From Snapshots to Flipbook—Resolving the Dynamics of Ribosome Biogenesis with Chemical Probes
Abstract
:1. Introduction
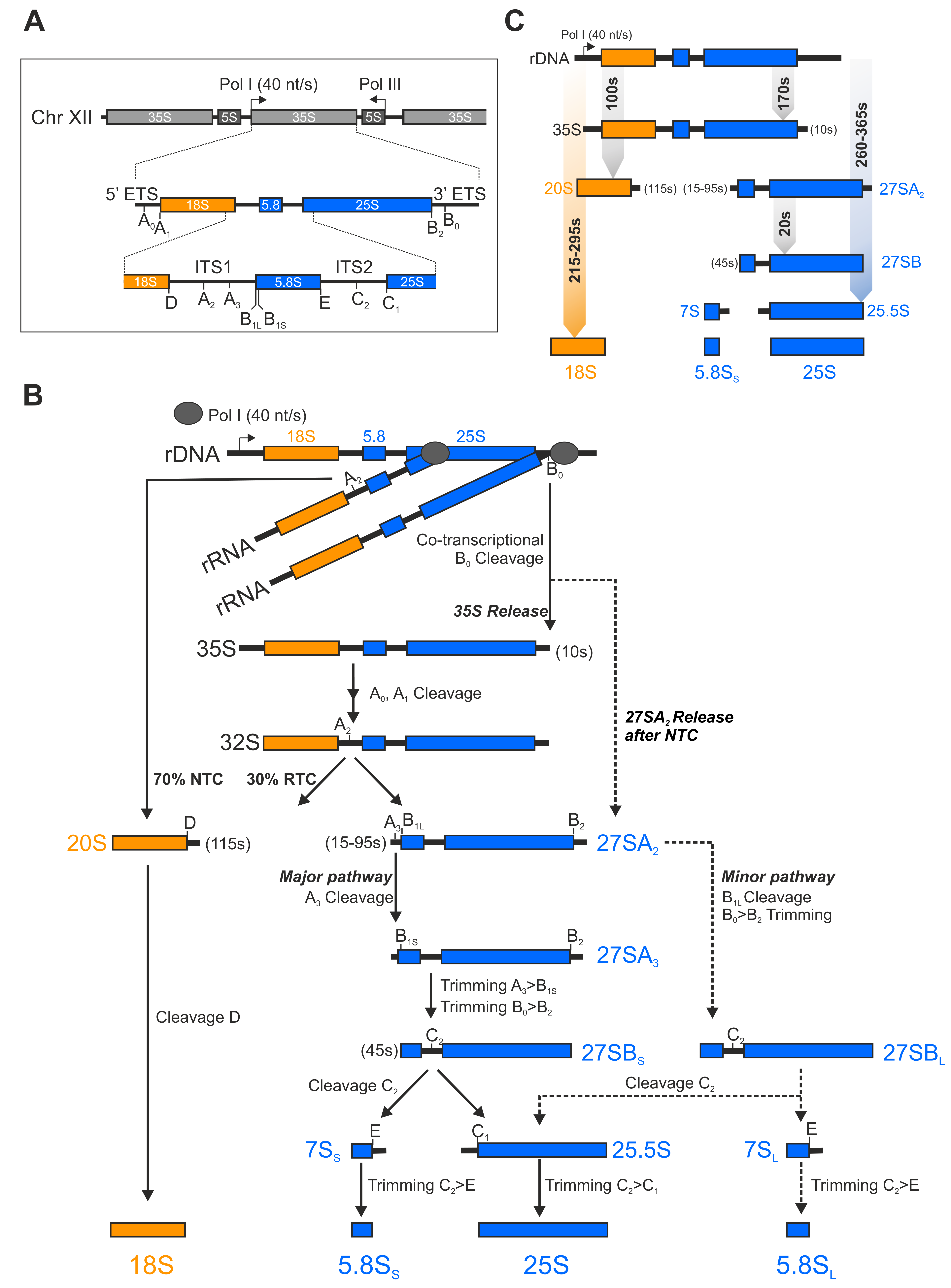
1.1. A Short Overview of the rRNA Processing Cascade in Yeast
1.2. 250 Assembly Factors Empower Pre-Ribosomal Particle Maturation
1.3. Ribosome Biogenesis Inhibitors—Chemical Probes to Unravel Ribosome Biogenesis
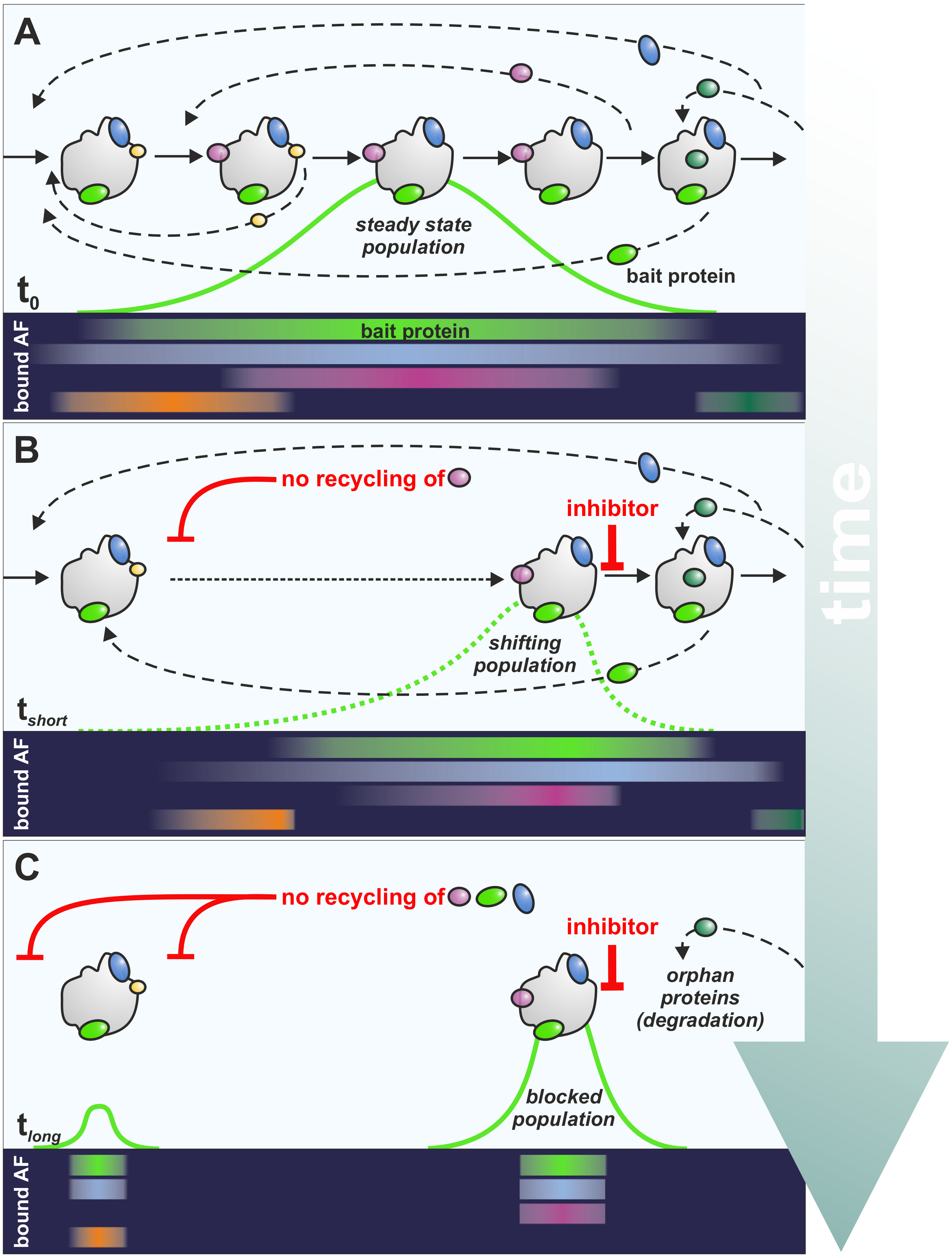
2. A Practical Guide to Read the Effects of Inhibitors on Ribosome Biogenesis
3. Examples for Ribosome Probing
3.1. Rbin-1 Blocks Rsa4 Release by Mdn1 (Rea1) in the Nucleoplasm
3.2. Dissecting the Complex Interwoven Pathway by the Help of Diazaborine
3.2.1. Tracing Pre-60S Particle Maturation from the Nucleolus to the Cytoplasm
3.2.2. Coordination of Particle Export and Downstream Maturation in the Cytoplasm
4. Cellular Response to Inhibition of Ribosome Biogenesis
4.1. Orphan Proteins Signal Stress Owing to Blocked Ribosome Biogenesis
4.2. Degradation of Aberrant Ribosomal Precursors
5. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baßler, J.; Hurt, E. Eukaryotic Ribosome Assembly. Annu. Rev. Biochem. 2019, 88, 281–306. [Google Scholar] [CrossRef] [PubMed]
- Klinge, S.; Woolford, J.L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, K.E.; Bohnsack, M.T. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef]
- Zaragoza, D.; Ghavidel, A.; Heitman, J.; Schultz, M.C. Rapamycin Induces the G0 Program of Transcriptional Repression in Yeast by Interfering with the TOR Signaling Pathway. Mol. Cell Biol. 1998, 18, 4463–4470. [Google Scholar] [CrossRef] [Green Version]
- Powers, T.; Walter, P. Regulation of Ribosome Biogenesis by the Rapamycin-sensitive TOR-signaling Pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 1999, 10, 987–1000. [Google Scholar] [CrossRef] [Green Version]
- Kos-Braun IC, Jung I, Koš, M. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLoS Biol. 2017, 15, e2000245. [Google Scholar] [CrossRef] [Green Version]
- Loewith, R.; Hallm, M.N. Target of Rapamycin (TOR) in Nutrient Signaling and Growth Control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [Green Version]
- De la Cruz, J.; Gómez-Herreros, F.; Rodríguez-Galán, O.; Begley, V.; de la Cruz Muñoz-Centeno, M.; Chávez, S. Feedback regulation of ribosome assembly. Curr. Genet. 2018, 64, 393–404. [Google Scholar] [CrossRef]
- Piazzi, M.; Bavelloni, A.; Gallo, A.; Faenza, I.; Blalock, W.L. Signal Transduction in Ribosome Biogenesis: A Recipe to Avoid Disaster. Int. J. Mol. Sci. 2019, 20, 2718. [Google Scholar] [CrossRef] [Green Version]
- Penzo, M.; Montanaro, L.; Treré, D.; Derenzini, M. The Ribosome Biogenesis—Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Aspesi, A.; Ellis, S.R. Rare ribosomopathies: Insights into mechanisms of cancer. Nat. Rev. Cancer 2019, 19, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.W.; Green, R. Ribosomopathies: There’s strength in numbers. Science 2017, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farley-Barnes, K.I.; Ogawa, L.M.; Baserga, S.J. Ribosomopathies: Old Concepts, New Controversies. Trends Genet. 2019, 35, 754–767. [Google Scholar] [CrossRef] [PubMed]
- Kressler, D.; Hurt, E.; Baβler, J. Driving ribosome assembly. Biochim. Biophys. Acta 2010, 1803, 673–683. [Google Scholar] [CrossRef] [Green Version]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. WIREs RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Pillet, B.; Mitterer, V.; Kressler, D.; Pertschy, B. Hold on to your friends: Dedicated chaperones of ribosomal proteins. BioEssays 2017, 39, e201600153. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Beckmann, R.; Cate, J.H.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014, 24, 165–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koš, M.; Tollervey, D. Yeast Pre-rRNA Processing and Modification Occur Cotranscriptionally. Mol. Cell 2010, 37, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Barandun, J.; Hunziker, M.; Klinge, S. Assembly and structure of the SSU processome—A nucleolar precursor of the small ribosomal subunit. Curr. Opin. Struct. Biol. 2018, 49, 85–93. [Google Scholar] [CrossRef]
- Chaker-Margot, M.; Barandun, J.; Hunziker, M.; Klinge, S. Architecture of the yeast small subunit processome. Science 2017, 355, eaal1880. [Google Scholar] [CrossRef]
- Chaker-Margot, M.; Hunziker, M.; Barandun, J.; Dill, B.D.; Klinge, S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol. 2015, 22, 920–923. [Google Scholar] [CrossRef]
- Cheng, J.; Kellner, N.; Berninghausen, O.; Hurt, E.; Beckmann, R. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017, 24, 954–964. [Google Scholar] [CrossRef]
- Sun, Q.; Zhu, X.; Qi, J.; An, W.; Lan, P.; Tan, D.; Chen, R.; Wang, B.; Zheng, S.; Zhang, C. Molecular architecture of the 90S small subunit pre-ribosome. eLife 2017, 6, e22086. [Google Scholar] [CrossRef]
- Kornprobst, M.; Turk, M.; Kellner, N.; Cheng, J.; Flemming, D.; Koš-Braun, I.; Koš, M.; Thoms, M.; Berninghausen, O.; Beckmann, R.; et al. Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell 2016, 166, 380–393. [Google Scholar] [CrossRef] [Green Version]
- Mougey, E.B.; O’Reilly, M.; Osheim, Y.; Miller, O.L.; Beyer, A.; Sollner-Webb, B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993, 7, 1609–1619. [Google Scholar] [CrossRef] [Green Version]
- Osheim, Y.N.; French, S.L.; Keck, K.M.; Champion, E.A.; Spasov, K.; Dragon, F.; Baserga, S.J.; Beyer, A.L. Pre-18S Ribosomal RNA Is Structurally Compacted into the SSU Processome Prior to Being Cleaved from Nascent Transcripts in Saccharomyces cerevisiae. Mol. Cell 2004, 16, 943–954. [Google Scholar] [CrossRef]
- Pillon, M.C.; Lo, Y.-H.; Stanley, R.E. IT’S 2 for the price of 1: Multifaceted ITS2 processing machines in RNA and DNA maintenance. DNA Rep. 2019, 81, 102653. [Google Scholar] [CrossRef]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.-F.; Chakraborty, A.; Gleizes P-E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef]
- Fernández-Pevida, A.; Kressler, D.; Cruz, J. Processing of preribosomal RNA in Saccharomyces cerevisiae. WIREs RNA 2015, 6, 191–209. [Google Scholar] [CrossRef]
- Nerurkar, P.; Altvater, M.; Gerhardy, S.; Schütz, S.; Fischer, U.; Weirich, C.; Panse, V.G. Eukaryotic Ribosome Assembly and Nuclear Export. Int. Rev. Cell Mol. Biol. 2015, 319, 107–140. [Google Scholar] [CrossRef]
- Hedges, J.; West, M.; Johnson, A.W. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005, 24, 567–579. [Google Scholar] [CrossRef] [Green Version]
- Menne, T.F.; Goyenechea, B.; Sánchez-Puig, N.; Wong, C.C.; Tonkin, L.M.; Ancliff, P.J.; Brost, R.L.; Costanzo, M.; Boone, C.; Warren, A.J. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 2007, 39, 486–495. [Google Scholar] [CrossRef]
- Altvater, M.; Chang, Y.; Melnik, A.; Occhipinti, L.; Schütz, S.; Rothenbusch, U.; Picotti, P.; Panse, V.G. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol. Syst. Biol. 2012, 8. [Google Scholar] [CrossRef]
- Weis, F.; Giudice, E.; Churcher, M.; Jin, L.; Hilcenko, C.; Wong, C.C.; Traynor, D.; Kay, R.R.; Warren, A.J. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015, 22, 914–919. [Google Scholar] [CrossRef] [Green Version]
- Greber, B.J.; Gerhardy, S.; Leitner, A.; Leibundgut, M.; Salem, M.; Boehringer, D.; Leulliot, N.; Aebersold, R.; Panse, V.G.; Ban, N. Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation. Cell 2016, 164, 91–102. [Google Scholar] [CrossRef]
- Patchett, S.; Musalgaonkar, S.; Malyutin, A.G.; Johnson, A.W. The T-cell leukemia related rpl10-R98S mutant traps the 60S export adapter Nmd3 in the ribosomal P site in yeast. PLoS Genet. 2017, 13, e1006894. [Google Scholar] [CrossRef]
- Kargas, V.; Castro-Hartmann, P.; Escudero-Urquijo, N.; Dent, K.; Hilcenko, C.; Sailer, C.; Zisser, G.; Marques-Carvalho, M.J.; Pellegrino, S.; Wawiórka, L.; et al. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. eLife 2019, 8, e44904. [Google Scholar] [CrossRef]
- Ma, C.; Wu, S.; Li, N.; Chen, Y.; Yan, K.; Li, Z.; Zheng, L.; Lei, J.; Woolford, J.L.; Gao, N. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat. Struct. Mol. Biol. 2017, 24, 214–220. [Google Scholar] [CrossRef]
- Lo, K.-Y.; Li, Z.; Bussiere, C.; Bresson, S.; Marcotte, E.M.; Johnson, A.W. Defining the Pathway of Cytoplasmic Maturation of the 60S Ribosomal Subunit. Mol. Cell 2010, 39, 196–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klingauf-Nerurkar, P.; Gillet, L.C.; Portugal-Calisto, D.; Oborská-Oplová, M.; Jäger, M.; Schubert, O.T.; Pisano, A.; Peña, C.; Rao, S.; Altvater, M.; et al. The GTPase Nog1 co-ordinates the assembly, maturation and quality control of distant ribosomal functional centers. eLife 2020, 9, e52474. [Google Scholar] [CrossRef]
- Biedka, S.; Micic, J.; Wilson, D.; Brown, H.; Diorio-Toth, L.; Woolford, J.L. Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J. Cell Biol. 2018, 217, 2503–2518. [Google Scholar] [CrossRef] [PubMed]
- Gamalinda, M.; Ohmayer, U.; Jakovljevic, J.; Kumcuoglu, B.; Woolford, J.; Mbom, B.; Lin, L.; Woolford, J.L. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 2014, 28, 198–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dez, C.; Houseley, J.; Tollervey, D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006, 25, 1534–1546. [Google Scholar] [CrossRef] [Green Version]
- Soudet, J.; Gélugne, J.-P.; Belhabich-Baumas, K.; Caizergues-Ferrer, M.; Mougin, A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010, 29, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Galán, O.; García-Gómez, J.J.; Kressler, D.; de la Cruz, J. Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae. RNA Biol. 2015, 12, 838–846. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Thoms, M.; Barrio-Garcia, C.; Thomson, E.; Flemming, D.; Beckmann, R.; Hurt, E. Preribosomes escaping from the nucleus are caught during translation by cytoplasmic quality control. Nat. Struct. Mol. Biol. 2017, 24, 1107–1115. [Google Scholar] [CrossRef]
- Harnpicharnchai, P.; Jakovljevic, J.; Horsey, E.; Miles, T.; Roman, J.; Rout, M.; Meagher, D.; Imai, B.; Guo, Y.; Brame, C.J.; et al. Composition and Functional Characterization of Yeast 66S Ribosome Assembly Intermediates. Mol. Cell 2001, 8, 505–515. [Google Scholar] [CrossRef]
- Nissan, T.A.; Baßler, J.; Petfalski, E.; Tollervey, D.; Hurt, E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002, 21, 5539–5547. [Google Scholar] [CrossRef]
- Baßler, J.; Grandi, P.; Gadal, O.; Leßmann, T.; Petfalski, E.; Tollervey, D.; Lechner, J.; Hurt, E. Identification of a 60S Preribosomal Particle that Is Closely Linked to Nuclear Export. Mol. Cell 2001, 8, 517–529. [Google Scholar] [CrossRef]
- Saveanu, C.; Bienvenu, D.; Namane, A.; Gleizes, P.-E.; Gas, N.; Jacquier, A.; Fromont-Racine, M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001, 20, 6475–6484. [Google Scholar] [CrossRef] [PubMed]
- Dragon, F.; Gallagher, J.E.G.; Compagnone-Post, P.A.; Mitchell, B.M.; Porwancher, K.A.; Wehner, K.A.; Wormsley, S.; Settlage, R.E.; Shabanowitz, J.; Osheim, Y. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002, 417, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Tollervey, D. Making ribosomes. Curr. Opin. Cell Biol. 2002, 14, 313–318. [Google Scholar] [CrossRef]
- Grandi, P.; Rybin, V.; Baßler, J.; Petfalski, E.; Strauß, D.; Marzioch, M.; Schäfer, T.; Kuster, B.; Tschochner, H.; Tollervey, D.; et al. 90S Pre-Ribosomes Include the 35S Pre-rRNA, the U3 snoRNP, and 40S Subunit Processing Factors but Predominantly Lack 60S Synthesis Factors. Mol. Cell 2002, 10, 105–115. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, X.; Zheng, S.; Tan, D.; Dong, M.-Q.; Ye, K. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell 2019, 10, 120–130. [Google Scholar] [CrossRef] [Green Version]
- Sanghai, Z.A.; Miller, L.; Molloy, K.R.; Barandun, J.; Hunziker, M.; Chaker-Margot, M.; Wang, J.; Chait, B.T.; Klinge, S. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 2018, 556, 126–129. [Google Scholar] [CrossRef]
- Wu, S.; Tutuncuoglu, B.; Yan, K.; Brown, H.; Zhang, Y.; Tan, D.; Gamalinda, M.; Yuan, Y.; Li, Z.; Jakovljevic, J.; et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 2016, 534, 133–137. [Google Scholar] [CrossRef]
- Schuller, J.M.; Falk, S.; Fromm, L.; Hurt, E.; Conti, E. Structure of the nuclear exosome captured on a maturing preribosome. Science 2018, 360, 219–222. [Google Scholar] [CrossRef] [Green Version]
- Kater, L.; Thoms, M.; Barrio-Garcia, C.; Cheng, J.; Ismail, S.; Ahmed, Y.L.; Bange, G.; Kressler, D.; Berninghausen, O.; Sinning, I.; et al. Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes. Cell 2017, 171, 1599–1610. [Google Scholar] [CrossRef] [Green Version]
- Leidig, C.; Thoms, M.; Holdermann, I.; Bradatsch, B.; Berninghausen, O.; Bange, G.; Sinning, I.; Hurt, E.; Beckmann, R. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 2014, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Barrio-Garcia, C.; Thoms, M.; Flemming, D.; Kater, L.; Berninghausen, O.; Baßler, J.; Beckmann, R.; Hurt, E. Architecture of the Rix1–Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat. Struct. Mol. Biol. 2016, 23, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Zisser, G.; Ohmayer, U.; Mauerhofer, C.; Mitterer, V.; Klein, I.; Rechberger, G.N.; Wolinski, H.; Prattes, M.; Pertschy, B.; Milkereit, P.; et al. Viewing pre-60S maturation at a minute’s timescale. Nucleic Acids Res. 2018, 46, 3140–3151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, B.; Kos-Braun, I.C.; Henras, A.K.; Dez, C.; Rueda, M.P.; Zhang, X.; Gadal, O.; Kos, M.; Shore, D. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. eLife 2019, 8, e45002. [Google Scholar] [CrossRef]
- Peltonen, K.; Colis, L.; Liu, H.; Jäämaa, S.; Moore, H.M.; Enbäck, J.; Laakkonen, P.; Vaahtokari, A.; Jones, R.J.; af Hällström, T.M.; et al. Identification of Novel p53 Pathway Activating Small-Molecule Compounds Reveals Unexpected Similarities with Known Therapeutic Agents. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer Activity of CX-3543: A Direct Inhibitor of rRNA Biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef] [Green Version]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Antonio, M.D.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Silvester, J.; Wei, V.; Garcia, J. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017, 8, 1–18. [Google Scholar] [CrossRef]
- Turi, Z.; Lacey, M.; Mistrik, M.; Moudry, P. Impaired ribosome biogenesis: Mechanisms and relevance to cancer and aging. Aging 2019, 11, 2512–2540. [Google Scholar] [CrossRef]
- Ferreira, R.; Schneekloth, J.S.; Panov, K.I.; Hannan, K.M.; Hannan, R.D. Targeting the RNA Polymerase I Transcription for Cancer Therapy Comes of Age. Cells 2020, 9, 266. [Google Scholar] [CrossRef] [Green Version]
- Pertschy, B.; Zisser, G.; Schein, H.; Köffel, R.; Rauch, G.; Grillitsch, K.; Morgenstern, C.; Durchschlag, M.; Högenauer, G.; Bergler, H. Diazaborine Treatment of Yeast Cells Inhibits Maturation of the 60S Ribosomal Subunit. Mol. Cell Biol. 2004, 24, 6476–6487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakalskiy, A.; Högenauer, G.; Ishikawa, T.; Wehrschütz-Sigl, E.; Wendler, F.; Teis, D.; Zisser, G.; Steven, A.C.; Bergler, H. Structural and Enzymatic Properties of the AAA Protein Drg1p fromSaccharomyces cerevisiae DECOUPLING OF INTRACELLULAR FUNCTION FROM ATPase ACTIVITY AND HEXAMERIZATION. J. Biol. Chem. 2002, 277, 26788–26795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loibl, M.; Klein, I.; Prattes, M.; Schmidt, C.; Kappel, L.; Zisser, G.; Gungl, A.; Krieger, E.; Pertschy, B.; Bergler, H. The Drug Diazaborine Blocks Ribosome Biogenesis by Inhibiting the AAA-ATPase Drg1. J. Biol. Chem. 2014, 289, 3913–3922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wendler, F.; Bergler, H.; Prutej, K.; Jungwirth, H.; Zisser, G.; Kuchler, K.; Högenauer, G. Diazaborine resistance in the yeast Saccharomyces cerevisiae reveals a link between YAP1 and the pleiotropic drug resistance genes PDR1 and PDR3. J. Biol. Chem. 1997, 272, 27091–27098. [Google Scholar] [CrossRef] [Green Version]
- Pertschy, B.; Saveanu, C.; Zisser, G.; Lebreton, A.; Tengg, M.; Jacquier, A.; Liebminger, E.; Nobis, B.; Kappel, L.; Klei, I.V.D.; et al. Cytoplasmic Recycling of 60S Preribosomal Factors Depends on the AAA Protein Drg1. Mol. Cell Biol. 2007, 27, 6581–6592. [Google Scholar] [CrossRef] [Green Version]
- Kappel, L.; Loibl, M.; Zisser, G.; Klein, I.; Fruhmann, G.; Gruber, C.; Unterweger, S.; Rechberger, G.; Pertschy, B.; Bergler, H. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 2012, 199, 771–782. [Google Scholar] [CrossRef] [Green Version]
- Kawashima, S.A.; Chen, Z.; Aoi, Y.; Patgiri, A.; Kobayashi, Y.; Nurse, P.; Kapoor, T.M. Potent, Reversible, and Specific Chemical Inhibitors of Eukaryotic Ribosome Biogenesis. Cell 2016, 167, 512–524. [Google Scholar] [CrossRef] [Green Version]
- Galani, K.; Nissan, T.A.; Petfalski, E.; Tollervey, D.; Hurt, E. Rea1, a Dynein-related Nuclear AAA-ATPase, Is Involved in Late rRNA Processing and Nuclear Export of 60 S Subunits. J. Biol. Chem. 2004, 279, 55411–55418. [Google Scholar] [CrossRef] [Green Version]
- Ulbrich, C.; Diepholz, M.; Baßler, J.; Kressler, D.; Pertschy, B.; Galani, K.; Böttcher, B.; Hurt, E. Mechanochemical Removal of Ribosome Biogenesis Factors from Nascent 60S Ribosomal Subunits. Cell 2009, 138, 911–922. [Google Scholar] [CrossRef] [Green Version]
- Baßler, J.; Kallas, M.; Pertschy, B.; Ulbrich, C.; Thoms, M.; Hurt, E. The AAA-ATPase Rea1 Drives Removal of Biogenesis Factors during Multiple Stages of 60S Ribosome Assembly. Mol. Cell 2010, 38, 712–721. [Google Scholar] [CrossRef] [Green Version]
- Awad, D.; Prattes, M.; Kofler, L.; Rössler, I.; Loibl, M.; Pertl, M.; Zisser, G.; Wolinski, H.; Pertschy, B.; Bergler, H. Inhibiting eukaryotic ribosome biogenesis. BMC Biol. 2019, 17. [Google Scholar] [CrossRef] [PubMed]
- Talkish, J.; Zhang, J.; Jakovljevic, J.; Horsey, E.W.; Woolford, J.L. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012, 40, 8646–8661. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Suzuki, H.; Kobayashi, Y.; Wang, A.C.; DiMaio, F.; Kawashima, S.A.; Walz, T.; Kapoor, T.M. Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis. Cell 2018, 175, 822–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prattes, L.; Bergler, S. Shaping the Nascent Ribosome: AAA-ATPases in Eukaryotic Ribosome Biogenesis. Biomolecules 2019, 9, 715. [Google Scholar] [CrossRef] [Green Version]
- Thoms, M.; Ahmed, Y.L.; Maddi, K.; Hurt, E.; Sinning, I. Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res. 2016, 44, 926–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinman, J.B.; Kapoor, T.M. Using chemical inhibitors to probe AAA protein conformational dynamics and cellular functions. Curr. Opin. Chem. Biol. 2019, 50, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Saveanu, C.; Namane, A.; Gleizes, P.-E.; Lebreton, A.; Rousselle, J.-C.; Noaillac-Depeyre, J.; Gas, N.; Jacquier, A.; Fromont-Racine, M. Sequential Protein Association with Nascent 60S Ribosomal Particles. Mol. Cell Biol. 2003, 23, 4449–4460. [Google Scholar] [CrossRef] [Green Version]
- Honma, Y.; Kitamura, A.; Shioda, R.; Maruyama, H.; Ozaki, K.; Oda, Y.; Mini, T.; Jenö, P.; Maki, Y.; Yonezawa, K.; et al. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J. 2006, 25, 3832–3842. [Google Scholar] [CrossRef] [Green Version]
- Jensen, B.C.; Wang, Q.; Kifer, C.T.; Parsons, M. The NOG1 GTP-binding Protein Is Required for Biogenesis of the 60 S Ribosomal Subunit. J. Biol. Chem. 2003, 278, 32204–32211. [Google Scholar] [CrossRef] [Green Version]
- Kallstrom, G.; Hedges, J.; Johnson, A. The Putative GTPases Nog1p and Lsg1p Are Required for 60S Ribosomal Subunit Biogenesis and Are Localized to the Nucleus and Cytoplasm, Respectively. Mol. Cell Biol. 2003, 23, 4344–4355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, M.; Hedges, J.B.; Chen, A.; Johnson, A.W. Defining the Order in Which Nmd3p and Rpl10p Load onto Nascent 60S Ribosomal Subunits. Mol. Cell Biol. 2005, 25, 3802–3813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, Y.; Granneman, S.; Thoms, M.; Manikas, R.-G.; Tollervey, D.; Hurt, E. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 2014, 505, 112–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, J.H.-N.; Kallstrom, G.; Johnson, A.W. Nmd3p Is a Crm1p-Dependent Adapter Protein for Nuclear Export of the Large Ribosomal Subunit. J. Cell Biol. 2000, 151, 1057–1066. [Google Scholar] [CrossRef]
- Thomas, F.; Kutay, U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003, 116, 2409–2419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trotta, C.R.; Lund, E.; Kahan, L.; Johnson, A.W.; Dahlberg, J.E. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003, 22, 2841–2851. [Google Scholar] [CrossRef] [Green Version]
- Tye, B.W.; Commins, N.; Ryazanova, L.V.; Wühr, M.; Springer, M.; Pincus, D.; Churchman, L.S. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. eLife 2019, 8, e43002. [Google Scholar] [CrossRef] [PubMed]
- Sung, M.-K.; Porras-Yakushi, T.R.; Reitsma, J.M.; Huber, F.M.; Sweredoski, M.J.; Hoelz, A.; Hess, S.; Deshaies, R.J. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife 2016, 5, e19105. [Google Scholar] [CrossRef]
- Li, J.; Labbadia, J.; Morimoto, R.I. Rethinking HSF1 in stress, development and organismal health. Trends Cell Biol. 2017, 27, 895–905. [Google Scholar] [CrossRef]
- Mahat, D.B.; Salamanca, H.H.; Duarte, F.M.; Danko, C.G.; Lis, J.T. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Mol. Cell 2016, 62, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Pincus, D.; Anandhakumar, J.; Thiru, P.; Guertin, M.J.; Erkine, A.M.; Gross, D.S. Genetic and epigenetic determinants establish a continuum of Hsf1 occupancy and activity across the yeast genome. Mol. Biol. Cell 2018, 29, 3168–3182. [Google Scholar] [CrossRef]
- Solís, E.J.; Pandey, J.P.; Zheng, X.; Jin, D.X.; Gupta, P.B.; Airoldi, E.M.; Pincus, D.; Denic, V. Defining the Essential Function of Yeast Hsf1 Reveals a Compact Transcriptional Program for Maintaining Eukaryotic Proteostasis. Mol. Cell 2016, 63, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Zinder, J.C.; Lima, C.D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017, 31, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Allmang, C.; Kufel, J.; Chanfreau, G.; Mitchell, P.; Petfalski, E.; Tollervey, D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999, 18, 5399–5410. [Google Scholar] [CrossRef] [Green Version]
- Briggs, M.W.; Burkard, K.T.D.; Butler, J.S. Rrp6p, the Yeast Homologue of the Human PM-Scl 100-kDa Autoantigen, Is Essential for Efficient 5.8 S rRNA,3′ End Formation. J. Biol. Chem. 1998, 273, 13255–13263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falk, S.; Tants, J.-N.; Basquin, J.; Thoms, M.; Hurt, E.; Sattler, M.; Conti, E. Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the preribosomal protein Nop53. RNA 2017, 23, 1780–1787. [Google Scholar] [CrossRef] [Green Version]
- Fromm, L.; Falk, S.; Flemming, D.; Schuller, J.M.; Thoms, M.; Conti, E.; Hurt, E. Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Wyers, F.; Rougemaille, M.; Badis, G.; Rousselle, J.-C.; Dufour, M.-E.; Boulay, J.; Régnault, B.; Devaux, F.; Namane, A.; Séraphin, B.; et al. Cryptic Pol II Transcripts Are Degraded by a Nuclear Quality Control Pathway Involving a New Poly(A) Polymerase. Cell 2005, 121, 725–737. [Google Scholar] [CrossRef] [Green Version]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA Degradation by the Exosome Is Promoted by a Nuclear Polyadenylation Complex. Cell 2005, 121, 713–724. [Google Scholar] [CrossRef]
- Wong, C.; Kewu, P.; Zhe, H.; Lee, J. Current perspectives on the role of TRAMP in nuclear RNA surveillance and quality control. Res. Rep. Biochem. 2015, 2015, 111. [Google Scholar] [CrossRef] [Green Version]
- Houseley, J.; LaCava, J.; Tollervey, D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006, 7, 529–539. [Google Scholar] [CrossRef]
- Bursać, S.; Brdovčak, M.C.; Pfannkuchen, M.; Orsolić, I.; Golomb, L.; Zhu, Y.; Katz, C.; Daftuar, L.; Grabušić, K.; Vukelić, I.; et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc. Natl. Acad. Sci. USA 2012, 109, 20467–20472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Q.; Wang, J.; Chen, Y.; Wang, S.; Cao, M.; Lu, H.; Zhou, X. Dual regulation of p53 by the ribosome maturation factor SBDS. Cell Death Dis. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, Z.-Q.; Li, F.-Q.; Chen, J.-N.; Gong, X.; Cao, B.-B.; Wang, W. Triptolide interrupts rRNA synthesis and induces the RPL23-MDM2-p53 pathway to repress lung cancer cells. Oncol. Rep. 2020, 43, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Bursac, S.; Brdovcak, M.C.; Donati, G.; Volarevic, S. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim. Biophys. Acta 2014, 1842, 817–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kofler, L.; Prattes, M.; Bergler, H. From Snapshots to Flipbook—Resolving the Dynamics of Ribosome Biogenesis with Chemical Probes. Int. J. Mol. Sci. 2020, 21, 2998. https://doi.org/10.3390/ijms21082998
Kofler L, Prattes M, Bergler H. From Snapshots to Flipbook—Resolving the Dynamics of Ribosome Biogenesis with Chemical Probes. International Journal of Molecular Sciences. 2020; 21(8):2998. https://doi.org/10.3390/ijms21082998
Chicago/Turabian StyleKofler, Lisa, Michael Prattes, and Helmut Bergler. 2020. "From Snapshots to Flipbook—Resolving the Dynamics of Ribosome Biogenesis with Chemical Probes" International Journal of Molecular Sciences 21, no. 8: 2998. https://doi.org/10.3390/ijms21082998
APA StyleKofler, L., Prattes, M., & Bergler, H. (2020). From Snapshots to Flipbook—Resolving the Dynamics of Ribosome Biogenesis with Chemical Probes. International Journal of Molecular Sciences, 21(8), 2998. https://doi.org/10.3390/ijms21082998





