Mol. Sci. 2016, 17, 1272. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-M.; Yang, Y.-P.; Chuang, J.-H.; Chuang, C.-M.; Lin, T.-W.; Wang, P.-H.; Yu, M.-H.; Chang, C.-C. Discovering the dysregulated molecular functions involved in malignant transformation of endometriosis to endometriosis-associated ovarian carcinoma using a data-driven, function-based analysis. Int. J. Mol. Sci. 2017, 18, 2345. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-M.; Wang, M.-L.; Lu, K.-H.; Yang, Y.-P.; Juang, C.-M.; Wang, P.-H.; Hsu, R.-J.; Yu, M.-H.; Chang, C.-C. Integrating the dysregulated inflammasome-based molecular functionome in the malignant transformation of endometriosis-associated ovarian carcinoma. Oncotarget 2018, 9, 3704. [Google Scholar] [CrossRef]
- Markiewski, M.M.; Lambris, J.D. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am. J. Pathol. 2007, 171, 715–727. [Google Scholar] [CrossRef]
- Bareke, H.; Akbuga, J. Complement system’s role in cancer and its therapeutic potential in ovarian cancer. Scand. J. Immunol. 2018, 88, E12672. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids. Res. 2015, 44, D733–D745. [Google Scholar] [CrossRef]
- Reis, E.S.; Mastellos, D.C.; Ricklin, D.; Mantovani, A.; Lambris, J.D. Complement in cancer: Untangling an intricate relationship. Nat. Rev. Immunol. 2018, 18, 5. [Google Scholar] [CrossRef]
- Fishelson, Z.; Kirschfink, M. Complement C5b-9 and cancer: Mechanisms of cell damage, cancer counteractions, and approaches for intervention. Front. Immunol. 2019, 10, 752. [Google Scholar] [CrossRef]
- Bjørge, L.; Hakulinen, J.; Vintermyr, O.; Jarva, H.; Jensen, T.S.; Iversen, O.E.; Meri, S. Ascitic complement system in ovarian cancer. Br. J. Cancer 2005, 92, 895. [Google Scholar] [CrossRef]
- Aslan, C.; Ak, H.; Askar, N.; Ozkaya, A.B.; Ergenoglu, A.M.; Yeniel, A.O.; Akdemir, A.; Aydin, H.H. Overexpression of complement C5 in endometriosis. Clin. Biochem. 2014, 47, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.S.; Vasquez, H.G.; Rupaimoole, R.; Pradeep, S.; Wu, S.; Zand, B.; Han, H.-D.; Rodriguez-Aguayo, C.; Bottsford-Miller, J.; Huang, J. Autocrine effects of tumor-derived complement. Cell. Rep. 2014, 6, 1085–1095. [Google Scholar] [CrossRef]
- Nunez-Cruz, S.; Gimotty, P.A.; Guerra, M.W.; Connolly, D.C.; Wu, Y.-Q.; DeAngelis, R.A.; Lambris, J.D.; Coukos, G.; Scholler, N. Genetic and pharmacologic inhibition of complement impairs endothelial cell function and ablates ovarian cancer neovascularization. Neoplasia 2012, 14, 994. [Google Scholar] [CrossRef]
- Vogt, L.; Schmitz, N.; Kurrer, M.O.; Bauer, M.; Hinton, H.I.; Behnke, S.; Gatto, D.; Sebbel, P.; Beerli, R.R.; Sonderegger, I. VSIG4, a B7 family–related protein, is a negative regulator of T cell activation. J. Clin. Investig. 2006, 116, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.M.; Jeong, D.H.; Choi, I.H.; Lee, D.S.; Kang, M.S.; Jung, K.O.; Jeon, Y.K.; Kim, Y.N.; Jung, E.J.; Lee, K.B. The significance of VSIG4 expression in ovarian cancer. Int. J. Gynecol. Cancer 2017, 27, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Investig. 2017, 127, 780–789. [Google Scholar] [CrossRef]
- Eddy, J.A.; Hood, L.; Price, N.D.; Geman, D. Identifying tightly regulated and variably expressed networks by differential rank conservation (Dirac). PLoS Comput. Biol. 2010, 6, e1000792. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. Proc: An open-source package for r and s+ to analyze and compare roc curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]



| Group of Stage | Sample | Control | Total | Case Mean (SD1) | Control Mean (SD1) | p-value |
| Early stage (stage I and II) | 27 | 136 | 163 | 0.7465(0.1114) | 0.7745(0.1284) | <0.05 |
| Advanced stage (stage III and IV) | 17 | 136 | 153 | 0.7309(0.1176) | 0.7744(0.1282) | <0.05 |
| N/A2 | 41 | 136 | 177 | 0.7374(0.1040) | 0.7745(0.1286) | <0.05 |
| Group of Stage | Sample | Control | Total | Case Mean (SD1) | Control Mean (SD1) | p-value |
| Early stage (stage I and II) | 27 | 136 | 163 | 0.7328(0.1045) | 0.7687(0.1239) | <0.05 |
| Advanced stage (stage III and IV) | 17 | 136 | 153 | 0.7255(0.1080) | 0.7687(0.1239) | <0.05 |
| N/A2 | 41 | 136 | 177 | 0.7269(0.0980) | 0.7682(0.1239) | <0.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, K.-M.; Lin, T.-W.; Liu, L.-C.; Yang, Y.-P.; Wang, M.-L.; Tsai, P.-H.; Wang, P.-H.; Yu, M.-H.; Chang, C.-M.; Chang, C.-C. The Potential Role of Complement System in the Progression of Ovarian Clear Cell Carcinoma Inferred from the Gene Ontology-Based Immunofunctionome Analysis. Int. J. Mol. Sci. 2020, 21, 2824. https://doi.org/10.3390/ijms21082824
Su K-M, Lin T-W, Liu L-C, Yang Y-P, Wang M-L, Tsai P-H, Wang P-H, Yu M-H, Chang C-M, Chang C-C. The Potential Role of Complement System in the Progression of Ovarian Clear Cell Carcinoma Inferred from the Gene Ontology-Based Immunofunctionome Analysis. International Journal of Molecular Sciences. 2020; 21(8):2824. https://doi.org/10.3390/ijms21082824
Chicago/Turabian StyleSu, Kuo-Min, Tzu-Wei Lin, Li-Chun Liu, Yi-Pin Yang, Mong-Lien Wang, Ping-Hsing Tsai, Peng-Hui Wang, Mu-Hsien Yu, Chia-Ming Chang, and Cheng-Chang Chang. 2020. "The Potential Role of Complement System in the Progression of Ovarian Clear Cell Carcinoma Inferred from the Gene Ontology-Based Immunofunctionome Analysis" International Journal of Molecular Sciences 21, no. 8: 2824. https://doi.org/10.3390/ijms21082824
APA StyleSu, K.-M., Lin, T.-W., Liu, L.-C., Yang, Y.-P., Wang, M.-L., Tsai, P.-H., Wang, P.-H., Yu, M.-H., Chang, C.-M., & Chang, C.-C. (2020). The Potential Role of Complement System in the Progression of Ovarian Clear Cell Carcinoma Inferred from the Gene Ontology-Based Immunofunctionome Analysis. International Journal of Molecular Sciences, 21(8), 2824. https://doi.org/10.3390/ijms21082824








