Abstract
Owing to a sessile lifestyle in nature, plants are routinely faced with diverse hostile environments such as various abiotic and biotic stresses, which lead to accumulation of free radicals in cells, cell damage, protein denaturation, etc., causing adverse effects to cells. During the evolution process, plants formed defense systems composed of numerous complex gene regulatory networks and signal transduction pathways to regulate and maintain the cell homeostasis. Among them, ubiquitin-proteasome pathway (UPP) is the most versatile cellular signal system as well as a powerful mechanism for regulating many aspects of the cell physiology because it removes most of the abnormal and short-lived peptides and proteins. In this system, the ubiquitin-conjugating enzyme (E2) plays a critical role in transporting ubiquitin from the ubiquitin-activating enzyme (E1) to the ubiquitin-ligase enzyme (E3) and substrate. Nevertheless, the comprehensive study regarding the role of E2 enzymes in plants remains unexplored. In this review, the ubiquitination process and the regulatory role that E2 enzymes play in plants are primarily discussed, with the focus particularly put on E2′s regulation of biological functions of the cell.
1. Introduction
Researchers have successfully extracted a small molecular protein (initially called APF-1 and later renamed as “Ubiquitin”) from reticulocytes, and this protein is a key factor in ATP-dependent substrate proteolysis and is universally present in many organisms [1,2,3,4]. Based on these experiments and later research findings, the Nobel Prize in Chemistry was awarded to Aaron Ciechanover, Avram Hershko and Irwin Rose in 2004, for their outstanding contributions to the discovery of ubiquitin-mediated protein degradation [5]. From then onwards, an increasing number of researchers devoted themselves to the ubiquitin-proteasome system (UPS). As a result, the cascade reaction process of this has been basically elucidated at present. Previous studies have shown that the ubiquitin-proteasome system regulates various biological functions of the plant as diverse as hormone responses (including auxin, jasmonic acid, gibberellins, ethylene, and abscisic acid) [6,7,8,9,10,11], abiotic stress response [12,13,14,15], plant growth and development [16,17], circadian rhythm [18], and plant immunity [19,20]. However, the nonproteolytic functions and mechanisms of most key enzymes (over 1600 loci in Arabidopsis thaliana genome) which are core components of the UPS (Ubiquitin-proteasome system) are still unclear, especially E3s (more than 1400 potential E3s are encoded in A. thaliana genome) and E2s (37 E2s are encoded in A. thaliana genome) [8,21]. Therefore, it will be an important task for future research to uncover the role of these enzymes.
2. Ubiquitin
Ubiquitin, a small protein of 76 amino acids acting as a tag in post-translational modification of proteins, is absolutely conserved in vertebrates and higher plants, and only two or three amino acid residues are different even among animals, plants, and fungi [22]. The high conservatism renders ubiquitin interchangeable and universal in different species [23]. Ubiquitin is a β-grasp fold protein consisting of a 3.5-turn α-helix, a short 310 helix against a five-strand mixed β-sheet and seven reverse turns [24]. Ubiquitin possesses two hydrophobic surfaces. One surface consists of Ile44, Leu8, Val70, and His68 [25], and the other focuses on Ile36 that can be recognized by HECT (Homology to E6-AP C terminus) E3s, DUBs (Deubiquitinating enzymes), and UBDs (Ubiquitin binding domains) [26,27,28]. Ubiquitin contains seven lysine residues, including Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, Lys-63 (K6, K11, K27, K29, K33, K48, and K63, respectively) [29,30]. Six of the seven Lys linkages (with a preference for Lys-48>Lys-63>Lys-11>>>Lys-33/Lys-29/Lys-6) were detected in A. thaliana [31,32]. Any one of the seven lysine residues or N-terminal Met1 residues in ubiquitin is likely to be involved in ubiquitin chain formation in vivo, and different ubiquitin linkages lead to the formation of different types of ubiquitin chains and induce diverse molecular signals in the cell [29,33,34,35,36]. Different ways of linking ubiquitin chains and substrates result in distinct fates of the substrates. It is well-known that K48-linked chains are mediators signaling proteasomal degradation and have diverse functions in the cell. For example, they respond to the abiotic/biotic environments [21,37,38]. Some researchers found that K11-linked chains also served as targeting signal for proteasomal degradation in vitro and in vivo [39]. Besides, it was observed in some studies that K63-linked chains not only served as a targeting signal for protein degradation [40], but also had non-degradative functions, such as DNA repair and NF-kb signaling, activation of protein kinases, and protein trafficking [37,41]. Lys-33-linked chains might have nothing to do with protein degradation because they did not increase if the proteasome was inhibited [37], However, Lys-33-linked chains were reported to have nonproteolytic functions, such as, negatively regulating the activity of AMPK (AMP-activated protein kinase) [42]. However, the biological functions of ubiquitin-ubiquitin linkages composed of K6, K27, and K29 are less understood. There is very limited information available on mechanisms and biological functions of different polyubiquitin linkages in prior studies, so researchers must focus on the mechanisms and action processes of polyubiquitination in the future.
In comparison, the small ubiquitin-related modifier (SUMO) which contains approximately 100–115 amino acids is similar to ubiquitin. The SUMO emerges as an influential class of conjugation of protein tags or a modifier to target proteins, and evidence proves that it plays a regulatory role in transcriptional control, subcellular trafficking, and cell cycle [43,44,45]. SUMOylation is driven by three components, which are heterodimeric SUMO-activating enzyme (E1), SUMO-conjugating enzyme (E2), and the substrate recognition factor (E3) [46,47]. SUMOylation is a reversible process, and the SUMO can be released from protein conjugates by SUMO proteases for SUMO recycling [45,48].
3. The Ubiquitin Enzymes and Ubiquitination Reactions
The ubiquitin-proteasome system/ubiquitin-proteasome pathway is a prevalent protein turnover system that can degrade or modify proteins in eukaryotic cells. The system can modulate numerous cellular processes, and is almost involved in all aspects of growth and development, so the modification of ubiquitination may be more important than modification of phosphorylation. The UPS consists of ubiquitin (Ub), ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin-ligase enzymes (E3), 26S proteasome, DUBs and target proteins [49,50,51], and it involves two consecutive processes. A chain of ubiquitin molecules selects and labels abnormal and short-lived peptides proteins and substrates at first, then the targets are broken down by the 26S proteasome, and finally the ubiquitin molecules are recycled for a further ubiquitination cycle.
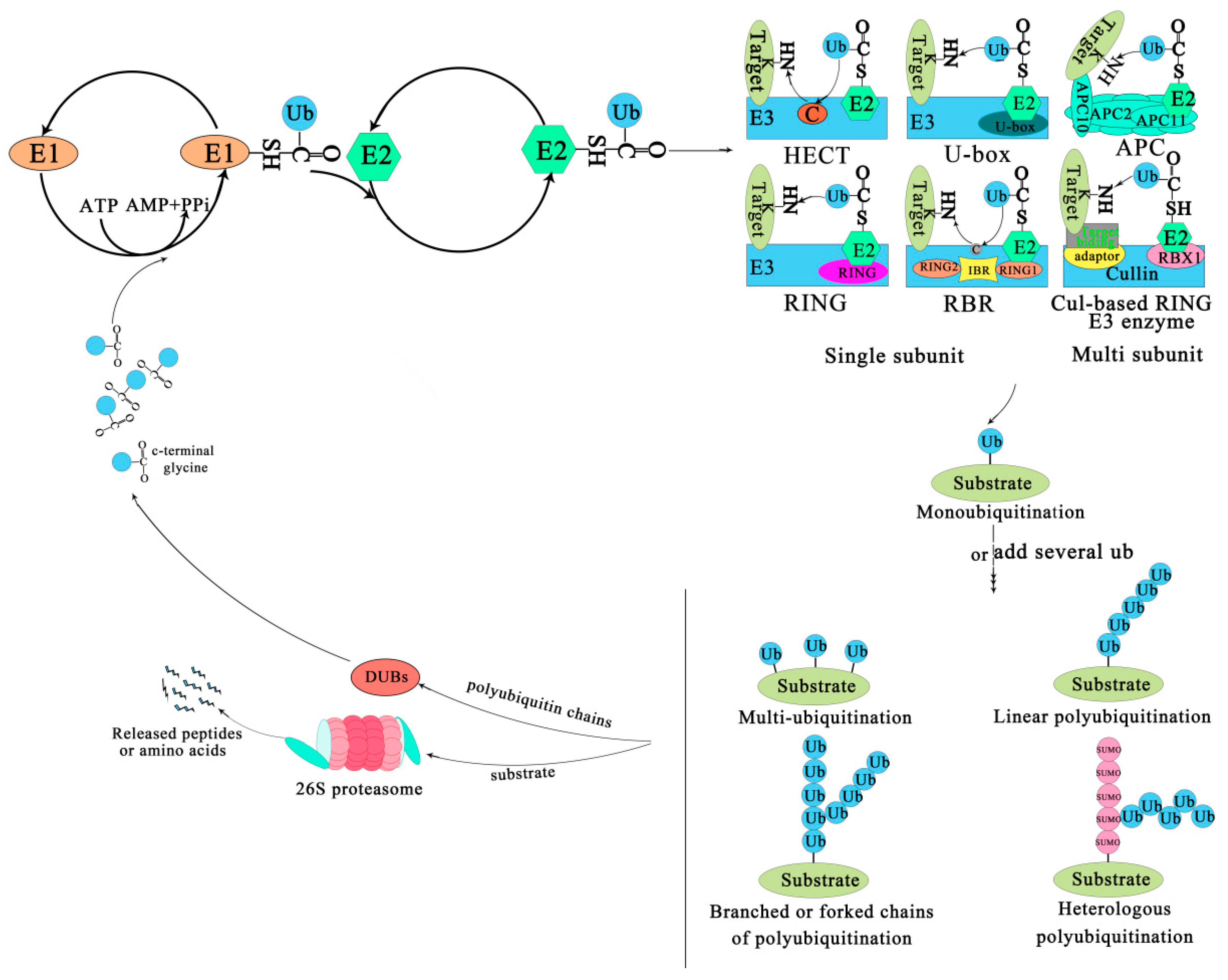
The covalent attachment of ubiquitin to specific target proteins is mainly accomplished by stepwise enzymatic cascade reactions, and ubiquitin is attached to the substrates via the concerted action of ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) [52,53,54]. The attachment of ubiquitin or ubiquitin chains to the substrate is a successive process (Figure 1). First, an E1-ubiquitin thioester bond is formed between the C-terminal Gly carboxyl group of ubiquitin and the active site Cys of the E1 enzyme by an ATP-dependent reaction. Then, the E1 transfers the activated ubiquitin to the Cys residue of the E2 enzyme to form an E2-ubiquitin thioester-linked intermediate by transesterification. Eventually the E2 transfers the ubiquitin to the substrate protein by E3 [55]. Ubiquitin is conjugated to the target protein through an isopeptide bond between its C-terminal glycine (Gly76) and the ε-amino group of a lysine residue [56]. There are three typical ways of linking the ubiquitin with the substrate. The first is called mono-ubiquitination, which refers to the modification of one site of the modification of a substrate by a single ubiquitin molecule. The second is multi-mono-ubiquitination, which means adding several ubiquitin molecules repetitively to distinct sites (multi-mono-ubiquitination). The third is called polyubiquitination (including linear polyubiquitination and branched polyubiquitination), in which ubiquitin molecules are added to the same site (polyubiquitination, including linear polyubiquitination and branched polyubiquitination) of a substrate. In the second and third ways of linking, the previously attached ubiquitin serves as the “acceptor” of subsequently added ubiquitin [51,54,57,58]. Of course, polyubiquitin chains linked by the same Lys are homogeneous, while those linked by different Lys are heterogeneous or mixed ones [59,60]. Subsequently, the substrate complex tagged by the ubiquitin is degraded by the 26S proteasome or executes nonproteolytic functions, such as some specific biological functions [41]. In most cases, the modified protein (polyubiquitination) is recognized and degraded by the 26S proteasome, and the ubiquitin or ubiquitin chain hydrolyzed and freed by deubiquitinating enzymes (DUBs) for further conjugation cycles after being removed from the substrate protein [61] (Figure 1). Nevertheless, the multi-mono-ubiquitination and mono-ubiquitination that can recruit binding partners, inhibit interactions, alter protein localization, or regulate protein activities [51,62] are necessary for different stages of the secretory/endocytic pathway when certain cargo proteins enter vesicles [51].
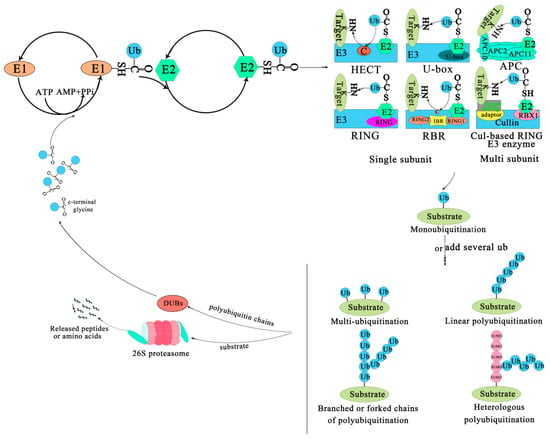
Figure 1.
The ubiquitin-proteasome system. The process of ubiquitination from activation to attachment to the substrate is catalyzed by three major enzymes. The substrates labeled by ubiquitin are degraded by the 26S proteasome or play a non-degradative role in other processes. Abbreviations: APC, Anaphase-promoting complex; DUBs, Deubiquitinating enzymes; E1, Ubiquitin-activating enzyme; E2, Ubiquitin-conjugating enzyme; E3, Ubiquitin-ligase enzyme; Cul-based, Cullin-RING box1-Ligase; HECT, Homology to E6-AP C Terminus; Ub, Ubiqitin; SUMO, Small ubiquitin-related modifier; RBX1, RING-Box 1; RING, Really interesting new gene; RBR, RING1-IBR(cysteine/histidine rich region)-RING2.
The family of E3 enzymes is large and diverse. Over 1400 E3s have been found in A. thaliana and they can be divided into U-box domain, RING (Really interesting new gene), HECT (Homologous to the E6AP carboxyl terminus) and Cullin–RING ligases (CRLs) [63,64]. CRLs are subdivided into SKP1-Cullin-F-box (SCF) type, broad complex/tramtrack/bric-a-brac (BTB) type, DDB1-binding/WD-40 domain-containing proteins (DWD) type, and anaphase-promoting complex (APC) [21]. An E3 is a protein or protein complex that conjugates to an E2 and a substrate. It can interact with the substrate directly or through substrate recognition modules of seven types of E3 ligases via the adaptor and target binding proteins (Figure 1). For example, the HECT enzyme transfers ubiquitin from an E2 to its Cys residues before it binds to the target protein NH2 conjugate [65,66]. The RING/U-box enzyme acts as a scaffold for the aggregation of an E2 and a substrate, directly transferring ubiquitin from an E2 to a substrate; Cul-based RING enzyme transfers ubiquitin from E2 to the substrate that binds with an accessory factor.
4. The Family of E2 Enzymes
The E2 enzyme is a key transfer site of ubiquitination, which interacts with E1 and E3 respectively. There are two puzzling questions about the ubiquitin-proteasome system. One is why E2 enzymes are present in all eukaryotes and the other is why we do not directly transfer ubiquitin from E1s to E3s. Some researchers believed that an E2 enzyme could combine with different types of E3s that recognized and selected distinct targets, and thereby played a decisive role in determining whether the labeled protein would be degraded or involved in nonproteolytic processes. In other words, E3s mainly select the substrate, while E2s determine the fate of the substrate [67]. Another possible and interesting consideration is that E2s promote ubiquitination independent of an E3 ligase, such as UBC22 [68,69].
Each E2 contains a core UBC domain composed of about 150–200 amino acid residues, in which the cysteine is an active site for the formation of thioester linkages [54,70]. The UBC domain has a structural conservation rate of 35% in different species [71], consisting mainly of four α-helices, an anti-parallel β-sheet and a short 310 helix [72,73,74]. Almost all UBC domains contain a conserved His-Pro-Asn tripeptide (HPN) located at about the tenth residue at the N-terminal of the Cys residue [2,75]. Aspartic acid in HPN catalyzes the formation of the isopeptide bond, histidine is essential for the structure, and proline promotes stable transition of these amino acid residues [75,76]. According to the location of the additional fragment in the UBC domain, the family of E2 enzymes can be categorized into four classes. Class I is present only in the UBC domain; class II is present in both N-terminal and UBC domain; class III is present in both C-terminal and UBC domain; and class IV is present in N-terminal, C-terminal and UBC domain [67]. E2 enzymes belong to the polygenic family, and the number of members varies from a dozen to dozens in different species. 20 E2 enzymes have been found in Caenorhabditis elegans [77], 40 in A. thaliana [68], 14 in Saccharomyces cerevisiae [78], 37 in human [67], 48 in rice [79], 75 in maize [80], 72 in banana [81], 43 in Vitis vinifera [82], 34 in Carica papaya [83], 59 in tomato [84], 40 in Dimocarpus longan Lour. [85], and 57 in potato [86].
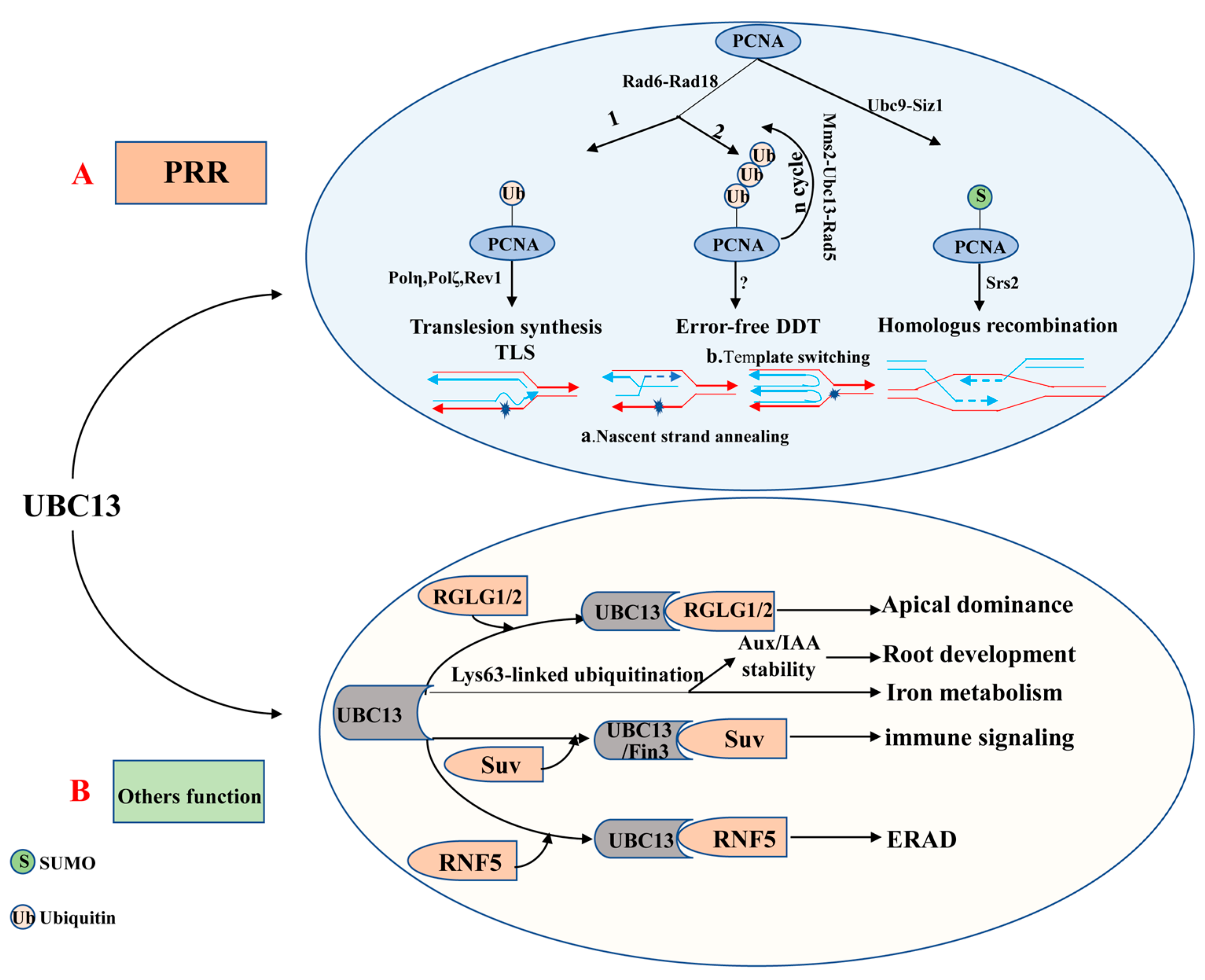
The ubiquitin E2 variant (UEV) is similar to the E2 enzyme in both sequence and structure, but it cannot form a thioester linkage with ubiquitin because of the lack of the catalytic cysteine residue [87,88]. The first UEV gene was initially defined as MMS2, which was obtained from budding yeast cells and associated with the error-free DNA-damage tolerance (DDT) pathway [87,89]. Although UEVs cannot conjugate with ubiquitin, UEV proteins form a stable heterodimer with E2s to facilitate the ubiquitination of a target protein [68,90,91]. It is clear that UBC13 usually combines with a heterologous UEV to form a complex which plays a critical role in DNA repair. The heterodimer-coupled E3 Rad5, for instance, can functionally remedy the corresponding ubc13 mutants’ defect in PRR (post-replication repair, also known as error-free DNA-damage tolerance) [87,92,93] (Figure 2A). The newly discovered expression of OsUEV1s is also capable of functionally rescuing the yeast mms2 mutant from death caused by DNA-damaging agents [94]. Much more importantly, the stable heterodimer formed by UBC13 and a UEV (from yeast or mammalian cells) is essential for assembly of Lys63-linked polyubiquitin chains [91,93,95]. 4 AtUev1s, 4 OsUev1s, and 3 BdUev1s have been found in A. thaliana, rice and Brachypodium distachyon, respectively [94,96,97]. COP10 is a type of UEV, which is essential to protein degradation [98]. Besides, COP10 plays a vital role in photo-morphogenesis and response against UV-B [90]. It remains unknown whether UEVs have other independent biological functions or play a role in regulating biological processes together with E2 or E3 enzymes. Therefore, this will be the direction for research in this field shortly.
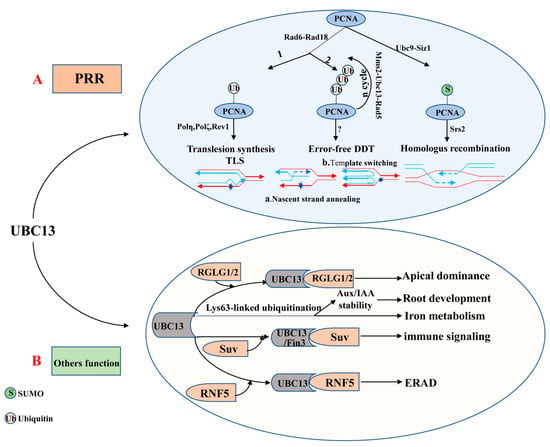
Figure 2.
The role of UBC13 in DNA damage responses and UBC13′s other functions. (A) UBC13 is involved in post-replication repair (PRR). (B) UBC13 participates in different biological processes by mediating Lys63-linked ubiquitination. Abbreviations: Aux/IAA, Auxin/indole-3-acetic acid; DDT, DNA damage tolerance; ERAD, ER-associated degradation; Mms2, A Ubc E2 variant; PRR, Postreplication repair; PCNA, Proliferating cell nuclear antigen; RGLG1/2, An E3 ligase; RNF5, RING finger protein 5; Rad5, An E3 enzyme; Rad6, An E2 enzyme; Rad18, An E3 enzyme; Suv, Fni3 (Fen-interacting protein, Fen is the host protein kinase) cofactor S. lycoperiscum Uev; Siz1, An E3 ligase; Srs2, A helicase; TLS, Translesion DNA synthesis; Ubc9, An E2 enzyme.
5. Functions of E2 Enzymes
The E2 enzymes are one of the key parts of the ubiquitin-proteasome system, having special significance and functions (Table 1).

Table 1.
Ubiquitin-conjugating enzymes with different known functions a.
5.1. Abiotic Stress Response
VrUBC1 is a ubiquitin-conjugating enzyme gene from mung bean, and responds to dehydration, high salinity and abscisic acid (ABA) treatment. VrUBC1-overexpressed plants are more sensitive to abscisic acid-mediated stomatal closure and have more tolerance to drought stress, indicating that VrUBC1 is a positive regulator under osmotic stress in A. thaliana [99]. OgUBC1 is a ubiquitin-conjugating enzyme gene from wild rice, triggered by salicylic acid in leaves [100]. Tobacco with overexpression of NtUBC1 exhibits increased resistance to cadmium (Cd) compared to WT [101]. A significantly increased expression of LeUBC1 under the conditions of heat shock and cadmium chloride (CdCl2) indicates that LeUBC1 can respond to these two stresses [102]. AhUBC2 is a ubiquitin-conjugating enzyme gene in peanuts, which can respond to PEG6000, high salinity, abscisic acid (ABA) and low temperature. Higher levels of P5CS1, RD29A, and KIN1 under a normal condition, and a higher level of proline under both soil-drought stress and control conditions are found in 35S:AhUBC2 transgenic plants, suggesting that the constitutive expression of AhUBC2 improves water-stress tolerance by activating an ABA-independent signaling pathway [103]. The GmUBC2 from soybean is the yeast homolog Rad6, capable of improving salt and drought tolerance, and it is involved in the regulation of ion homeostasis, osmolyte synthesis, and oxidative stress responses in A. thaliana [104]. Similarly, the increased expression of CmUBC in transcript level in melon plants under drought and salinity stresses indicates that CmUBC can respond to physiological water stress [105]. The Rad6 gene of Hevea brasiliensis shares a similarity of over 96% with OsRad6 in rice and AtUBC2 in A. thaliana, and its expression can be markedly induced by the latex stimulator ethephon (ET) and methyl jasmonate (MeJA) [106]. RCE1 from A. thaliana is homologous with the human UBC12 gene, and its conjugation with the ubiquitin-like protein RUB1 that combines with AtCUL1 affects the functions of SCF E3s and thus influences the auxin response [107,108]. A study showed stable and high expression of OsUBC13 under different biotic and abiotic stresses, suggesting OsUBC13 as a housekeeping gene [109]. Recent research has shown that UBC13 is a key regulator of A. thaliana response to low temperature [110] (Figure 2B). UBC18 can interact with ethylene response factor 1 (ERF1), and the differences in abundance of ERF1 between UBC18 mutants and UBC18 overexpression lines are caused by ubiquitination. Down-regulation of UBC18 leads to increased ERF1 and proline accumulation. Besides, UBC18 negatively regulates responses to drought and salt stresses [111]. AtUBC32, AtUBC33, and AtUBC34 can interact with PUB19, a negative regulator of the drought stress response. Repression of AtUBC32, AtUBC33, and AtUBC34 expression in both single and triple mutant lines can promote abscisic acid-mediated stomatal closure and improve tolerance to drought stress, indicating that AtUBC32, AtUBC33, and AtUBC34 play a negative regulatory role in drought stress through mediation of the abscisic acid [112]. AtUBC32, AtUBC33, and AtUBC34 belong to group XIV UBCs, and UBC32-GFP is located in ER membranes [113]. UBC32 can interact with the RING ligase DOA10B (At4g32670) which is remarkably similar to yeast Doa10 (Degradation of Alpha2). UBC32 mutants are extremely sensitive to paraquat treatment compared with overexpression plants, suggesting that UBC32 is an essential ERAD component and plays an important part in the response to oxidative stress and salt stress [114]. Although changes in expression of UBC genes in A. thaliana and rice are not induced by cold stress treatment [103], some reports have found that the UBCs gene in other species have responses to cold and heat stresses [80,82]. UBC24 (pho2) is regulated by microRNA399 (miR399) to control inorganic phosphate (Pi) homeostasis and Pi translocation and remobilization [115,116,117,118].
5.2. Plant Immune Responses
Fni3, Lys-63-linked ubiquitination, is a homolog of the UBC13-type ubiquitin-conjugating enzyme. A decreased expression of Fni3 or the ubiquitin E2 variant Suv can affect Fen-mediated immunity and cell death caused by several other resistance (R) proteins and their cognate effectors (Figure 2B). This study suggests that Fni3/Sl-Ubc13-2 and Suv positively regulate plant immunity [128]. By knocking down the Group III E2 genes in Nicotiana benthamiana via VIGS (gene silencing), it is found that these genes are necessary for plant development, immunity-associated ROS production, and suppression of immunity-associated PCD (programmed cell death) by AvrPtoB [134,135]. Besides, knocking down the expression of TaU4 gene which encodes a ubiquitin-conjugating enzyme 4 in Triticum aestivum by VIGS can slow down the development of disease symptoms and reduce Septoria sporulation (Mycosphaerella graminicola, commonly known as Septoria), indicating that TaU4 is a negative regulator for defense against the phytopathogen Septoria in wheat [123]. The group III E2 genes act together with SlPUB13 on the ubiquitination of FLS2 in vitro and regulate FLS2-mediated immune signaling [136]. Heterologous expression of the OgUBC1 in A. thaliana defends plants from UV-B-mediated cell damage and enables them to resist Botrytis cinerea [100].
5.3. Plant Growth and Development UBC13 Catalyzes Non-Canonical Lys63-Linked Ubiquitin Chains
A. thaliana carrying UBC13 mutants can display strong phenotypes and is unable to grow root hairs in case of iron deficiency. Especially, A. thaliana containing UBC13A and UBC13B mutants have a significantly reduced root-hair density. Therefore, it is concluded that UBC13A/B is involved in epidermal cell differentiation and iron deficiency responses [117,137]. UBC13A/B mutants of A. thaliana can influence primary and lateral roots [137,138] (Figure 2B). A VIGS analysis showed that SlUBC32 and SlUBC41 were implicated in the regulation of fruit ripening [139]. UBC22 is the sole member of subfamily X, and can catalyze the assembly of ubiquitin chains in the absence of E3 in vitro [68]. Silique length and the seed numbers are significantly decreased and nearly 90% of the ovules are aborted after knocking out UBC22 gene mutants in A. thaliana, indicating that UBC22 is required for the development of female gametophytes [140]. The RING finger protein TaRF1 can interact with TaUBC28, which potentially plays a role in spike development of wheat [131]. UBC21 is better known as PEROXIN 4 (PEX4). Pex4/ubc21 mutants have reduced root elongation because of their resistance to indole butyric acid (IBA), while the major auxin indoleacetic acid (IAA) processed from IBA can significantly promote the root growth of pex4 mutants [129]. There are two E3 homologs (HUB1 and HUB2) and three RAD6 homologs (UBC1, UBC2, and UBC3) in A. thaliana [120]. As reported by researchers previously, HUB1 and HUB2 were involved in regulating seed dormancy, leaf and root growth [141,142]. The ubc1 and ubc2 mutants displayed an early-flowering phenotype, and they worked together with HUB1 and HUB2 to control the flowering time of A. thaliana [141,143,144]. Similar studies involving AtUBC1/AtUBC2 and HUB1/HUB2 also have shown that these four genes mediate the H2B ubiquitination, thereby activating FLC expression and repressing flowering of the plant [119].
5.4. Error-Free DNA-Damage Tolerance and Repair
To date, UBC13 is the only E2 known to be able to catalyze K63-linked polyubiquitin chain assembly in eukaryotes [93,145,146]. In most cases, the K63-linked polyubiquitin chain assembled combines with a UEV (e.g., MMS2) as a cofactor [93,95,147]. Evidence is accumulating to suggest that the UBC13-MMS2 complex is involved in DTT (also known as DNA post-replication repair) and DNA repair [147,148,149,150] (Figure 2A). DNA radiation repair pathways are mainly divided into nucleotide excision repair, homologous recombination repair, and the damage tolerance pathway initially. Interestingly, the damage tolerance pathway is different from the other two major pathways as it does not remove or repair the original lesion but bypass the lesion [151]. It is now clear that PRR includes two branches, namely the error-prone (mutagenesis) branch and the error-free branch [87,152,153]. The error-prone branch is also called DNA translation synthesis (TLS), which is mediated by non-essential DNA polymerases including Polη (Rev3+Rev7), Polζ, and Rev1. By contrast, the error-free branch is mediated by the current model Ubc13-Mms2 [92,154,155]. These two independent sub-pathways typically rely on RAD6 and RAD18 [149,156], which are recruited to the stalled replication fork by RPA to facilitate mono-ubiquitination of the proliferating cell nuclear antigen (PCNA). Rad6 from Saccharomyces cerevisiae is a ubiquitin-conjugating enzyme (E2), which is proven to play a role in DNA repair, damage-induced mutagenesis, and sporulation [157,158]. In fact, for the former, PCNA encoded by Pol30 at the K164 is mono-ubiquitinated by the Rad6-Rad18 to promote TLS, while for the latter, mono-ubiquitin-POL30 is further poly-ubiquitinated by the Mms2-Ubc13-Rad5 complex to facilitate error-free damage tolerance [159,160]. Besides, the PCNA can also be modified by the SUMO at K164 under the catalysis of the E2-E3 complex Siz1-Ubc9 [159,161]. The DNA helicase Srs2 enters the stalled replication forks under the mediation effect of the SUMOylated PCNA, suppressing the redundant and harmful homologous recombination [162,163,164].
6. Concluding Remarks and Future Perspectives
E2 enzymes are the intersection between E1 and E3 enzymes and determine the ubiquitination of specific target proteins by tagging different types of E3 enzymes. This review elaborates on some of the reported functions of E2 enzymes. Since the direct interaction between E3 enzymes and substrates results in the substrate ubiquitination, previous studies mainly focused on the mechanisms and functions of E3 enzymes. E3-mediated substrate degradation has always been the concern of research, but E2 enzymes that play a crucial role in this mechanism have not gotten much attention. The present model of ubiquitination only expounds the process of Ub transferring. The findings on the driving mechanism of E2 enzymes recognizing diverse E3 and specific substrates are very limited. Moreover, it is wondered whether E2 enzymes have other biological functions independent of the ubiquitin-proteasome system, such as regulating other signaling pathways to regulate the expression of downstream genes through signaling pathways. Much more importantly, only a few studies have investigated the role of E2 enzymes, especially in plants. Therefore, future research should focus on clarifying above important issues and explore new concerned research directions.
Author Contributions
Writing—original draft preparation, W.L., X.T., and X.Q., X.F.; writing—review and editing, S.G., R.M., S.L., N.Z., H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by National Natural Science Foundation of China (31860399) and Potato Industry Technology System of Gansu Province (GARS-03-P1).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ciehanover, A.; Hod, Y.; Hershko, A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem. Biophys. Res. Commun. 1978, 81, 1100–1105. [Google Scholar] [CrossRef]
- Callis, J. The ubiquitination machinery of the ubiquitin system. Arab. Book Am. Soc. Plant Biol. 2014, 12, e0174. [Google Scholar] [CrossRef]
- Ciechanover, A.; Heller, H.; Elias, S.; Haas, A.L.; Hershko, A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. USA 1980, 77, 1365. [Google Scholar] [CrossRef]
- Ciechanover, A.; Elias, S.; Heller, H.; Ferber, S.; Hershko, A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J. Biol. Chem. 1980, 255, 7525–7528. [Google Scholar]
- Wilkinson, K.D. The discovery of ubiquitin-dependent proteolysis. Proc. Natl. Acad. Sci. USA 2005, 102, 15280. [Google Scholar] [CrossRef]
- Chen, H.; Ma, B.; Zhou, Y.; He, S.-J.; Tang, S.-Y.; Lu, X.; Xie, Q.; Chen, S.-Y.; Zhang, J.-S. E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc. Natl. Acad. Sci. USA 2018, 115, 4513. [Google Scholar] [CrossRef]
- Nagels Durand, A.; Pauwels, L.; Goossens, A. The ubiquitin system and jasmonate signaling. Plants 2016, 5, 6. [Google Scholar] [CrossRef]
- Wang, F.; Deng, X.W. Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Res. 2011, 21, 1286–1294. [Google Scholar] [CrossRef]
- He, F.; Wang, H.-L.; Li, H.-G.; Su, Y.; Li, S.; Yang, Y.; Feng, C.-H.; Yin, W.; Xia, X. PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant Biotechnol. J. 2018, 16, 1514–1528. [Google Scholar] [CrossRef]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef]
- Qiao, H.; Chang, K.N.; Yazaki, J.; Ecker, J.R. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009, 23, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Jang, I.-C.; Seo, H.S. COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity. Front. Plant Sci. 2016, 7, 1182. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, W.T. Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 2013, 587, 2584–2590. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci 2014, 5, 135. [Google Scholar] [CrossRef]
- Cho, S.K.; Ryu, M.Y.; Seo, D.H.; Kang, B.G.; Kim, W.T. The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol. 2011, 157, 2240. [Google Scholar] [CrossRef]
- Koops, P.; Pelser, S.; Ignatz, M.; Klose, C.; Marrocco-Selden, K.; Kretsch, T. EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 5547–5560. [Google Scholar] [CrossRef] [PubMed]
- Gil, K.-E.; Kim, W.-Y.; Lee, H.-J.; Faisal, M.; Saquib, Q.; Alatar, A.A.; Park, C.-M. ZEITLUPE contributes to a thermoresponsive protein quality control system in Arabidopsis. Plant Cell 2017, 29, 2882. [Google Scholar] [CrossRef]
- Lin, S.-S.; Martin, R.; Mongrand, S.; Vandenabeele, S.; Chen, K.-C.; Jang, I.-C.; Chua, N.-H. RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J. 2008, 56, 550–561. [Google Scholar] [CrossRef]
- Luo, H.; Laluk, K.; Lai, Z.; Veronese, P.; Song, F.; Mengiste, T. The Arabidopsis botrytis susceptible1 interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 2010, 154, 1766. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Callis, J.; Carpenter, T.; Sun, C.W.; Vierstra, R.D. Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 1995, 139, 921. [Google Scholar]
- Ling, R.; Colón, E.; Dahmus, M.E.; Callis, J. Histidine-tagged ubiquitin substitutes for wild-type ubiquitin in saccharomyces cerevisiae and facilitates isolation and identification of in vivo substrates of the ubiquitin pathway. Anal. Biochem. 2000, 282, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Vijay-Kumar, S.; Bugg, C.E.; Cook, W.J. Structure of ubiquitin refined at 1.8Åresolution. J. Mol. Biol. 1987, 194, 531–544. [Google Scholar] [CrossRef]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains—From structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef]
- Kamadurai, H.B.; Souphron, J.; Scott, D.C.; Duda, D.M.; Miller, D.J.; Stringer, D.; Piper, R.C.; Schulman, B.A. Insights into ubiquitin transfer cascades from a structure of a UbcH5B∼ubiquitin-HECTNEDD4L complex. Mol. Cell 2009, 36, 1095–1102. [Google Scholar] [CrossRef]
- Hu, M.; Li, P.; Li, M.; Li, W.; Yao, T.; Wu, J.-W.; Gu, W.; Cohen, R.E.; Shi, Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 2002, 111, 1041–1054. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Horton, J.R.; Mullally, J.E.; Heroux, A.; Cheng, X.; Wilkinson, K.D. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 2006, 124, 1197–1208. [Google Scholar] [CrossRef]
- Ikeda, F.; Dikic, I. Atypical ubiquitin chains: New molecular signals. EMBO Rep. 2008, 9, 536–542. [Google Scholar] [CrossRef]
- Iwai, K.; Tokunaga, F. Linear polyubiquitination: A new regulator of NF-κB activation. EMBO Rep. 2009, 10, 706–713. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 2013, 25, 1523. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Vert, G. Unraveling K63 polyubiquitination networks by sensor-based proteomics. Plant Physiol. 2016, 171, 1808. [Google Scholar] [CrossRef] [PubMed]
- Trempe, J.-F. Reading the ubiquitin postal code. Curr. Opin. Struct. Biol. 2011, 21, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Dikic, I. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Baumann, K. Lys33-linked ubiquitin in post-Golgi transport. Nat. Rev. Mol. Cell Biol. 2014, 15, 365. [Google Scholar] [CrossRef]
- Kristariyanto, Y.A.; Choi, S.-Y.; Rehman, S.A.A.; Ritorto, M.S.; Campbell, D.G.; Morrice, N.A.; Toth, R.; Kulathu, Y. Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations. Biochem. J. 2015, 467, 345–352. [Google Scholar] [CrossRef]
- Xu, P.; Duong, D.M.; Seyfried, N.T.; Cheng, D.; Xie, Y.; Robert, J.; Rush, J.; Hochstrasser, M.; Finley, D.; Peng, J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009, 137, 133–145. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef]
- Saeki, Y.; Kudo, T.; Sone, T.; Kikuchi, Y.; Yokosawa, H.; Toh-e, A.; Tanaka, K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009, 28, 359–371. [Google Scholar] [CrossRef]
- Chen, Z.J.; Sun, L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 2009, 33, 275–286. [Google Scholar] [CrossRef]
- Alhakim, A.K.; Zagorska, A.; Chapman, L.; Deak, M.; Peggie, M.; Alessi, D.R. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 2008, 411, 249–260. [Google Scholar] [CrossRef]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512. [Google Scholar] [CrossRef]
- Miller, M.J.; Scalf, M.; Rytz, T.C.; Hubler, S.L.; Smith, L.M.; Vierstra, R.D. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol. AMP Cell. Proteom. 2013, 12, 449. [Google Scholar] [CrossRef]
- Park, H.C.; Choi, W.; Park, H.J.; Cheong, M.S.; Koo, Y.D.; Shin, G.; Chung, W.S.; Kim, W.-Y.; Kim, M.G.; Bressan, R.A.; et al. Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol. Cells 2011, 32, 143–151. [Google Scholar] [CrossRef]
- Johnson, E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004, 73, 355–382. [Google Scholar] [CrossRef]
- Castro, P.H.; Tavares, R.M.; Bejarano, E.R.; Azevedo, H. SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life Sci. 2012, 69, 3269–3283. [Google Scholar] [CrossRef]
- Colby, T.; Matthäi, A.; Boeckelmann, A.; Stuible, H.-P. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006, 142, 318. [Google Scholar] [CrossRef]
- Xu, F.-Q.; Xue, H.-W. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin–proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef]
- Mukhopadhyay, D.; Riezman, H.J.S. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007, 315, 201–205. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A.P. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Schulman, B.A.; Wade Harper, J. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta Mol. Cell Res. 2004, 1695, 55–72. [Google Scholar] [CrossRef]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Sci. 2016, 7, 806. [Google Scholar] [CrossRef]
- Behrends, C.; Harper, J.W. Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 2011, 18, 520–528. [Google Scholar] [CrossRef]
- Genschik, P. RPN10: A case study for ubiquitin binding proteins and more. Plant Cell 2019, 31, 1398–1399. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, K.P.; Lledias, F.; Kisselev, A.F.; Scaglione, K.M.; Skowyra, D.; Gygi, S.P.; Goldberg, A.L. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007, 282, 17375–17386. [Google Scholar] [CrossRef]
- Komander, D. Mechanism, specificity and structure of the deubiquitinases. In Conjugation and Deconjugation of Ubiquitin Family Modifiers; Springer: New York, NY, USA, 2010; Volume 54, pp. 69–87. [Google Scholar]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L. Chapter three-role of the ubiquitin proteasome system in plant response to abiotic stress. In International Review of Cell and Molecular Biology; Galluzzi, L., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 343, pp. 65–110. [Google Scholar]
- Kelley, D.R. E3 ubiquitin ligases: Key regulators of hormone signaling in plants. Mol. AMP Cell. Proteom. 2018, 17, 1047. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Parry, G.; Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell 2004, 16, 3181. [Google Scholar] [CrossRef]
- van Wijk, S.J.L.; Timmers, H.T.M. The family of ubiquitin-conjugating enzymes (E2s): Deciding between life and death of proteins. FASEB J. 2009, 24, 981–993. [Google Scholar] [CrossRef]
- Kraft, E.; Stone, S.L.; Ma, L.; Su, N.; Gao, Y.; Lau, O.-S.; Deng, X.-W.; Callis, J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 2005, 139, 1597. [Google Scholar] [CrossRef]
- van Nocker, S.; Walker, J.M.; Vierstra, R.D. The Arabidopsis thaliana UBC7/13/14 genes encode a family of multiubiquitin chain-forming E2 enzymes. J. Biol. Chem. 1996, 271, 12150–12158. [Google Scholar] [CrossRef]
- Hofmann, R.M.; Pickart, C.M. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 2001, 276, 27936–27943. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Jaffee, M.; Iyer, L.M.; Aravind, L. Anatomy of the E2 ligase fold: Implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. J. Struct. Biol. 2008, 162, 205–218. [Google Scholar] [CrossRef]
- Okamoto, Y.; Ozaki, T.; Miyazaki, K.; Aoyama, M.; Miyazaki, M.; Nakagawara, A. UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res. 2003, 63, 4167. [Google Scholar]
- Özkan, E.; Yu, H.; Deisenhofer, J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA 2005, 102, 18890. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, D.M.; Stoll, K.E.; Klevit, R.E. E2s: Structurally economical and functionally replete. Biochem. J. 2011, 433, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-Y.; Hanlon, M.; Eddins, M.; Tsui, C.; Rogers, R.S.; Jensen, J.P.; Matunis, M.J.; Weissman, A.M.; Wolberger, C.P.; Pickart, C.M. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003, 22, 5241–5250. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.T.; Ayrault, O.; Hunt, H.W.; Taherbhoy, A.M.; Duda, D.M.; Scott, D.C.; Borg, L.A.; Neale, G.; Murray, P.J.; Roussel, M.F.; et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 2009, 33, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.; Crowe, E.; Stevens, T.A.; Candido, E.P.M. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: Ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 2001, 3, research0002.1. [Google Scholar] [CrossRef]
- Michelle, C.; Vourc’h, P.; Mignon, L.; Andres, C.R. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 2009, 68, 616–628. [Google Scholar] [CrossRef]
- Bae, H.; Kim, W.T. Classification and interaction modes of 40 rice E2 ubiquitin-conjugating enzymes with 17 rice ARM-U-box E3 ubiquitin ligases. Biochem. Biophys. Res. Commun. 2014, 444, 575–580. [Google Scholar] [CrossRef]
- Jue, D.; Sang, X.; Lu, S.; Dong, C.; Zhao, Q.; Chen, H.; Jia, L. Genome-wide identification, phylogenetic and expression analyses of the ubiquitin-conjugating enzyme gene family in maize. PLoS ONE 2015, 10, e0143488. [Google Scholar] [CrossRef]
- Dong, C.; Hu, H.; Jue, D.; Zhao, Q.; Chen, H.; Xie, J.; Jia, L. The banana E2 gene family: Genomic identification, characterization, expression profiling analysis. Plant Sci. 2016, 245, 11–24. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Y.; Xin, H.; Li, S.; Liang, Z. Involvement of ubiquitin-conjugating enzyme (E2 gene Family) in ripening process and response to cold and heat stress of Vitis vinifera. Sci. Rep. 2017, 7, 13290. [Google Scholar] [CrossRef] [PubMed]
- Jue, D.; Sang, X.; Shu, B.; Liu, L.; Wang, Y.; Jia, Z.; Zou, Y.; Shi, S. Characterization and expression analysis of genes encoding ubiquitin conjugating domain-containing enzymes in Carica papaya. PLoS ONE 2017, 12, e0171357. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Bhatt, T.K. Genome-wide identification and expression analysis of E2 ubiquitin-conjugating enzymes in tomato. Sci. Rep. 2017, 7, 8613. [Google Scholar] [CrossRef] [PubMed]
- Jue, D.; Sang, X.; Liu, L.; Shu, B.; Wang, Y.; Xie, J.; Liu, C.; Shi, S. The ubiquitin-conjugating enzyme gene family in Longan (Dimocarpus longan Lour.): Genome-wide identification and gene expression during flower induction and abiotic stress responses. Molecules 2018, 23, 662. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tang, X.; Zhu, X.; Qi, X.; Zhang, N.; Si, H. Genome-wide identification and expression analysis of the E2 gene family in potato. Mol. Biol. Rep. 2019, 46, 777–791. [Google Scholar] [CrossRef]
- Broomfield, S.; Chow, B.L.; Xiao, W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5678. [Google Scholar] [CrossRef] [PubMed]
- Sancho, E.; Vilá, M.R.; Sánchez-Pulido, L.; Lozano, J.J.; Paciucci, R.; Nadal, M.; Fox, M.; Harvey, C.; Bercovich, B.; Loukili, N.; et al. Role of UEV-1, an inactive variant of the E2 ubiquitin conjugating enzymes, in in vitro differentiation and cell cycle behavior of HT-29-M6 intestinal mucosecretory cells. Mol. Cell. Biol. 1998, 18, 576. [Google Scholar] [CrossRef]
- Xiao, W.; Broomfield, S.; Chow, B.L.; Lin, S.L.; Wei, Y.-F. The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved Ubc-like protein family. Nucleic Acids Res. 1998, 26, 3908–3914. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, Y.; Sullivan, J.A.; Komatsu, S.; Gusmaroli, G.; Suzuki, G.; Yin, J.; Ishibashi, T.; Saijo, Y.; Rubio, V.; Kimura, S.; et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 2004, 18, 2172–2181. [Google Scholar] [CrossRef]
- Pastushok, L.; Moraes, T.F.; Ellison, M.J.; Xiao, W. A single Mms2 “key” residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. J. Biol. Chem. 2005, 280, 17891–17900. [Google Scholar] [CrossRef]
- Ulrich, H.D.; Jentsch, S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000, 19, 3388–3397. [Google Scholar] [CrossRef]
- Hofmann, R.M.; Pickart, C.M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 1999, 96, 645–653. [Google Scholar] [CrossRef]
- Wang, Q.; Zang, Y.; Zhou, X.; Xiao, W. Characterization of four rice UEV1 genes required for Lys63-linked polyubiquitination and distinct functions. BMC Plant Biol. 2017, 17, 126. [Google Scholar] [CrossRef]
- McKenna, S.; Spyracopoulos, L.; Moraes, T.; Pastushok, L.; Ptak, C.; Xiao, W.; Ellison, M.J. Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J. Biol. Chem. 2001, 276, 40120–40126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wen, P.; Lau, O.-S.; Deng, X.-W. Characterization of the ubiquitin E2 enzyme variant gene family in Arabidopsis. J. Integr. Plant Biol. 2007, 49, 120–126. [Google Scholar] [CrossRef]
- Guo, H.; Wen, R.; Wang, Q.; Datla, R.; Xiao, W. Three Brachypodium distachyon Uev1s promote Ubc13-mediated Lys63-linked polyubiquitination and confer different functions. Front. Plant Sci. 2016, 7, 1551. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lau, O.S.; Deng, X.W. Effect of Arabidopsis COP10 ubiquitin E2 enhancement activity across E2 families and functional conservation among its canonical homologues. Biochem. J. 2009, 418, 683–690. [Google Scholar] [CrossRef]
- Chung, E.; Cho, C.-W.; So, H.-A.; Kang, J.-S.; Chung, Y.S.; Lee, J.-H. Overexpression of VrUBC1, a mung bean E2 ubiquitin-conjugating enzyme, enhances osmotic stress tolerance in Arabidopsis. PLoS ONE 2013, 8, e66056. [Google Scholar] [CrossRef]
- Jeon, E.H.; Pak, J.H.; Kim, M.J.; Kim, H.J.; Shin, S.H.; Lee, J.H.; Kim, D.H.; Oh, J.S.; Oh, B.-J.; Jung, H.W.; et al. Ectopic expression of ubiquitin-conjugating enzyme gene from wild rice, OgUBC1, confers resistance against UV-B radiation and Botrytis infection in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2012, 427, 309–314. [Google Scholar] [CrossRef]
- Bahmani, R.; Kim, D.; Lee, B.D.; Hwang, S. Over-expression of tobacco UBC1 encoding a ubiquitin-conjugating enzyme increases cadmium tolerance by activating the 20S/26S proteasome and by decreasing Cd accumulation and oxidative stress in tobacco (Nicotiana tabacum). Plant Mol. Biol. 2017, 94, 433–451. [Google Scholar] [CrossRef]
- Feussner, K.; Feussner, I.; Leopold, I.; Wasternack, C. Isolation of a cDNA coding for an ubiquitin-conjugating enzyme UBC1 of tomato—the first stress-induced UBC of higher plants 1 2. FEBS Lett. 1997, 409, 211–215. [Google Scholar] [CrossRef]
- Wan, X.; Mo, A.; Liu, S.; Yang, L.; Li, L. Constitutive expression of a peanut ubiquitin-conjugating enzyme gene in Arabidopsis confers improved water-stress tolerance through regulation of stress-responsive gene expression. J. Biosci. Bioeng. 2011, 111, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.-A.; Chang, R.-Z.; Qiu, L.-J. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Mol. Biol. 2010, 72, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Baloglu, M.C.; Patir, M.G. Molecular characterization, 3D model analysis, and expression pattern of the CmUBC gene encoding the melon ubiquitin-conjugating enzyme under drought and salt stress conditions. Biochem. Genet. 2014, 52, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Qin, B. The function of Rad6 gene in Hevea brasiliensis extends beyond DNA repair. Plant Physiol. Biochem. 2013, 66, 134–140. [Google Scholar] [CrossRef]
- del Pozo, J.C.; Estelle, M. The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 1999, 96, 15342. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Mergner, J. The NEDD8 modification pathway in plants. Front. Plant Sci. 2014, 5, 103. [Google Scholar]
- Zang, Y.; Wang, Q.; Xue, C.; Li, M.; Wen, R.; Xiao, W. Rice UBC13, a candidate housekeeping gene, is required for K63-linked polyubiquitination and tolerance to DNA damage. Rice 2012, 5, 24. [Google Scholar] [CrossRef]
- Wang, L.; Wen, R.; Wang, J.; Xiang, D.; Wang, Q.; Zang, Y.; Wang, Z.; Huang, S.; Li, X.; Datla, R.; et al. Arabidopsis UBC13 differentially regulates two programmed cell death pathways in responses to pathogen and low-temperature stress. New Phytol. 2019, 221, 919–934. [Google Scholar] [CrossRef]
- Cheng, M.-C.; Kuo, W.-C.; Wang, Y.-M.; Chen, H.-Y.; Lin, T.-P. UBC18 mediates ERF1 degradation under light–dark cycles. New Phytol. 2017, 213, 1156–1167. [Google Scholar] [CrossRef]
- Ahn, M.Y.; Oh, T.R.; Seo, D.H.; Kim, J.H.; Cho, N.H.; Kim, W.T. Arabidopsis group XIV ubiquitin-conjugating enzymes AtUBC32, AtUBC33, and AtUBC34 play negative roles in drought stress response. J. Plant Physiol. 2018, 230, 73–79. [Google Scholar] [CrossRef]
- Cui, F.; Liu, L.; Zhao, Q.; Zhang, Z.; Li, Q.; Lin, B.; Wu, Y.; Tang, S.; Xie, Q. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 2012, 24, 233. [Google Scholar] [CrossRef] [PubMed]
- Feng; Cui; Lijing; Liu; Qingliang; Li; Chengwei; Yang; Qi; Xie, UBC32 mediated oxidative tolerance in Arabidopsis. J. Genet. Genom. 2012, 39, 415–417. [CrossRef]
- Aung, K.; Lin, S.-I.; Wu, C.-C.; Huang, Y.-T.; Su, C.-l.; Chiou, T.-J. Pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a MicroRNA399 target gene. Plant Physiol. 2006, 141, 1000. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Datt Pant, B.; Stitt, M.; Scheible, W.-R. PHO2, MicroRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Seo, J.S.; Chua, N.-H. NITROGEN LIMITATION ADAPTATION Recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 2014, 26, 454. [Google Scholar] [CrossRef]
- Liu, T.-Y.; Huang, T.-K.; Tseng, C.-Y.; Lai, Y.-S.; Lin, S.-I.; Lin, W.-Y.; Chen, J.-W.; Chiou, T.-J. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 2012, 24, 2168. [Google Scholar] [CrossRef]
- Xu, L.; Ménard, R.; Berr, A.; Fuchs, J.; Cognat, V.; Meyer, D.; Shen, W.-H. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009, 57, 279–288. [Google Scholar] [CrossRef]
- Cao, Y.; Dai, Y.; Cui, S.; Ma, L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 2008, 20, 2586. [Google Scholar] [CrossRef]
- Imura, Y.; Molho, M.; Chuang, C.; Nagy, P.D. Cellular Ubc2/Rad6 E2 ubiquitin-conjugating enzyme facilitates tombusvirus replication in yeast and plants. Virology 2015, 484, 265–275. [Google Scholar] [CrossRef]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef]
- Millyard, L.; Lee, J.; Zhang, C.; Yates, G.; Sadanandom, A. The ubiquitin conjugating enzyme, TaU4 regulates wheat defence against the phytopathogen Zymoseptoria tritici. Sci. Rep. 2016, 6, 35683. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.; Wiener, R.; Reiss, Y.; Ravid, T. Distinct activation of an E2 ubiquitin-conjugating enzyme by its cognate E3 ligases. Proc. Natl. Acad. Sci. USA 2015, 112, E625. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Newton, L.; Li, G.; Wang, H.; Xiao, W. Arabidopsis thaliana UBC13: Implication of error-free DNA damage tolerance and lys63-linked polyubiquitylation in plants. Plant Mol. Biol. 2006, 61, 241–253. [Google Scholar] [CrossRef]
- Li, J.; Wen, R.; Andersen, P.; Liang, Y.; Li, Q.; Xiao, W.; Cui, Z. Zebrafish Ubc13 is required for Lys63-linked polyubiquitination and DNA damage tolerance. Mol. Cell. Biochem. 2010, 343, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Grisvard, J.; Aubusson-Fleury, A.; Baroin-Tourancheau, A. Multiple uses of lys63-polyubiquitination in the ciliate Sterkiella histriomuscorum. Protist 2010, 161, 479–488. [Google Scholar] [CrossRef]
- Mural, R.V.; Liu, Y.; Rosebrock, T.R.; Brady, J.J.; Hamera, S.; Connor, R.A.; Martin, G.B.; Zeng, L. The tomato Fni3 Lysine-63–specific ubiquitin-conjugating enzyme and Suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell 2013, 25, 3615. [Google Scholar] [CrossRef]
- Zolman, B.K.; Monroe-Augustus, M.; Silva, I.D.; Bartel, B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 2005, 17, 3422. [Google Scholar] [CrossRef]
- Wiborg, J.; O’Shea, C.; Skriver, K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 2008, 413, 447–457. [Google Scholar] [CrossRef]
- Hong, M.J.; Seo, Y.W. A potential role of UBC28 interacting RING finger protein TaRF1 in spike development of wheat. J. Plant Biochem. Biotechnol. 2014, 23, 421–429. [Google Scholar] [CrossRef]
- Liu, L.; Cui, F.; Li, Q.; Yin, B.; Zhang, H.; Lin, B.; Wu, Y.; Xia, R.; Tang, S.; Xie, Q. The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 2011, 21, 957–969. [Google Scholar] [CrossRef]
- Chen, Q.; Zhong, Y.; Wu, Y.; Liu, L.; Wang, P.; Liu, R.; Cui, F.; Li, Q.; Yang, X.; Fang, S.; et al. HRD1-mediated ERAD tuning of ER-bound E2 is conserved between plants and mammals. Nat. Plants 2016, 2, 16094. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lin, N.-C.; Martin, G.B. Two distinct pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 2002, 109, 589–598. [Google Scholar] [CrossRef]
- Zhou, B.; Zeng, L. Elucidating the role of highly homologous Nicotiana benthamiana ubiquitin E2 gene family members in plant immunity through an improved virus-induced gene silencing approach. Plant Methods 2017, 13, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zeng, L. The tomato U-box type E3 Ligase PUB13 acts with group III ubiquitin E2 enzymes to modulate FLS2-mediated immune signaling. Front. Plant Sci. 2018, 9, 615. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Schmidt, W. A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 2010, 62, 330–343. [Google Scholar] [CrossRef]
- Wen, R.; Wang, S.; Xiang, D.; Venglat, P.; Shi, X.; Zang, Y.; Datla, R.; Xiao, W.; Wang, H. UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J. 2014, 80, 424–436. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, W.; Cai, J.; Zhang, Y.; Qin, G.; Tian, S. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol. 2014, 15, 548. [Google Scholar] [CrossRef]
- Wang, S.; Cao, L.; Wang, H. Arabidopsis ubiquitin-conjugating enzyme UBC22 is required for female gametophyte development and likely involved in Lys11-linked ubiquitination. J. Exp. Bot. 2016, 67, 3277–3288. [Google Scholar] [CrossRef]
- Liu, Y.; Koornneef, M.; Soppe, W.J.J. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 2007, 19, 433. [Google Scholar] [CrossRef]
- Fleury, D.; Himanen, K.; Cnops, G.; Nelissen, H.; Boccardi, T.M.; Maere, S.; Beemster, G.T.S.; Neyt, P.; Anami, S.; Robles, P.; et al. The Arabidopsis thaliana homolog of yeast BRE1 Has a function in cell cycle regulation during early leaf and root growth. Plant Cell 2007, 19, 417. [Google Scholar] [CrossRef]
- Xu, L.; Shen, W.-H. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 2008, 18, 1966–1971. [Google Scholar] [CrossRef]
- Gu, X.; Jiang, D.; Wang, Y.; Bachmair, A.; He, Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009, 57, 522–533. [Google Scholar] [PubMed]
- Galan, J.M.; Haguenauer-Tsapis, R. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. Embo J. 1997, 16, 5847–5854. [Google Scholar] [PubMed]
- Spence, J.; Gali, R.R.; Dittmar, G.; Sherman, F.; Karin, M.; Finley, D. Cell cycle–regulated modification of the ribosome by a variant multiubiquitin chain. Cell 2000, 102, 67–76. [Google Scholar] [PubMed]
- Wen, R.; Torres-Acosta, J.A.; Pastushok, L.; Lai, X.; Pelzer, L.; Wang, H.; Xiao, W. Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response. Plant Cell 2008, 20, 213. [Google Scholar]
- Berndsen, C.E.; Wiener, R.; Yu, I.W.; Ringel, A.E.; Wolberger, C. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat. Chem. Biol. 2013, 9, 154–156. [Google Scholar]
- Andersen, P.L.; Xu, F.; Xiao, W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008, 18, 162–173. [Google Scholar]
- Eddins, M.J.; Carlile, C.M.; Gomez, K.M.; Pickart, C.M.; Wolberger, C. Mms2–Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006, 13, 915–920. [Google Scholar]
- Broomfield, S.; Hryciw, T.; Xiao, W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. DNA Repair 2001, 486, 167–184. [Google Scholar] [CrossRef]
- Xiao, W.; Chow, B.L.; Fontanie, T.; Ma, L.; Silvia, B.; Hryciw, T.; Broomfield, S. Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res. DNA Repair 1999, 435, 1–11. [Google Scholar]
- Zhang, W.; Qin, Z.; Zhang, X.; Xiao, W. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Lett. 2011, 585, 2786–2794. [Google Scholar] [PubMed]
- Xiao, W.; Chow, B.L.; Broomfield, S.; Hanna, M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics 2000, 155, 1633. [Google Scholar]
- Hedglin, M.; Benkovic, S.J. Regulation of Rad6/Rad18 activity during DNA damage tolerance. Annu. Rev. Biophys. 2015, 44, 207–228. [Google Scholar] [PubMed]
- Prakash, L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. MGG 1981, 184, 471–478. [Google Scholar] [PubMed]
- Koken, M.H.; Reynolds, P.; Jaspers-Dekker, I.; Prakash, L.; Prakash, S.; Bootsma, D.; Hoeijmakers, J.H. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. USA 1991, 88, 8865. [Google Scholar]
- Yamamoto, T.; Mori, Y.; Ishibashi, T.; Uchiyama, Y.; Sakaguchi, N.; Furukawa, T.; Hashimoto, J.; Kimura, S.; Sakaguchi, K. Characterization of Rad6 from a higher plant, rice (Oryza sativa L.) and its interaction with Sgt1, a subunit of the SCF ubiquitin ligase complex. Biochem. Biophys. Res. Commun. 2004, 314, 434–439. [Google Scholar]
- Hoege, C.; Pfander, B.; Moldovan, G.-L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar]
- Xu, X.; Blackwell, S.; Lin, A.; Li, F.; Qin, Z.; Xiao, W. Error-free DNA-damage tolerance in Saccharomyces cerevisiae. Mutat. Res. Rev. Mutat. Res. 2015, 764, 43–50. [Google Scholar]
- Stelter, P.; Ulrich, H.D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 2003, 425, 188–191. [Google Scholar]
- Papouli, E.; Chen, S.; Davies, A.A.; Huttner, D.; Krejci, L.; Sung, P.; Ulrich, H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 2005, 19, 123–133. [Google Scholar]
- Pfander, B.; Moldovan, G.-L.; Sacher, M.; Hoege, C.; Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005, 436, 428–433. [Google Scholar] [PubMed]
- Watts, F.Z. Sumoylation of PCNA: Wrestling with recombination at stalled replication forks. DNA Repair 2006, 5, 399–403. [Google Scholar] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).