The Dark Side of Orchid Symbiosis: Can Tulasnella calospora Decompose Host Tissues?
Abstract
:1. Introduction
2. Results
2.1. Seed Germination and Microscopy Observations
2.2. CAZymes Profiles in T. calospora and Other Basidiomycetes
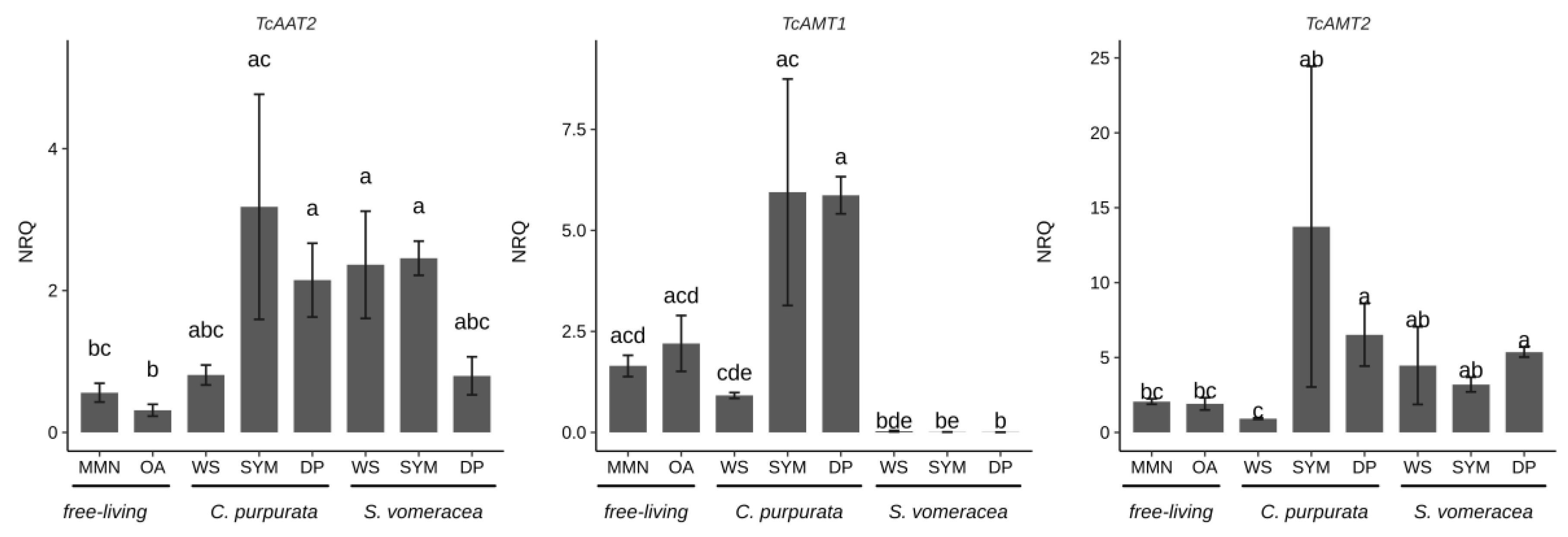
2.3. CAZymes Expression in the Free-Living Mycelium and in Plant Tissues
3. Discussion
4. Materials and Methods
4.1. Fungal Material and Growth as Free-living Mycelium
4.2. Orchid Seed Germination and Symbiotic Fungal Growth
4.3. Microscopy
4.4. RNA Extraction and cDNA Synthesis
4.5. Primers Design and RT-qPCR
4.6. Statistical Analysis
4.7. RNA-seq Expression Profiles of CAZymes in T. calospora and Other Basidiomycetes Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OM | Orchid Mycorrhiza |
| OMF | Orchid Mycorrhizal Fungi |
| PCW | Plant Cell Wall |
| CAZyme | Carbohydrate-active enzyme |
| GH | Glycoside-Hydrolase |
| GT | Glycosyl Transferase |
| PL | Polysaccharide Lyase |
| CE | Carbohydrate Esterase |
| AA | Auxiliary Activities |
| LPMO | Lytic Polysaccharide Monooxygenase |
| ECM | Ectomycorrhiza |
| WGA | Wheat Germ Agglutinin |
References
- Brundrett, M.C. Understanding the roles of multifunctional mycorrhizal and endophytic fungi. In Microbial Root Endophytes; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 281–298. [Google Scholar]
- Dearnaley, J.D.W.; Martos, F.; Selosse, M.-A. Orchid mycorrhizas: Molecular ecology, physiology, evolution and conservation aspects. In Fungal Associations; Hock, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 207–230. [Google Scholar]
- Dearnaley, J.D.W. Further advances in orchid mycorrhizal research. Mycorrhiza 2007, 17, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Girlanda, M.; Segreto, R.; Cafasso, D.; Liebel, H.T.; Rodda, M.; Ercole, E.; Cozzolino, S.; Gebauer, G.; Perotto, S. Photosynthetic Mediterranean meadow orchids feature partial mycoheterotrophy and specific mycorrhizal associations. Am. J. Bot. 2011, 98, 1148–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veldre, V.; Abarenkov, K.; Bahram, M.; Martos, F.; Selosse, M.A.; Tamm, H.; Kõljalg, U.; Tedersoo, L. Evolution of nutritional modes of Ceratobasidiaceae (Cantharellales, Basidiomycota) as revealed from publicly available ITS sequences. Fungal Ecol. 2013, 6, 256–268. [Google Scholar] [CrossRef]
- Weiß, M.; Waller, F.; Zuccaro, A.; Selosse, M.-A. Sebacinales—One thousand and one interactions with land plants. New Phytol. 2016, 211, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Martos, F.; Dulormne, M.; Pailler, T.; Bonfante, P.; Faccio, A.; Fournel, J.; Dubois, M.P.; Selosse, M.A. Independent recruitment of saprotrophic fungi as mycorrhizal partners by tropical achlorophyllous orchids. New Phytol. 2009, 184, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, K.; Matsubayashi, J.; Tayasu, I. Some mycoheterotrophic orchids depend on carbon from dead wood: Novel evidence from a radiocarbon approach. New Phytol. 2020. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Nurfadilah, S.; Swarts, N.D.; Dixon, K.W.; Lambers, H.; Merritt, D.J. Variation in nutrient-acquisition patterns by mycorrhizal fungi of rare and common orchids explains diversification in a global biodiversity hotspot. Ann. Bot. 2013, 111, 1233–1241. [Google Scholar] [CrossRef] [Green Version]
- Bahram, M.; Peay, K.G.; Tedersoo, L. Local-scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytol. 2015, 205, 1454–1463. [Google Scholar] [CrossRef]
- Voyron, S.; Ercole, E.; Ghignone, S.; Perotto, S.; Girlanda, M. Fine-scale spatial distribution of orchid mycorrhizal fungi in the soil of host-rich grasslands. New Phytol. 2017, 213, 1428–1439. [Google Scholar] [CrossRef]
- Egidi, E.; May, T.W.; Franks, A.E. Seeking the needle in the haystack: Undetectability of mycorrhizal fungi outside of the plant rhizosphere associated with an endangered Australian orchid. Fungal Ecol. 2018, 33, 13–23. [Google Scholar] [CrossRef]
- Harley, J.L.; Smith, S.E. Mycorrhizal Symbiosis; Academic Press Inc.: Cambridge, MA, USA, 1983. [Google Scholar]
- Leake, J.R. The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 1994, 127, 171–216. [Google Scholar] [CrossRef]
- Fesel, P.H.; Zuccaro, A. Dissecting endophytic lifestyle along the parasitism/mutualism continuum in Arabidopsis. Curr. Opin. Microbiol. 2016, 32, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veneault-Fourrey, C.; Commun, C.; Kohler, A.; Morin, E.; Balestrini, R.; Plett, J.; Danchin, E.; Coutinho, P.; Wiebenga, A.; de Vries, R.P.; et al. Genomic and transcriptomic analysis of Laccaria bicolor CAZome reveals insights into polysaccharides remodelling during symbiosis establishment. Fungal Genet. Biol. 2014, 72, 168–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sillo, F.; Fangel, J.U.; Henrissat, B.; Faccio, A.; Bonfante, P.; Martin, F.; Willats, W.G.T.; Balestrini, R. Understanding plant cell-wall remodelling during the symbiotic interaction between Tuber melanosporum and Corylus avellana using a carbohydrate microarray. Planta 2016, 244, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Balestrini, R.; Bonfante, P. Cell wall remodeling in mycorrhizal symbiosis: A way towards biotrophism. Front. Plant. Sci. 2014, 5, 237. [Google Scholar] [CrossRef] [Green Version]
- Williamson, B.; Hadley, G. Penetration and infection of orchid protocorms by Thanatephorus cucumeris. Pathology 1970, 60, 1092–1096. [Google Scholar]
- Shimura, H.; Koda, Y. Enhanced symbiotic seed germination of Cypripedium macranthos var. rebunense following inoculation after cold treatment. Physiol. Plant. 2005, 123, 281–287. [Google Scholar] [CrossRef]
- Cig, A.; Yılmaz, H. In vitro symbiotic culture studies of some orchid species. J. Agric. Sci. 2017, 23, 453–463. [Google Scholar]
- Perotto, S.; Rodda, M.; Benetti, A.; Sillo, F.; Ercole, E.; Rodda, M.; Girlanda, M.; Murat, C.; Balestrini, R. Gene expression in mycorrhizal orchid protocorms suggests a friendly plant–fungus relationship. Planta 2014, 239, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Fochi, V.; Chitarra, W.; Kohler, A.; Voyron, S.; Singan, V.R.; Lindquist, E.A.; Barry, K.W.; Girlanda, M.; Grigoriev, I.V.; Martin, F.; et al. Fungal and plant gene expression in the Tulasnella calospora–Serapias vomeracea symbiosis provides clues about nitrogen pathways in orchid mycorrhizas. New Phytol. 2017, 213, 365–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fochi, V.; Falla, N.; Girlanda, M.; Perotto, S.; Balestrini, R. Cell-specific expression of plant nutrient transporter genes in orchid mycorrhizae. Plant. Sci. 2017, 263, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Kämper, J.; Kahmann, R.; Bölker, M.; Ma, L.J.; Brefort, T.; Saville, B.J.; Banuett, F.; Kronstad, J.W.; Gold, S.E.; Müller, O.; et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago Maydis. Nature 2006, 444, 97–101. [Google Scholar]
- Ohm, R.A.; De Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; De Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E.; et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 28, 957–963. [Google Scholar] [CrossRef] [Green Version]
- Ohm, R.A.; Riley, R.; Salamov, A.; Min, B.; Choi, I.G.; Grigoriev, I.V. Genomics of wood-degrading fungi. Fungal Genet. Biol. 2014, 72, 82–90. [Google Scholar] [CrossRef]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.L.; Hacquard, S.; Amselem, J.; Cantarel, B.L.; et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef] [Green Version]
- Cuomo, C.A.; Bakkeren, G.; Khalil, H.B.; Panwar, V.; Joly, D.; Linning, R.; Sakthikumar, S.; Song, X.; Adiconis, X.; Fan, L.; et al. Comparative analysis highlights variable genome content of wheat rusts and divergence of the mating loci. G3 Genes Genomes Genet. 2017, 7, 361–376. [Google Scholar] [CrossRef] [Green Version]
- Salcedo, A.; Rutter, W.; Wang, S.; Akhunova, A.; Bolus, S.; Chao, S.; Anderson, N.; De Soto, M.F.; Rouse, M.; Szabo, L.; et al. Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 2017, 358, 1604–1606. [Google Scholar] [CrossRef] [Green Version]
- Lanver, D.; Müller, A.N.; Happel, P.; Schweizer, G.; Haas, F.B.; Franitza, M.; Pellegrin, C.; Reissmann, S.; Altmüller, J.; Rensing, S.A.; et al. The biotrophic development of Ustilago maydis studied by RNA-Seq analysis. Plant. Cell 2018, 30, 300–323. [Google Scholar] [CrossRef] [Green Version]
- Krizsán, K.; Almási, É.; Merényi, Z.; Sahu, N.; Virágh, M.; Kószó, T.; Mondo, S.; Kiss, B.; Bálint, B.; Kües, U.; et al. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc. Natl. Acad. Sci. USA 2019, 116, 7409–7418. [Google Scholar]
- Balestrini, R.; Nerva, L.; Sillo, F.; Girlanda, M.; Perotto, S. Plant and fungal gene expression in mycorrhizal protocorms of the orchid Serapias vomeracea colonized by Tulasnella calospora. Plant. Signal. Behav. 2014, 9, e977707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, C.-M.; Chung, K.M.; Liang, C.-K.; Tsai, W.-C. New insights into the symbiotic relationship between orchids and fungi. Appl. Sci. 2019, 9, 585. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Anasontzis, G.E.; Labourel, A.; Champion, C.; Haon, M.; Kemppainen, M.; Commun, C.; Deveau, A.; Pardo, A.; Veneault-Fourrey, C.; et al. The ectomycorrhizal basidiomycete Laccaria bicolor releases a secreted β-1,4 endoglucanase that plays a key role in symbiosis development. New Phytol. 2018, 220, 1309–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, P.L.; Lahiri, S. Lusianthrin and lusianthridin, two stilbenoids from the orchid Lusia indivisa. Phytochemistry 1990, 29, 621–624. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy data-base to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Ercole, E.; Rodda, M.; Girlanda, M.; Perotto, S. Establishment of a symbiotic in vitro system between a green meadow orchid and a Rhizoctonia-like fungus. Bioprotocol 2015, 5, e1482. [Google Scholar] [CrossRef]
- Calevo, J.; Giovannini, A.; Cornara, L.; Peccenini, S. Asymbiotic seed germination of hand-pollinated terrestrial orchids. Acta Hort. 2017, 1155, 415–418. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Otero, J.T.; Ackerman, J.D.; Bayman, P. Differences in mycorrhizal preferences between two tropical orchids. Mol. Ecol. 2004, 13, 2393–2404. [Google Scholar] [CrossRef]
- Gueddari, N.E.E.; Rauchhaus, U.; Moerschbacher, B.M.; Deising, H.B. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 2002, 156, 103–112. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant. Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Guether, M.; Balestrini, R.; Hannah, M.; He, J.; Udvardi, M.K.; Bonfante, P. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 2009, 182, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Berruti, A.; Christiaens, A.; De Keyser, E.; Van Labeke, M.-C.; Scariot, V. Cold treatment breaks dormancy but jeopardizes flower quality in Camellia japonica L. Front. Plant. Sci. 2015, 6, 983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- Ogle, D.H.; Wheeler, P.; Dinno, A. FSA: Fisheries Stock Analysis. R Package Version 0.8.26. Available online: https://github.com/droglenc/FSA (accessed on 23 December 2019).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active en-zyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
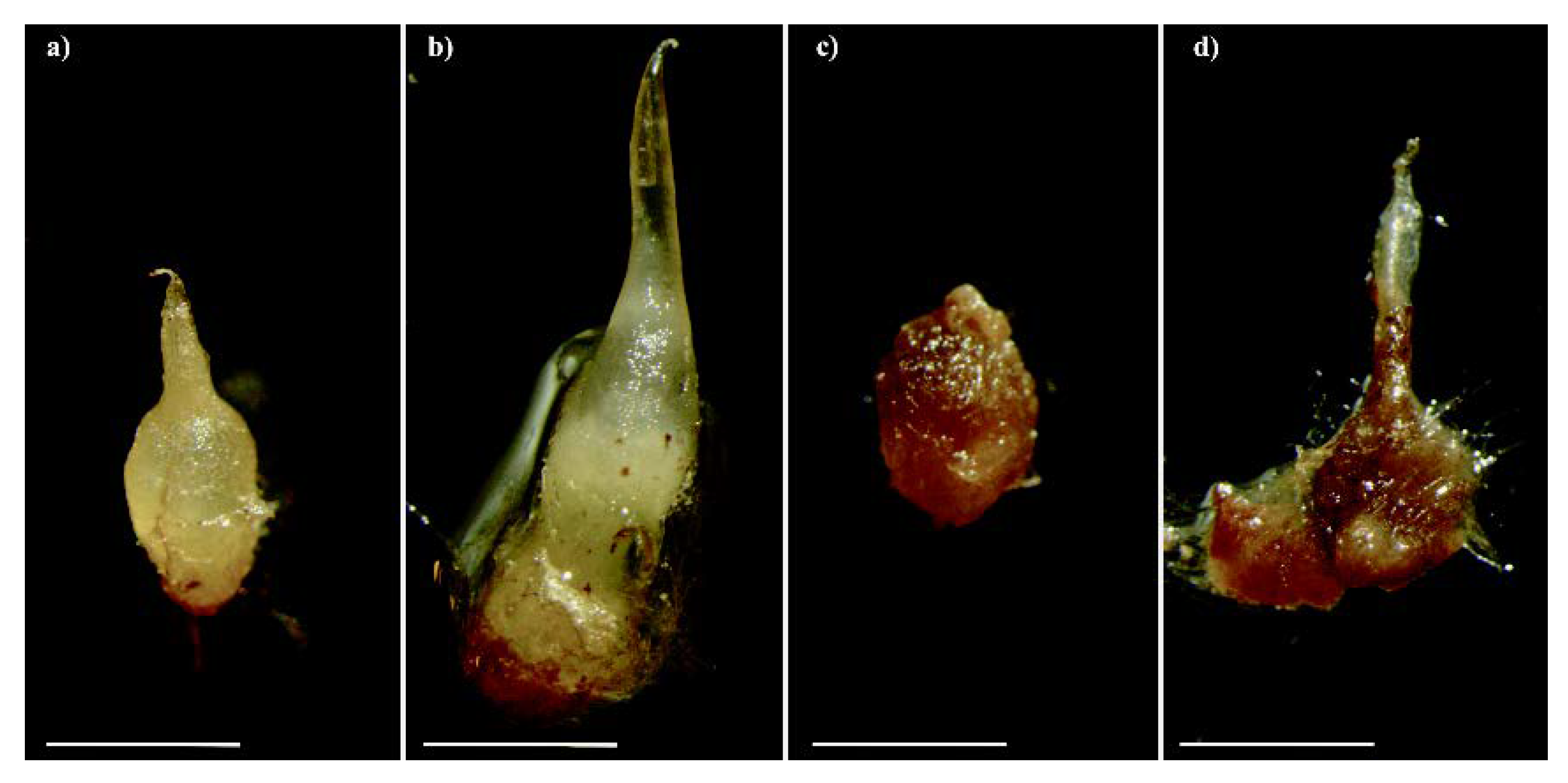

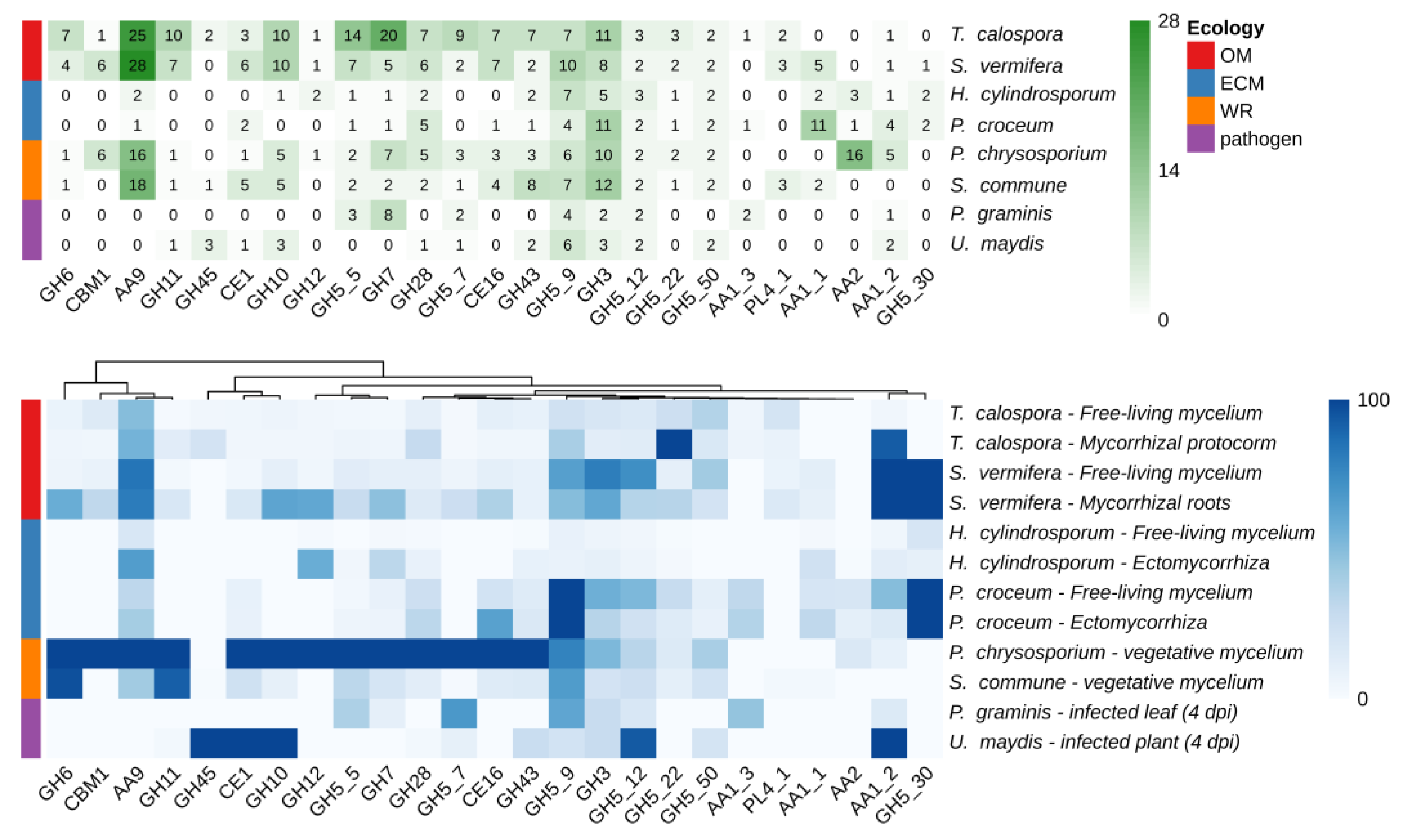

| Experimental Condition | ID | Description |
|---|---|---|
| Symbiotic roots/protocorms | SYM | Roots of C. purpurata seedlings or S. vomeracea protocorms colonized by T. calospora |
| Dark colonized protocorm/seedlings | DP | Protocorms/young seedlings of C. purpurata or protocorms of S. vomeracea, colonized by T. calospora, turned brown/black |
| Free-living fungus on dead plant tissues | WS | Dead leaves obtained from asymbiotic growth, then aseptically dried and inoculated with T. calospora |
| Free-living fungus on OA | OA | Free-living T. calospora grown on a complex Oat-meal Agar medium |
| Free-living fungus on MMN | MMN | Free-living T. calospora grown on mineral Melin-Norkins modified medium |
| Gene Name | Putative Function | Transcript ID † | MYC Normalized Reads ‡ | FLMNormalized Reads ‡ |
|---|---|---|---|---|
| Tulasnella calospora | ||||
| TcGH6 | cellobiohydrolase | 69053 | 2.71 | 12.98 |
| TcGH10a | endo-1,4-β-xylanase | 14789 | 3.37 | 24.59 |
| TcGH11 | endo-1,4-β-xylanase | 80414 | 5.43 | 0.62 |
| TcGH45* | endoglucanase | 224031 | 26.58 | 6.49 |
| TcAA9a | Lytic polysaccharide monooxygenase (LPMO) | 75481 | 3.45 | 12.18 |
| TcAA9b | LPMO | 6298 | 102.01 | 61.82 |
| TcAA9f | LPMO | 4643 | 16.35 | 0.33 |
| TcAMT1 | ammonium transporter | 241330 | 584.63 | 324.12 |
| TcAMT2 | ammonium transporter | 183841 | 525.27 | 184.32 |
| TcAAT2 | amino acid transporter | 81605 | 363.85 | 77.20 |
| Serapias vomeracea | ||||
| SvNod1 | early nodulin 55-2, putative | DN89686_c0_g1_i1 | - | - |
| SvEXO | exocyst subunit exo70 | DN73752_c2_g2_i1 | 42.61 | 7.38 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adamo, M.; Chialva, M.; Calevo, J.; De Rose, S.; Girlanda, M.; Perotto, S.; Balestrini, R. The Dark Side of Orchid Symbiosis: Can Tulasnella calospora Decompose Host Tissues? Int. J. Mol. Sci. 2020, 21, 3139. https://doi.org/10.3390/ijms21093139
Adamo M, Chialva M, Calevo J, De Rose S, Girlanda M, Perotto S, Balestrini R. The Dark Side of Orchid Symbiosis: Can Tulasnella calospora Decompose Host Tissues? International Journal of Molecular Sciences. 2020; 21(9):3139. https://doi.org/10.3390/ijms21093139
Chicago/Turabian StyleAdamo, Martino, Matteo Chialva, Jacopo Calevo, Silvia De Rose, Mariangela Girlanda, Silvia Perotto, and Raffaella Balestrini. 2020. "The Dark Side of Orchid Symbiosis: Can Tulasnella calospora Decompose Host Tissues?" International Journal of Molecular Sciences 21, no. 9: 3139. https://doi.org/10.3390/ijms21093139
APA StyleAdamo, M., Chialva, M., Calevo, J., De Rose, S., Girlanda, M., Perotto, S., & Balestrini, R. (2020). The Dark Side of Orchid Symbiosis: Can Tulasnella calospora Decompose Host Tissues? International Journal of Molecular Sciences, 21(9), 3139. https://doi.org/10.3390/ijms21093139







