C5-Substituted 2-Selenouridines Ensure Efficient Base Pairing with Guanosine; Consequences for Reading the NNG-3′ Synonymous mRNA Codons
Abstract
:1. Introduction
2. Results
2.1. Chemistry
2.2. Physicochemical Analysis
2.2.1. Effect of pH on the Structure of 2-Selenouridines 1–3 and Their Thio-Precursors 5–7
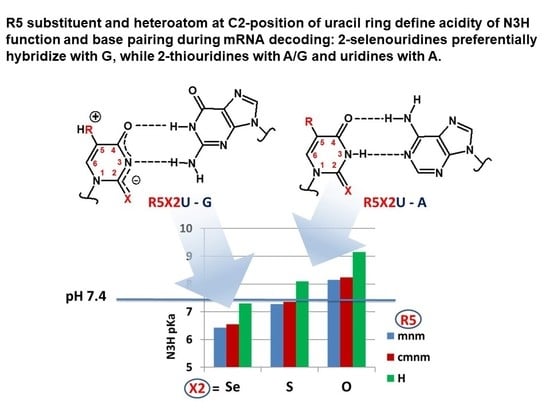
2.2.2. pKa Determination and Analysis of the Content of the Ionized Fraction of 1–3
2.3. Structural Analysis
2.3.1. Se2U Crystal Structure
2.3.2. Structural Analysis of Sugar Residues of 1–3 in Solution
2.4. Molecular Modeling
2.4.1. Structural Analysis of m1Se2Ura and m1mnm5Se2Ura Tautomers
2.4.2. Gibbs Free Energies of the Tautomers of 1-Methyl-2-Selenouracil and 1-Methyl-5-Methylaminomethyl-2-Selenouracil
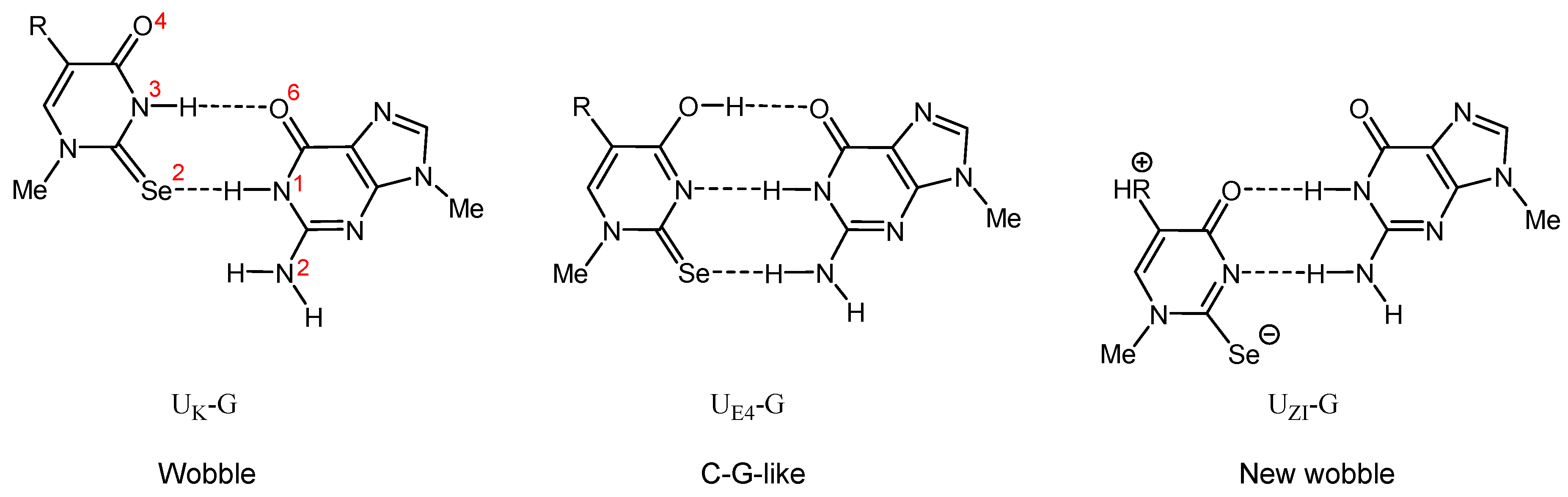
2.4.3. Enthalpies of Base Complexation (Pairing)
2.4.4. Atomic Charge Distribution (ESP, Merz–Kollman Scheme) in Water
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.2. Physicochemical Studies
4.2.1. The UV Measurements and Extinction Coefficients of 1, 2, and 3
4.2.2. Potentiometric Measurements
4.3. Crystallographic Analysis
4.4. Quantum Mechanical Calculations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2017, 46, D303–D307. [Google Scholar] [CrossRef]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosjean, H.; de Crécy-Lagard, V.; Marck, C. Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010, 584, 252–264. [Google Scholar] [CrossRef]
- Pollo-Oliveira, L.; de Crecy-Lagard, V. Can protein expression be regulated by modulation of tRNA modification profiles. Biochemistry 2019, 58, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Krutyhołowa, R.; Zakrzewski, K.; Glatt, S. Charging the code—tRNA modification complexes. Curr. Opin. Struct. Biol. 2019, 55, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.M.; Alexander, R.W. Bacterial wobble modifications of NNA decoding tRNAs. IUBMB Life 2019, 71, 1158–1166. [Google Scholar] [CrossRef]
- Schaffrath, R.; Leidel, S.A. Wobble uridine modifications—A reason to live, a reason to die?! RNA Biol. 2017, 14, 1209–1222. [Google Scholar] [CrossRef]
- Ching, W.M. Characterization of selenium-containing tRNAGlu from Clostridium sticklandii. Arch. Biochem. Biophys. 1986, 244, 137–146. [Google Scholar] [CrossRef]
- Wittwer, A.J.; Stadtman, T.C. Biosynthesis of 5-methylaminomethyl-2-selenouridine, a naturally occurring nucleoside in Escherichia coli tRNA. Arch. Biochem. Biophys. 1986, 248, 540–550. [Google Scholar] [CrossRef]
- Wittwer, A.J.; Tsai, L.; Ching, W.M.; Stadtman, T.C. Identification and synthesis of a naturally occurring selenonucleoside in bacterial tRNAs: 5-[(methylamino)methyl]-2-selenouridine. Biochemistry 1984, 23, 4650–4655. [Google Scholar] [CrossRef]
- Wolfe, M.D.; Ahmed, F.; Lacourciere, G.M.; Lauhon, C.T.; Stadtman, T.C.; Larson; T. Functional diversity of the rhodanese homology domain: The Escherichia coli ybbB gene encodes a selenophosphate-dependent tRNA 2-selenouridine synthase. J. Biol. Chem. 2004, 279, 1801–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrot, B.; Sochacka, E.; Duchler, M. tRNA structural and functional changes induced by oxidative stress. Cell. Mol. Life Sci. 2011, 68, 4023–4032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ching, W.M. Occurrence of selenium-containing tRNAs in mouse leukemia cells. Proc. Natl. Acad. Sci. USA 1984, 81, 3010–3013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizutani, T.; Watanabe, T.; Kanaya, K.; Nakagawa, Y.; Fujiwara, T. Trace 5-methylaminomethyl-2-selenouridine in bovine tRNA and the selenouridine synthase activity in bovine liver. Mol. Biol. Rep. 1999, 26, 167–172. [Google Scholar] [CrossRef]
- Ching, W.M.; Wittwer, A.J.; Tsai, L.; Stadtman, T.C. Distribution of two selenonucleosides among the selenium-containing tRNAs from Methanococcus vannielii. Proc. Natl. Acad. Sci. USA 1984, 81, 57–60. [Google Scholar] [CrossRef] [Green Version]
- Politino, M.; Tsai, L.; Veres, Z.; Stadtman, T.C. Biosynthesis of selenium-modified tRNAs in Methanococcus vannielii. Proc. Natl. Acad. Sci. USA 1990, 87, 6345–6348. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.X.; An, Y.X.; Chen, Z.X.; Xu, H.B. Isolation and partial characterization of selenium-containing tRNA from germinating barley. Biol. Trace Elem. Res. 2001, 82, 247–257. [Google Scholar] [CrossRef]
- Wen, T.N.; Li, C.; Chen, C.S. Ubiquity of selenium-containing tRNA in plants. Plant Sci. 1988, 57, 185–193. [Google Scholar]
- Mihara, H.; Kato, S.; Lacourciere, G.M.; Stadtman, T.C.; Kennedy, R.A.; Kurihara, T.; Tokumoto, U.; Takahashi, Y.; Esaki, N. The iscS gene is essential for the biosynthesis of 2-selenouridine in tRNA and the selenocysteine-containing formate dehydrogenase H. Proc. Natl. Acad. Sci. USA 2002, 99, 6679–6683. [Google Scholar] [CrossRef] [Green Version]
- Shigi, N. Biosynthesis and functions of sulfur modifications in tRNA. Front Genet. 2014, 5, 67. [Google Scholar] [CrossRef] [Green Version]
- Jäger, G.; Chen, P.; Björk, G.R. Transfer RNA bound to MnmH protein is enriched with geranylated tRNA—A possible intermediate in its selenation? PLoS ONE 2016, 11, e0153488. [Google Scholar] [CrossRef] [Green Version]
- Su, D.; Ojo, T.T.; Söll, D.; Hohn, M.J. Selenomodification of tRNA in archaea requires a bipartite rhodanese enzyme. FEBS Lett. 2012, 586, 717–721. [Google Scholar] [CrossRef] [Green Version]
- Dumelin, C.E.; Chen, Y.; Leconte, A.M.; Chen, Y.G.; Liu, D.R. Discovery and biological characterization of geranylated RNA in bacteria. Nat. Chem. Biol. 2012, 8, 913–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartos, P.; Maciaszek, A.; Rosinska, A.; Sochacka, E.; Nawrot, B. Transformation of a wobble 2-thiouridine to 2-selenouridine via S-geranyl-2-thiouridine as a possible cellular pathway. Bioorg. Chem. 2014, 56, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Sierant, M.; Leszczynska, G.; Sadowska, K.; Komar, P.; Radzikowska-Cieciura, E.; Sochacka, E.; Nawrot, B. Escherichia coli tRNA 2-selenouridine synthase (SelU) converts S2U-RNA to Se2U-RNA via S-geranylated-intermediate. FEBS Lett. 2018, 592, 2248–2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Crain, P.F.; Näsvall, S.J.; Pomerantz, S.C.; Björk, G.R. A “gain of function” mutation in a protein mediates production of novel modified nucleosides. EMBO J. 2005, 24, 1842–1851. [Google Scholar] [CrossRef] [Green Version]
- Wise, D.S.; Townsend, L.B. Synthesis of the selenopyrimidine nucleosides 2-seleno- and 4-selenouridine. J. Hetrocycl. Chem. 1972, 9, 1461–1462. [Google Scholar] [CrossRef]
- Shiue, C.h.-Y.; Chu, S.-H. A facile synthesis of 1-β-D-arabinofuranosyl-2-seleno- and -4-selenouracil and related compounds. J. Org. Chem. 1975, 40, 2971–2974. [Google Scholar] [CrossRef]
- Sun, H.; Sheng, J.; Hassan, A.E.; Jiang, S.; Gan, J.; Huang, Z. Novel RNA base pair with higher specificity using single selenium atom. Nucleic Acids Res. 2012, 40, 5171–5179. [Google Scholar] [CrossRef] [Green Version]
- Kogami, M.; Davis, D.R.; Koketsu, M. An efficient synthesis of 2-selenouridine and its phosphoramidite precursor. Heterocycles 2016, 92, 64–74. [Google Scholar]
- Caton-Williams, J.; Huang, Z. Biochemistry of Selenium-Derivatized Naturally Occurring and Unnatural Nucleic Acids. Chem. Biodivers. 2008, 5, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jiang, S.; Caton-Williams, J.; Liu, H.; Huang, Z. 2-Selenouridine triphosphate synthesis and Se-RNA transcription. RNA 2013, 19, 1309–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sierzputowska-Gracz, H.; Sochacka, E.; Malkiewicz, A.; Kuo, K.; Gehrke, C.W.; Agris, P.F. Chemistry and structure of modified uridines in the anticodon, wobble position of transfer RNA are determined by thiolation. J. Am. Chem. Soc. 1987, 109, 7171–7177. [Google Scholar] [CrossRef]
- Testa, S.M.; Disney, M.D.; Turner, D.H.; Kierzek, R. Thermodynamics of RNA-RNA duplexes with 2- or 4-thiouridines: Implications for antisense design and targeting a group I intron. Biochemistry 1999, 38, 16655–16662. [Google Scholar] [CrossRef]
- Bartos, P.; Ebenryter-Olbinska, K.; Sochacka, E.; Nawrot, B. The influence of the C5 substituent on the 2-thiouridine desulfuration pathway and the conformational analysis of the resulting 4-pyrimidinone products. Bioorg. Med. Chem. 2015, 23, 5587–5594. [Google Scholar] [CrossRef]
- Zhang, R.B.; Eriksson, L.A. Theoretical study on conformational preferences of ribose in 2-thiouridine--the role of the 2’OH group. Phys. Chem. Chem. Phys. 2010, 12, 3690–3697. [Google Scholar] [CrossRef]
- Agris, P.F.; Sierzputowska-Gracz, H.; Smith, W.; Malkiewicz, A.; Sochacka, E.; Nawrot, B. Thiolation of uridine carbon-2 restricts the motional dynamics of the transfer RNA wobble position nucleoside. J. Am. Chem. Soc. 1992, 114, 2652–2656. [Google Scholar] [CrossRef]
- Krüger, M.K.; Pedersen, S.; Hagervall, T.G.; Sørensen, M.A. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 1998, 284, 621–631. [Google Scholar] [CrossRef]
- Hagervall, T.G.; Pomerantz, S.C.; McCloskey, J.A. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypermodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol. 1998, 284, 33–42. [Google Scholar] [CrossRef]
- Murphy, F.V.; Ramakrishnan, V.; Malkiewicz, A.; Agris, P.F. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004, 12, 1186–1191. [Google Scholar] [CrossRef]
- Vendeix, F.A.P.; Murphy, F.V.; Gustilo, E.; Graham, W.; Cantora, W.; Leszczynska, G.; Sproat, B.; Malkiewicz, A.; Agris, P. Human tRNALys3UUU is pre-structured by natural modifications for cognate and wobble codon binding through keto–enol tautomerism. J. Mol. Biol. 2012, 416, 467–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weixlbaumer, A.; Murphy, F.V.; Dziergowska, A.; Malkiewicz, A.; Vendix, F.A.P.; Agris, P.F.; Ramakrishnan, V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007, 14, 498–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozov, A.; Demeshkina, N.; Khusainov, I.; Westhof, E.; Yusupov, M.; Yusupova, G. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 2016, 7, 10457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westhof, E.; Yusupov, M.; Yusupova, G. The multiple flavors of GoU pairs in RNA. J. Mol. Recognit. 2019, 32, e2782. [Google Scholar] [CrossRef] [Green Version]
- Sochacka, E.; Lodyga-Chruscinska, E.; Pawlak, J.; Cypryk, M.; Bartos, P.; Ebenryter-Olbinska, K.; Leszczynska, G.; Nawrot, B. C5-substituents of uridines and 2-thiouridines present at the wobble position of tRNA determine the formation of their keto-enol or zwitterionic forms—A factor important for accuracy of reading of guanosine at the 3΄-end of the mRNA codons. Nucleic Acids Res. 2017, 45, 4825–4836. [Google Scholar] [CrossRef] [Green Version]
- Stadtman, T.C. Specific occurrence of selenium in enzymes and amino acid tRNAs. FASEB J. 1987, 1, 375–379. [Google Scholar] [CrossRef]
- Wittwer, A.J.; Ching, W.M. Selenium-containing tRNA(Glu) and tRNA(Lys) from Escherichia coli: Purification, codon specificity and translational activity. Biofactors 1989, 2, 27–34. [Google Scholar]
- Leszczynska, G.; Pieta, J.; Leonczak, P.; Tomaszewska, A.; Malkiewicz, A. Site-specific incorporation of 5-methylaminomethyl-2-thiouridine and 2-thiouridine(s) into RNA sequences. Tetrahedron Lett. 2012, 53, 1214–1217. [Google Scholar] [CrossRef]
- Sundaram, M.; Crain, P.F.; Davis, D.R. Synthesis and characterization of the native anticodon domain of E. coli tRNALys: Simultaneous incorporation of modified nucleosides mnm5s2U, t6A, and pseudouridine using phosphoramidite chemistry. J. Org. Chem. 2000, 65, 5609–5614. [Google Scholar] [CrossRef]
- Leszczynska, G.; Pieta, J.; Wozniak, K.; Malkiewicz, A. Site-selected incorporation of 5-carboxymethylaminomethyl-2-thiouridine into RNA sequences by phosphoramidite chemistry. Org. Biomol. Chem. 2014, 12, 1052–1056. [Google Scholar] [CrossRef] [Green Version]
- Leszczynska, G.; Leonczak, P.; Dziergowska, A.; Malkiewicz, A. mt-tRNA components: Synthesis of (2-thio)uridines modified with blocked glycine/taurine moieties at C-5,1. Nucleosides Nucleotides Nucleic Acids 2013, 32, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.R.; Bajji, A.C. Introduction of hypermodified nucleotides in RNA. In Methods in Molecular Biology; Herdewijn, P., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2005; pp. 18–204. [Google Scholar]
- Klayman, D.L.; Griffin, T.S. Reaction of selenium with sodium borohydride in protic solvents. A facile method for the introduction of selenium into organic molecules. J. Am. Chem. Soc. 1973, 95, 197–199. [Google Scholar] [CrossRef]
- Hassan, A.E.A.; Sheng, J.; Zhang, W.; Huang, Z. High fidelity of base pairing by 2-selenothymidine in DNA. J. Am. Chem. Soc. 2010, 132, 2120–2121. [Google Scholar] [CrossRef] [PubMed]
- Kaburagi, Y.; Kishi, Y. Operationally simple and efficient workup procedure for TBAF-mediated desilylation: Application to halichondrin synthesis. Org. Lett. 2007, 9, 723–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gans, P.; Vacca, A.; Sabatini, A. SUPERQUAD: An improved general program for computation of formation constants from potentiometric data. J. Chem. Soc. Dalton Trans. 1985, 6, 1195–1200. [Google Scholar] [CrossRef]
- De Levie, R. The Henderson-Hasselbalch Equation: Its History and Limitations. J. Chem. Educ. 2003, 80, 146. [Google Scholar] [CrossRef] [Green Version]
- Takai, K.; Yokoyama, S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003, 31, 6383–6391. [Google Scholar] [CrossRef] [Green Version]
- Hawkinson, S.W. The crystal and molecular structure of 2-thiouridine. Acta Cryst. B 1977, 33, 80–85. [Google Scholar] [CrossRef]
- Green, E.A.; Rosenstein, R.D.; Shiono, R.; Abraham, D.J.; Trus, B.L.; Marsh, R.E. The crystal structure of uridine. Acta Cryst. B 1975, 31, 102–107. [Google Scholar] [CrossRef]
- Landry, V.K.; Minoura, M.; Pang, K.; Buccella, D.; Kelly, B.V.; Parkin, G. Synthesis and structural characterization of 1-mesityl-1,3-dihydro-imidazole-2-selone and bis(1-mesitylimidazol-2-yl)diselenide: Experimental evidence that the selone is more stable than the selenol tautomer. J. Am. Chem. Soc. 2006, 128, 12490–12497. [Google Scholar] [CrossRef]
- Altona, C.; Sundaraligham, M. Conformational analysis of the sugar ring in nucleosides and nucleotides. New description using the concept of pseudorotation. J. Am. Chem. Soc. 1972, 94, 8205–8212. [Google Scholar] [CrossRef] [PubMed]
- Saenger, W. Principles of Nucleic Acids Structure; Springer Verlag: New York, NY, USA, 1984; pp. 1–84. [Google Scholar]
- Davis, D.R. Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies). In Modification and Editing of RNA; Grosjean, H., Benne, R., Eds.; ASM Press: Washington, DC, USA, 1998; chapter 5; pp. 85–102. [Google Scholar]
- Larsen, A.T.; Fahrenbach, A.C.; Sheng, J.; Pian, J.; Szostak, J.W. Thermodynamic insight into 2-thiouridine-enhanced RNA hybridization. Nucleic Acids Res. 2015, 43, 7675–7687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanus, M.; Ryjácek, F.; Kabelác, M.; Kubar, T.; Bogdan, T.V.; Trygubenko, S.A.; Hobza, P. Correlated ab initio study of nucleic acid bases and their tautomers in the gas phase, in a microhydrated environment and in aqueous solution. Guanine: Surprising stabilization of rare tautomers in aqueous solution. J. Am. Chem. Soc. 2003, 125, 7678–7688. [Google Scholar] [PubMed]
- Wang, W.; Hellinga, H.W.; Beese, L.S. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Nat. Acad. Sci. USA 2011, 108, 17644–17648. [Google Scholar] [CrossRef] [Green Version]
- Sponer, J.; Jurecka, P.; Hobza, P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J. Am. Chem. Soc. 2004, 126, 10142–10151. [Google Scholar] [CrossRef]
- Mizuno, H.; Sundaralingam, M. Stacking of Crick wobble pair and Watson-Crick pair: Stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon-anticodon wobble interaction. Nucleic Acids Res. 1978, 15, 4451–4461. [Google Scholar] [CrossRef]
- Singh, U.C.; Kollman, P.A. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 1984, 5, 129–145. [Google Scholar] [CrossRef]
- Duchler, M.; Leszczynska, G.; Sochacka, E.; Nawrot, B. Nucleoside modifications in the regulation of gene expression: Focus on tRNA. Cell. Mol. Life Sci. 2016, 73, 3075–3095. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Vazquez, J.; Vargas-Perez, I.; Sanso, M.; Buhne, K.; Carmona, M.; Paulo, E.; Hermand, D.; Rodríguez-Gabriel, M.; Ayté, J.; Leidel, S.; et al. Modification of tRNA(Lys)UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 2013, 9, e1003647. [Google Scholar] [CrossRef] [Green Version]
- Kurata, S.; Weixlbaumer, A.; Ohtsuki, T.; Shimazaki, T.; Wada, T.; Kirino, Y.; Takai, K.; Watanabe, K.; Ramakrishnan, V.; Suzuki, T. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding. J. Biol. Chem. 2008, 283, 18801–18811. [Google Scholar] [CrossRef] [Green Version]
- Crick, F.H. Codon-anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef]
- Westhof, E.; Yusupov, M.; Yusupova, G. Recognition of Watson-Crick base pairs: Constraints and limits due to geomet ric selection and tautomerism. F1000Prime Rep. 2014, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Leszczynska, G.; Sadowska, K.; Bartos, P.; Nawrot, B.; Sochacka, E. S-Geranylated 2-thiouridines of bacterial tRNAs: Chemical synthesis and physicochemical properties. Eur. J. Org. Chem. 2016, 21, 3482–3485. [Google Scholar] [CrossRef]
- Sochacka, E.; Szczepanowski, R.H.; Cypryk, M.; Sobczak, M.; Janicka, J.; Kraszewska, K.; Bartos, P.; Chwialkowska, A.; Nawrot, B. 2-Thiouracil deprived of thiocarbonyl function preferentially base pairs with guanine rather than adenine in RNA and DNA duplexes. Nucleic Acids Res. 2015, 43, 2499–2512. [Google Scholar] [CrossRef] [Green Version]
- Rajur, S.B.; McLaughlin, L.W. The synthesis of oligodeoxynucleotides containing 2-thiothymine and 5-methyl-4-pyrimidinone base analogues. Tetrahedron Lett. 1992, 33, 6081–6084. [Google Scholar] [CrossRef]
- Collins English Dictionary, 13th ed.; HarperCollins.: New York, NY, USA, 16 November 2018.
- Reich, H.J.; Hondal, R.J. Why Nature chose selenium. ACS Chem. Biol. 2016, 11, 821–841. [Google Scholar] [CrossRef]
- Sugiura, Y.; Hojo, Y.; Tamai, Y.; Tanaka, H. Selenium protection against mercury toxicity. Binding of methylmercury by the selenohydryl-containing ligand. J. Am. Chem. Soc. 1976, 98, 2339–2341. [Google Scholar] [CrossRef]
- Huber, R.E.; Criddle, R.S. Comparison of the chemical properties of selenocysteine and selenocystine with their sulfur analog. Arch. Biochem. Biophys. 1967, 122, 164–173. [Google Scholar] [CrossRef]
- Sierant, M.; Leszczynska, G.; Sadowska, K.; Dziergowska, A.; Rozanski, M.; Sochacka, E.; Nawrot, B. S-Geranyl-2-thiouridine wobble nucleosides of bacterial tRNAs; chemical and enzymatic synthesis of S-geranylated-RNAs and their physicochemical characterization. Nucleic Acids Res. 2016, 44, 10986–10998. [Google Scholar] [CrossRef] [Green Version]
- Guerra, C.F.; Bickelhaupt, F.M. Substituent effects on hydrogen bonds in DNA. In Computational Studies of RNA and DNA in Šponer, F. Lankaš Computational Studies of RNA and DNA; Springer: Dordrecht, The Netherlands, 2006; pp. 463–484S. [Google Scholar]
- Van der Lubbe, S.C.C.; Guerra, C.F. Hydrogen-bond strength of CC and GG pairs determined by steric repulsion: Electrostatics and charge transfer overruled. Chem. Eur. J. 2017, 23, 10249–10253. [Google Scholar] [CrossRef] [Green Version]
- Agris, P.F.; Eruysal, E.R.; Narendran, A.; Väre, V.Y.P.; Vangaveti, S.; Ranganathan, S.V. Celebrating wobble decoding: Half a century and still much is new. RNA Biol. 2018, 15, 537–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea RJHoward, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystal. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parameterization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104–154119. [Google Scholar] [CrossRef] [Green Version]
- Antony, J.; Grimme, S. Density functional theory including dispersion corrections for intermolecular interactions in a large benchmark set of biologically relevant molecules. Phys. Chem. Chem. Phys. 2006, 8, 5287–5293. [Google Scholar] [CrossRef] [PubMed]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J. Comput. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Cordero, B.; Gomez, V.; Platero-Prats, A.E.; Reves, M.; Echeverria, J.; Cremades, E.; Barragan, F.; Alvarez, S. Covalent radii revisited. Dalton Trans. 2008, 21, 2832–2838. [Google Scholar] [CrossRef] [PubMed]
- Boys, S.F.; Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970, 19, 553–566. [Google Scholar] [CrossRef]









| Comp. Number | Abbreviation Name | λmax (nm) | ε1 (dm3·mol−1·cm−1) | ||
|---|---|---|---|---|---|
| pH 3 | pH 7 | pH 8 | |||
| 1 | mnm5Se2U | 223, 314 | 231,302 | 240, 298 | 12669 (303 nm), 6901 (260 nm) |
| 2 | cmnm5Se2U | 223, 314 | 231,302 | 240, 298 | 12695 (310 nm), 4951 (260 nm) |
| 3 | Se2U | 223, 309 | 225,306 | 240, 295 | 13992 (308 nm), 2687 (260 nm) |
| 5 | mnm5S2U | 219, 274 | 221,275 | 240, 276 | 10411 (276 nm), 7283 (260 nm) |
| 6 | cmnm5S2U | 221, 275 | 221,275 | 223, 240, 277 | 12104 (276 nm), 7645 (260 nm) |
| 7 | S2U | 217, 276 | 220,276 | 221, 240, 274 | 14410 (276 nm), 9160 (260 nm) |
| R (Abbreviated Name of the Substituent) | Donor/Acceptor | Nucleoside1 | ||
|---|---|---|---|---|
 |  |  | ||
| CH3NHCH2 (mnm) | N3H | 6.43 (>90, 782) | 7.28 (57, 342) | 8.15 (15) |
| NHCH2 | 9.36 | 9.51 | 10.02 | |
| HOOC-CH2NHCH2 (cmnm) | N3H | 6.55 (89, 742) | 7.36 (52, 302) | 8.24 (13) |
| NHCH2 | 8.89 | 9.10 | 10.13 | |
| COOH | 2.26 | 2.50 | 3.05 | |
| H | N3H | 7.303 (58, 342) | 8.09 (17) | 9.15 (2) |
| Conformational Parameters | Se2U (1) | S2U (7) | U (10) | |
|---|---|---|---|---|
| A | B | |||
| P [o] | 14.4 (N) | 9.7 (N) | 3.8 (N) | 13.9 (N) |
| ψm [o] | 34.7 | 36.5 | 39.6 | 41.6 |
| Sugar moiety | C3′-endo | C3′-endo | C3′-endo C2′-exo | C3′-endo |
| C4′-C5′ bond | trans | trans | gauche(+) | gauche(+) |
| C1′-N1 bond | anti | anti | anti | anti |
| Compound Number | Abbreviation Name | C3′-endo (%) |
|---|---|---|
| 1 | mnm5Se2U | 72 |
| 2 | cmnm5Se2U | 79 |
| 3 | Se2U | 80 |
| 5 | mnm5S2U | 76 1 |
| 6 | cmnm5S2U | 82 1 |
| 7 | S2U | 71 1 |
| 8 | mnm5U | 57 1 |
| 9 | cmnm5U | 58 1 |
| 10 | U | 53 1 |
| Base Pair Mode | ΔH298 of a Base Pair of m9Gua with m1R5X2Ura Component (kcal/mol) | ||||||
|---|---|---|---|---|---|---|---|
| R5 | H | mnm | |||||
| X | O1 | S1 | Se | O1 | S1 | Se | |
| UK-G (wobble) | −10.0 | −8.1 | −7.9 | −10.2 | −8.4 | −8.4 | |
| UE4-G (C–U-TG-like) | −7.8 | −6.6 | −6.2 | −7.6 | −6.4 | −6.8 | |
| UZI-G (new wobble) | - | - | - | −5.9 | −7.3 | −8.6 | |
| X | O | S | Se |
|---|---|---|---|
| Charge transfer [e] | 0.093 | 0.170 | 0.243 |
| Δq = qX-qO [e] | 0 | 0.077 | 0.150 (qSe-qO) 0.073 (qSe-qS) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leszczynska, G.; Cypryk, M.; Gostynski, B.; Sadowska, K.; Herman, P.; Bujacz, G.; Lodyga-Chruscinska, E.; Sochacka, E.; Nawrot, B. C5-Substituted 2-Selenouridines Ensure Efficient Base Pairing with Guanosine; Consequences for Reading the NNG-3′ Synonymous mRNA Codons. Int. J. Mol. Sci. 2020, 21, 2882. https://doi.org/10.3390/ijms21082882
Leszczynska G, Cypryk M, Gostynski B, Sadowska K, Herman P, Bujacz G, Lodyga-Chruscinska E, Sochacka E, Nawrot B. C5-Substituted 2-Selenouridines Ensure Efficient Base Pairing with Guanosine; Consequences for Reading the NNG-3′ Synonymous mRNA Codons. International Journal of Molecular Sciences. 2020; 21(8):2882. https://doi.org/10.3390/ijms21082882
Chicago/Turabian StyleLeszczynska, Grazyna, Marek Cypryk, Bartlomiej Gostynski, Klaudia Sadowska, Paulina Herman, Grzegorz Bujacz, Elzbieta Lodyga-Chruscinska, Elzbieta Sochacka, and Barbara Nawrot. 2020. "C5-Substituted 2-Selenouridines Ensure Efficient Base Pairing with Guanosine; Consequences for Reading the NNG-3′ Synonymous mRNA Codons" International Journal of Molecular Sciences 21, no. 8: 2882. https://doi.org/10.3390/ijms21082882
APA StyleLeszczynska, G., Cypryk, M., Gostynski, B., Sadowska, K., Herman, P., Bujacz, G., Lodyga-Chruscinska, E., Sochacka, E., & Nawrot, B. (2020). C5-Substituted 2-Selenouridines Ensure Efficient Base Pairing with Guanosine; Consequences for Reading the NNG-3′ Synonymous mRNA Codons. International Journal of Molecular Sciences, 21(8), 2882. https://doi.org/10.3390/ijms21082882