Digital PCR: A Reliable Tool for Analyzing and Monitoring Hematologic Malignancies
Abstract
1. Introduction
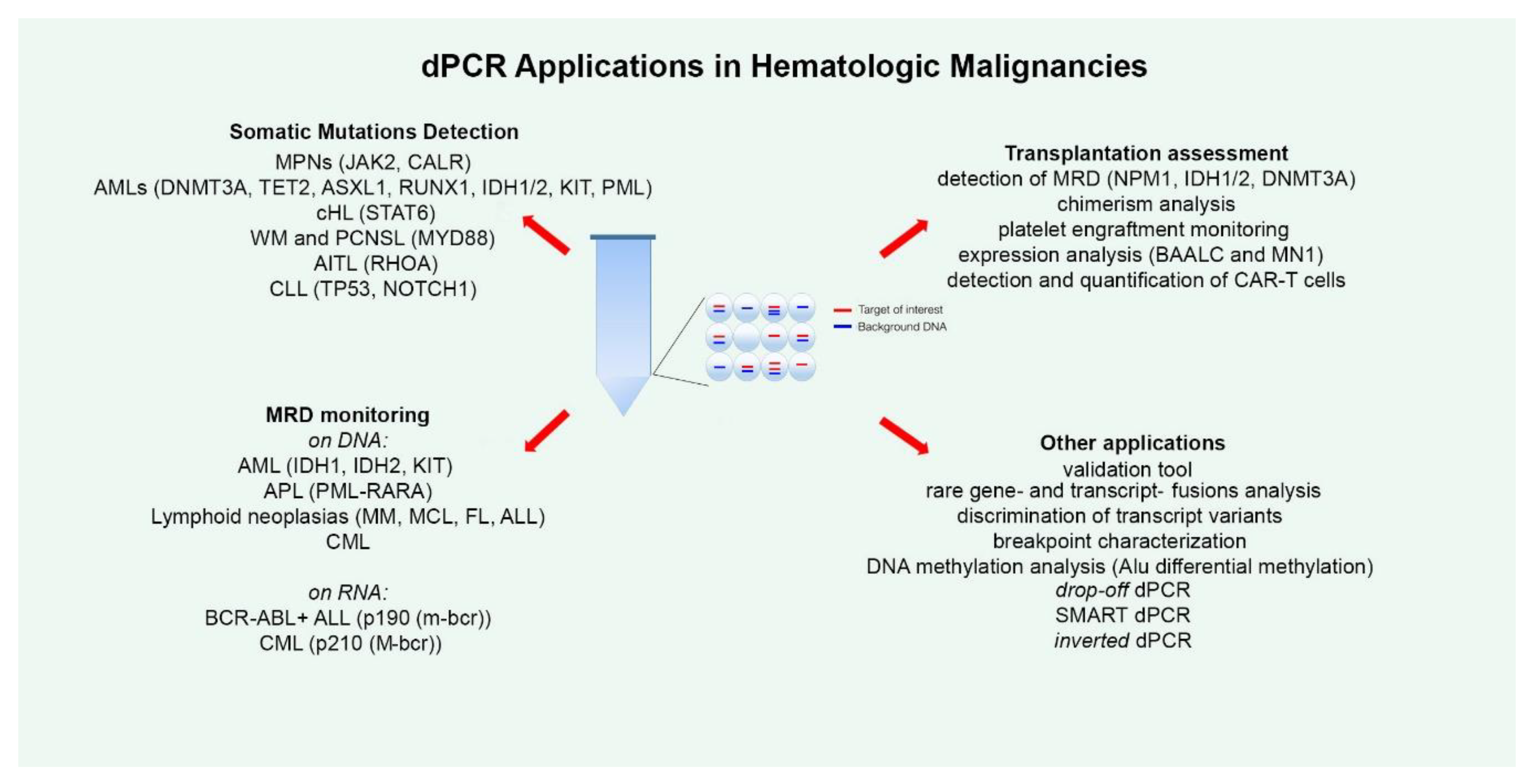
2. dPCR for Detecting Somatic Mutations
3. dPCR for MRD Monitoring
4. dPCR and Transplantation
5. Other dPCR Applications and Evolution
6. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AITL | angioimmunoblastic T-cell lymphoma |
| Allo-HSCT | allogeneic-HSCT |
| AMLs | Acute Myeloid Leukemias |
| APL | acute promyelocytic leukemia |
| BM | bone marrow |
| CAR-T | chimeric antigen receptor T |
| CBF | core binding factor |
| cfDNA | cell-free DNA |
| cHL | Classical Hodgkin lymphoma |
| CLL | Chronic Lymphocytic Leukemia |
| CML | Chronic myeloid leukemia |
| CNS | central nervous system |
| CR | complete remission |
| DLBCL | Diffuse Large B-Cell Lymphomas |
| dPCR | digital PCR |
| ddPCR | droplet digital PCR |
| ET | essential thrombocythemia |
| FA | fractional abundance |
| FFPE | formalin-fixed paraffin-embedded |
| FISH | fluorescence in situ hybridization |
| FL | Follicular Lymphoma |
| FLA | Fragment length analysis |
| FU | follow-up |
| HSCT | hematopoietic stem cell transplantation |
| IgH | immunoglobulin heavy chain |
| MC | mixed chimerism |
| MCL | Mantle Cell Lymphoma |
| MF | Myelofibrosis |
| MLPA | Multiplex ligation-dependent probe amplification |
| MM | Multiple Myeloma |
| MRD | Minimal Residual Disease |
| NGS | Next-Generation Sequencing |
| OS | Overall survival |
| PAI | preferential allelic imbalance |
| PB | peripheral blood |
| PC | plasma cells |
| PCNSL | central nervous system lymphomas |
| PFS | progression-free survival |
| Ph- MPNs | Philadelphia negative chronic Myeloproliferative Neoplasms |
| PMBCL | primary mediastinal large B cell lymphoma |
| PNA-LNA | peptide nucleic acid-locked nucleic acid |
| PNQ | positive not-quantifiable |
| PTCL | peripheral T-cell lymphoma |
| PV | polycythemia vera |
| qPCR | Real-time Quantitative PCR |
| SMART-ddPCR | Somatic Mutation Allelic Ratio Test ddPCR |
| SNP | single nucleotide polymorphism |
| SS | sanger sequencing |
| STRs | short tandem repeats |
| TDS | targeted deep sequencing |
| TKI | tyrosine kinase inhibitors |
| VAF | variant allele fractions |
| WM | Waldenström Macroglobulinemia |
References
- Morley, A.A. Digital PCR: A brief history. Biomol. Detect. Quantif. 2014, 1, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Jeffreys, A.J.; Neumann, R.; Wilson, V. Repeat unit sequence variation in minisatellites: A novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell 1990, 60, 473–485. [Google Scholar] [CrossRef]
- Ruano, G.; Kidd, K.K.; Stephens, J.C. Haplotype of multiple polymorphisms resolved by enzymatic amplification of single DNA molecules. Proc. Natl. Acad. Sci. USA 1990, 87, 6296–6300. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Balfe, P.; Peutherer, J.F.; Ludlam, C.A.; Bishop, J.O.; Brown, A.J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J. Virol. 1990, 64, 864–872. [Google Scholar] [CrossRef]
- Brisco, M.J.; Condon, J.; Sykes, P.J.; Neoh, S.H.; Morley, A.A. Detection and quantitation of neoplastic cells in acute lymphoblastic leukaemia, by use of the polymerase chain reaction. Br. J. Haematol. 1991, 79, 211–217. [Google Scholar] [CrossRef]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992, 13, 444–449. [Google Scholar]
- Huggett, J.F.; Cowen, S.; Foy, C.A. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin. Chem. 2015, 61, 79–88. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Dressman, D.; Yan, H.; Traverso, G.; Kinzler, K.W.; Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. USA 2003, 100, 8817–8822. [Google Scholar] [CrossRef]
- Goh, H.G.; Lin, M.; Fukushima, T.; Saglio, G.; Kim, D.; Choi, S.Y.; Kim, S.H.; Lee, J.; Lee, Y.S.; Oh, S.M.; et al. Sensitive quantitation of minimal residual disease in chronic myeloid leukemia using nanofluidic digital polymerase chain reaction assay. Leuk. Lymphoma 2011, 52, 896–904. [Google Scholar] [CrossRef]
- Alikian, M.; Whale, A.S.; Akiki, S.; Piechocki, K.; Torrado, C.; Myint, T.; Cowen, S.; Griffiths, M.; Reid, A.G.; Apperley, J.; et al. RT-qPCR and RT-digital PCR: A comparison of different platforms for the evaluation of residual disease in chronic myeloid leukemia. Clin. Chem. 2017, 63, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.J.; George, D.; Czech, J.; Yu, M.; Joseph, L. Detection and quantification of BCR-ABL1 fusion transcripts by droplet digital PCR. J. Mol. Diagn. 2014, 16, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Madic, J.; Zocevic, A.; Senlis, V.; Fradet, E.; Andre, B.; Muller, S.; Dangla, R.; Droniou, M.E. Three-color crystal digital PCR. Biomol. Detect. Quantif. 2016, 10, 34–46. [Google Scholar] [CrossRef]
- Low, H.; Chan, S.-J.; Soo, G.-H.; Ling, B.; Tan, E.-L. ClarityTM digital PCR system: A novel platform for absolute quantification of nucleic acids. Anal. Bioanal. Chem. 2017, 409, 1869–1875. [Google Scholar] [CrossRef]
- Thress, K.S.; Brant, R.; Carr, T.H.; Dearden, S.; Jenkins, S.; Brown, H.; Hammett, T.; Cantarini, M.; Barrett, J.C. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015, 90, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Balaj, L.; Liau, L.M.; Samuels, M.L.; Kotsopoulos, S.K.; Maguire, C.A.; LoGuidice, L.; Soto, H.; Garrett, M.; Zhu, L.D.; et al. Beaming and droplet digital pcr analysis of mutant idh1 mrna in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol. Ther. Nucleic Acids 2013, 2, e109. [Google Scholar] [CrossRef] [PubMed]
- Olmedillas-López, S.; García-Arranz, M.; García-Olmo, D. Current and emerging applications of droplet digital PCR in oncology. Mol. Diagn. Ther. 2017, 21, 493–510. [Google Scholar] [CrossRef]
- Voso, M.T.; Ottone, T.; Lavorgna, S.; Venditti, A.; Maurillo, L.; Lo-Coco, F.; Buccisano, F. MRD in AML: The role of new techniques. Front. Oncol. 2019, 9, 655. [Google Scholar] [CrossRef]
- Cilloni, D.; Petiti, J.; Rosso, V.; Andreani, G.; Dragani, M.; Fava, C.; Saglio, G. Digital PCR in myeloid malignancies: Ready to replace quantitative PCR? Int. J. Mol. Sci. 2019, 20, 2249. [Google Scholar] [CrossRef]
- Takamatsu, H.; Takamatsu, H. Comparison of minimal residual disease detection by multiparameter flow cytometry, ASO-qPCR, droplet digital PCR, and deep sequencing in patients with multiple myeloma who underwent autologous stem cell transplantation. J. Clin. Med. 2017, 6, 91. [Google Scholar] [CrossRef]
- Della Starza, I.; Chiaretti, S.; De Propris, M.S.; Elia, L.; Cavalli, M.; De Novi, L.A.; Soscia, R.; Messina, M.; Vitale, A.; Guarini, A.; et al. Minimal residual disease in acute lymphoblastic leukemia: Technical and clinical advances. Front. Oncol. 2019, 9, 726. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Cui, X.; Hu, J.; Li, Z.; Choi, J.R.; Yang, Q.; Lin, M.; Li, Y.H.; Xu, F. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens. Bioelectron. 2017, 90, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Sauzade, M.; Brouzes, E. DPCR: A technology review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed]
- Dube, S.; Qin, J.; Ramakrishnan, R. Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS ONE 2008, 3, e2876. [Google Scholar] [CrossRef]
- Whale, A.S.; Cowen, S.; Foy, C.A.; Huggett, J.F. Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS ONE 2013, 8, e58177. [Google Scholar] [CrossRef]
- Williams, R.; Peisajovich, S.G.; Miller, O.J.; Magdassi, S.; Tawfik, D.S.; Griffiths, A.D. Amplification of complex gene libraries by emulsion PCR. Nat. Methods 2006, 3, 545–550. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Kalinina, O.; Lebedeva, I.; Brown, J.; Silver, J. Nanoliter scale PCR with TaqMan detection. Nucleic Acids Res. 1997, 25, 1999–2004. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. 1), S49–S52. [Google Scholar]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Koren-Michowitz, M.; Shimoni, A.; Daraio, F.; Crasto, F.; Lorenzatti, R.; Volchek, Y.; Amariglio, N.; Gottardi, E.; Saglio, G.; Nagler, A. Sensitive replicate real-time quantitative PCR of BCR-ABL shows deep molecular responses in long-term post-allogeneic stem cell transplantation chronic myeloid leukemia patients. Biol. Blood Marrow Transpl. 2015, 21, 1852–1855. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, E.; Lee, K.J.; Park, H.; Chung, J.Y.; Lee, M.N.; Chang, M.H.; Yoo, J.; Lee, H.; Kong, S.Y.; Eom, H.S. Clinical implications of quantitative JAK2 V617F analysis using droplet digital PCR in myeloproliferative neoplasms. Ann. Lab. Med. 2018, 38, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Klampfl, T.; Gisslinger, H.; Harutyunyan, A.S.; Nivarthi, H.; Rumi, E.; Milosevic, J.D.; Them, N.C.C.; Berg, T.; Gisslinger, B.; Pietra, D.; et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N. Engl. J. Med. 2013, 369, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- Storlazzi, C.T.; Albano, F.; Locunsolo, C.; Lonoce, A.; Funes, S.; Guastadisegni, M.C.; Cimarosto, L.; Impera, L.; D’Addabbo, P.; Panagopoulos, I.; et al. t(3;12)(q26;q14) in polycythemia vera is associated with upregulation of the HMGA2 gene. Leukemia 2006, 20, 2190–2192. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fontanelli, G.; Baratè, C.; Ciabatti, E.; Guerrini, F.; Grassi, S.; Del Re, M.; Morganti, R.; Petrini, I.; Arici, R.; Barsotti, S.; et al. Real-time PCR and droplet digital PCR: Two techniques for detection of the JAK2V617F mutation in Philadelphia-negative chronic myeloproliferative neoplasms. Int. J. Lab. Hematol. 2015, 37, 766–773. [Google Scholar] [CrossRef]
- Waterhouse, M.; Follo, M.; Pfeifer, D.; von Bubnoff, N.; Duyster, J.; Bertz, H.; Finke, J. Sensitive and accurate quantification of JAK2 V617F mutation in chronic myeloproliferative neoplasms by droplet digital PCR. Ann. Hematol. 2016, 95, 739–744. [Google Scholar] [CrossRef]
- Link-Lenczowska, D.; Pallisgaard, N.; Cordua, S.; Zawada, M.; Czekalska, S.; Krochmalczyk, D.; Kanduła, Z.; Sacha, T. A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann. Hematol. 2018, 97, 2299–2308. [Google Scholar] [CrossRef]
- Kröger, N.; Badbaran, A.; Holler, E.; Hahn, J.; Kobbe, G.; Bornhäuser, M.; Reiter, A.; Zabelina, T.; Zander, A.R.; Fehse, B. Monitoring of the JAK2-V617F mutation by highly sensitive quantitative real-time PCR after allogeneic stem cell transplantation in patients with myelofibrosis. Blood 2007, 109, 1316–1321. [Google Scholar] [CrossRef]
- Nystrand, C.F.; Ghanima, W.; Waage, A.; Jonassen, C.M. JAK2 V617F mutation can be reliably detected in serum using droplet digital PCR. Int. J. Lab. Hematol. 2018, 40, 181–186. [Google Scholar] [CrossRef]
- Anelli, L.; Zagaria, A.; Coccaro, N.; Tota, G.; Minervini, A.; Casieri, P.; Impera, L.; Minervini, C.F.; Brunetti, C.; Ricco, A.; et al. Droplet digital PCR assay for quantifying of CALR mutant allelic burden in myeloproliferative neoplasms. Ann. Hematol. 2016, 95, 1559–1560. [Google Scholar] [CrossRef]
- Mansier, O.; Migeon, M.; Saint-Lézer, A.; James, C.; Verger, E.; Robin, M.; Socié, G.; Bidet, A.; Mahon, F.X.; Cassinat, B.; et al. Quantification of the mutant CALR allelic burden by digital PCR: Application to minimal residual disease evaluation after bone marrow transplantation. J. Mol. Diagn. 2016, 18, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Badbaran, A.; Fehse, B.; Christopeit, M.; Aranyossy, T.; Ayuk, F.A.; Wolschke, C.; Kröger, N. Digital-PCR assay for screening and quantitative monitoring of calreticulin (CALR) type-2 positive patients with myelofibrosis following allogeneic stem cell transplantation. Bone Marrow Transpl. 2016, 51, 872–873. [Google Scholar] [CrossRef] [PubMed]
- Handschuh, L.; Kazmierczak, M.; Milewski, M.C.; Goralski, M.; Luczak, M.; Wojtaszewska, M.; Uszczynska-Ratajczak, B.; Lewandowski, K.; Komarnicki, M.; Figlerowicz, M. Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR. Int. J. Oncol. 2018, 52, 656–678. [Google Scholar] [CrossRef] [PubMed]
- Parkin, B.; Londoño-Joshi, A.; Kang, Q.; Tewari, M.; Rhim, A.D.; Malek, S.N. Ultrasensitive mutation detection identifies rare residual cells causing acute myelogenous leukemia relapse. J. Clin. Investig. 2017, 127, 3484–3495. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, Z.; Wang, W.; Zhu, G.; Guo, J.; Chen, X.; Zheng, C.; Xu, Z.; Chang, J.; Ren, F.; et al. Monitoring of clonal evolution of double C-KIT exon 17 mutations by droplet digital PCR in patients with core-binding factor acute myeloid leukemia. Leuk. Res. 2018, 69, 89–93. [Google Scholar] [CrossRef]
- Alfonso, V.; Iaccarino, L.; Ottone, T.; Cicconi, L.; Lavorgna, S.; Divona, M.; Cairoli, R.; Cristiano, A.; Ciardi, C.; Travaglini, S.; et al. Early and sensitive detection of PML-A216V mutation by droplet digital PCR in ATO-resistant acute promyelocytic leukemia. Leukemia 2019, 33, 1527–1530. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Lee Harris, N.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Tiacci, E.; Ladewig, E.; Schiavoni, G.; Penson, A.; Fortini, E.; Pettirossi, V.; Wang, Y.; Rosseto, A.; Venanzi, A.; Vlasevska, S.; et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood 2018, 131, 2454–2465. [Google Scholar] [CrossRef]
- Reichel, J.; Chadburn, A.; Rubinstein, P.G.; Giulino-Roth, L.; Tam, W.; Liu, Y.; Gaiolla, R.; Eng, K.; Brody, J.; Inghirami, G.; et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015, 125, 1061–1072. [Google Scholar] [CrossRef]
- Camus, V.; Stamatoullas, A.; Mareschal, S.; Viailly, P.J.; Sarafan-Vasseur, N.; Bohers, E.; Dubois, S.; Picquenot, J.M.; Ruminy, P.; Maingonnat, C.; et al. Detection and prognostic value of recurrent exportin 1 mutations in tumor and cell-free circulating DNA of patients with classical Hodgkin lymphoma. Haematologica 2016, 101, 1094–1101. [Google Scholar] [CrossRef]
- Spina, V.; Bruscaggin, A.; Cuccaro, A.; Martini, M.; Di Trani, M.; Forestieri, G.; Manzoni, M.; Condoluci, A.; Arribas, A.; Terzi-Di-Bergamo, L.; et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood 2018, 131, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, L.; Noerenberg, D.; Young, E.; Mylonas, E.; Abdulla, M.; Frick, M.; Asmar, F.; Ljungström, V.; Schneider, M.; Yoshida, K.; et al. Frequent NFKBIE deletions are associated with poor outcome in primary mediastinal B-cell lymphoma. Blood 2016, 128, 2666–2670. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Hansmann, M.L.; Bohle, V.; Martin-Subero, J.I.; Hartmann, S.; Mechtersheimer, G.; Klapper, W.; Vater, I.; Giefing, M.; Gesk, S.; et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 2009, 206, 981–989. [Google Scholar] [CrossRef]
- Van Roosbroeck, K.; Cox, L.; Tousseyn, T.; Lahortiga, I.; Gielen, O.; Cauwelier, B.; De Paepe, P.; Verhoef, G.; Marynen, P.; Vandenberghe, P.; et al. JAK2 rearrangements, including the novel SEC31A-JAK2 fusion, are recurrent in classical Hodgkin lymphoma. Blood 2011, 117, 4056–4064. [Google Scholar] [CrossRef]
- Mottok, A.; Renné, C.; Seifert, M.; Oppermann, E.; Bechstein, W.; Hansmann, M.L.; Küppers, R.; Bräuninger, A. Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood 2009, 114, 4503–4506. [Google Scholar] [CrossRef]
- Weniger, M.A.; Melzner, I.; Menz, C.K.; Wegener, S.; Bucur, A.J.; Dorsch, K.; Mattfeldt, T.; Barth, T.F.E.; Möller, P. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006, 25, 2679–2684. [Google Scholar] [CrossRef]
- Gunawardana, J.; Chan, F.C.; Telenius, A.; Woolcock, B.; Kridel, R.; Tan, K.L.; Ben-Neriah, S.; Mottok, A.; Lim, R.S.; Boyle, M.; et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat. Genet. 2014, 46, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Kleppe, M.; Tousseyn, T.; Geissinger, E.; Atak, Z.K.; Aerts, S.; Rosenwald, A.; Wlodarska, I.; Cools, J. Mutation analysis of the tyrosine phosphatase PTPN2 in Hodgkin’s lymphoma and T-cell non-Hodgkin’s lymphoma. Haematologica 2011, 96, 1723–1727. [Google Scholar] [CrossRef] [PubMed]
- Vandenberghe, P.; Wlodarska, I.; Tousseyn, T.; Dehaspe, L.; Dierickx, D.; Verheecke, M.; Uyttebroeck, A.; Bechter, O.; Delforge, M.; Vandecaveye, V.; et al. Non-invasive detection of genomic imbalances in Hodgkin/Reed-Sternberg cells in early and advanced stage Hodgkin’s lymphoma by sequencing of circulating cell-free DNA: A technical proof-of-principle study. Lancet. Haematol. 2015, 2, e55–e65. [Google Scholar] [CrossRef]
- Bessi, L.; Viailly, P.J.; Bohers, E.; Ruminy, P.; Maingonnat, C.; Bertrand, P.; Vasseur, N.S.; Beaussire, L.; Cornic, M.; Etancelin, P.; et al. Somatic mutations of cell-free circulating DNA detected by targeted next-generation sequencing and digital droplet PCR in classical Hodgkin lymphoma. Leuk. Lymphoma 2019, 60, 498–502. [Google Scholar] [CrossRef]
- Campo, E.; Swerdlow, S.H.; Harris, N.L.; Pileri, S.; Stein, H.; Jaffe, E.S. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood 2011, 117, 5019–5032. [Google Scholar] [CrossRef]
- Dimopoulos, M.A.; Kastritis, E. How I treat Waldenström macroglobulinemia. Blood 2019, 134, 2022–2035. [Google Scholar] [CrossRef]
- Treon, S.P.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Sheehy, P.; Manning, R.J.; Patterson, C.J.; Tripsas, C.; et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N. Engl. J. Med. 2012, 367, 826–833. [Google Scholar] [CrossRef]
- Yang, G.; Zhou, Y.; Liu, X.; Xu, L.; Cao, Y.; Manning, R.J.; Patterson, C.J.; Buhrlage, S.J.; Gray, N.; Tai, Y.T.; et al. A mutation in MYD88 (L265P) supports the survival of lymphoplasmacytic cells by activation of Bruton tyrosine kinase in Waldenström macroglobulinemia. Blood 2013, 122, 1222–1232. [Google Scholar] [CrossRef]
- Drandi, D.; Genuardi, E.; Dogliotti, I.; Ferrante, M.; Jiménez, C.; Guerrini, F.; Lo Schirico, M.; Mantoan, B.; Muccio, V.; Lia, G.; et al. Highly sensitive MYD88 l265p mutation detection by droplet digital polymerase chain reaction in waldenström macroglobulinemia. Haematologica 2018, 103, 1029–1037. [Google Scholar] [CrossRef]
- Lo Schirico, M.; Ferrante, M.; Dogliotti, I.; Zamò, A.; Ferrero, B.; Bertuzzo, D.; Benevolo, G.; Omedè, P.; Cavallo, F.; Ladetto, M.; et al. Droplet digital PCR assay for MYD88L265P. HemaSphere 2020, 4, e324. [Google Scholar] [CrossRef]
- Hattori, K.; Sakata-Yanagimoto, M.; Suehara, Y.; Yokoyama, Y.; Kato, T.; Kurita, N.; Nishikii, H.; Obara, N.; Takano, S.; Ishikawa, E.; et al. Clinical significance of disease-specific MYD88 mutations in circulating DNA in primary central nervous system lymphoma. Cancer Sci. 2018, 109, 225–230. [Google Scholar] [CrossRef]
- Zorofchian, S.; Lu, G.; Zhu, J.J.; Duose, D.Y.; Windham, J.; Esquenazi, Y.; Ballester, L.Y. Detection of the MYD88p.L265P mutation in the CSF of a patient with secondary central nervous system lymphoma. Front. Oncol. 2018, 8, 382. [Google Scholar] [CrossRef]
- Tanzima Nuhat, S.; Sakata-Yanagimoto, M.; Komori, D.; Hattori, K.; Suehara, Y.; Fukumoto, K.; Fujisawa, M.; Kusakabe, M.; Matsue, K.; Wakamatsu, H.; et al. Droplet digital polymerase chain reaction assay and peptide nucleic acid-locked nucleic acid clamp method for RHOA mutation detection in angioimmunoblastic T-cell lymphoma. Cancer Sci. 2018, 109, 1682–1689. [Google Scholar] [CrossRef]
- Raponi, S.; Del Giudice, I.; Marinelli, M.; Wang, J.; Cafforio, L.; Ilari, C.; Piciocchi, A.; Messina, M.; Bonina, S.; Tavolaro, S.; et al. Genetic landscape of ultra-stable chronic lymphocytic leukemia patients. Ann. Oncol. 2018, 9, 966–972. [Google Scholar] [CrossRef]
- Amin, N.A.; Seymour, E.; Saiya-Cork, K.; Parkin, B.; Shedden, K.; Malek, S.N. A quantitative analysis of subclonal and clonal gene mutations before and after therapy in chronic lymphocytic leukemia. Clin. Cancer Res. 2016, 22, 4525–4535. [Google Scholar] [CrossRef]
- Minervini, A.; Minervini, C.F.; Anelli, L.; Zagaria, A.; Casieri, P.; Coccaro, N.; Cumbo, C.; Tota, G.; Impera, L.; Orsini, P.; et al. Droplet digital PCR analysis of NOTCH1 gene mutations in chronic lymphocytic leukemia. Oncotarget 2016, 7, 86469–86479. [Google Scholar] [CrossRef][Green Version]
- Bacher, U.; Dicker, F.; Haferlach, C.; Alpermann, T.; Rose, D.; Kern, W.; Haferlach, T.; Schnittger, S. Quantification of rare NPM1 mutation subtypes by digital PCR. Br. J. Haematol. 2014, 167, 710–714. [Google Scholar] [CrossRef]
- Petrova, L.; Vrbacky, F.; Lanska, M.; Zavrelova, A.; Zak, P.; Hrochova, K. IDH1 and IDH2 mutations in patients with acute myeloid leukemia: Suitable targets for minimal residual disease monitoring? Clin. Biochem. 2018, 61, 34–39. [Google Scholar] [CrossRef]
- Grassi, S.; Guerrini, F.; Ciabatti, E.; Salehzadeh, S.; Domenichini, C.; Di vita, A.; Tarrini, G.; Metelli, M.R.; Benedetti, E.; Caracciolo, F.; et al. IDH2 gene mutations detection in acute myeloid leukemia: Screening and MRD monitoring by “drop-off” ddPCR. HemaSphere 2019, 3, 454. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, W.; Zheng, C.; Tan, Y.; Chen, X.; Xu, J.; Xu, Z.; Ren, F.; Zhang, Y.; Li, G.; et al. Clinical significance of droplet digital PCR quantitative monitoring of KIT gene mutation levels in core binding factor leukemia. Int. J. Lab. Hematol. 2018, 40, 124–126. [Google Scholar] [CrossRef]
- Albano, F.; Zagaria, A.; Anelli, L.; Coccaro, N.; Tota, G.; Brunetti, C.; Minervini, C.F.; Impera, L.; Minervini, A.; Cellamare, A.; et al. Absolute quantification of the pretreatment PML-RARA transcript defines the relapse risk in acute promyelocytic leukemia. Oncotarget 2015, 6, 13269–13277. [Google Scholar] [CrossRef]
- Brunetti, C.; Anelli, L.; Zagaria, A.; Minervini, A.; Minervini, C.F.; Casieri, P.; Coccaro, N.; Cumbo, C.; Tota, G.; Impera, L.; et al. Droplet digital PCR is a reliable tool for monitoring minimal residual disease in acute promyelocytic leukemia. J. Mol. Diagn. 2017, 19, 437–444. [Google Scholar] [CrossRef][Green Version]
- Yuan, D.; Cui, M.; Yu, S.; Wang, H.; Jing, R. Droplet digital PCR for quantification of PML-RARA in acute promyelocytic leukemia: A comprehensive comparison with real-time PCR. Anal. Bioanal. Chem. 2019, 411, 895–903. [Google Scholar] [CrossRef]
- Ferrero, S.; Ladetto, M.; Drandi, D.; Cavallo, F.; Genuardi, E.; Urbano, M.; Caltagirone, S.; Grasso, M.; Rossini, F.; Guglielmelli, T.; et al. Long-term results of the GIMEMA VEL-03-096 trial in MM patients receiving VTD consolidation after ASCT: MRD kinetics’ impact on survival. Leukemia 2015, 29, 689–695. [Google Scholar] [CrossRef]
- Pott, C. Minimal residual disease detection in mantle cell lymphoma: Technical aspects and clinical relevance. Semin. Hematol. 2011, 48, 172–184. [Google Scholar] [CrossRef]
- Ladetto, M.; Pagliano, G.; Ferrero, S.; Cavallo, F.; Drandi, D.; Santo, L.; Crippa, C.; De Rosa, L.; Pregno, P.; Grasso, M.; et al. Major tumor shrinking and persistent molecular remissions after consolidation with bortezomib, thalidomide, and dexamethasone in patients with autografted myeloma. J. Clin. Oncol. 2010, 28, 2077–2084. [Google Scholar] [CrossRef]
- Pott, C.; Hoster, E.; Delfau-Larue, M.H.; Beldjord, K.; Böttcher, S.; Asnafi, V.; Plonquet, A.; Siebert, R.; Callet-Bauchu, E.; Andersen, N.; et al. Molecular remission is an independent predictor of clinical outcome in patients with mantle cell lymphoma after combined immunochemotherapy: A European MCL intergroup study. Blood 2010, 115, 3215–3223. [Google Scholar] [CrossRef]
- Ladetto, M.; Lobetti-Bodoni, C.; Mantoan, B.; Ceccarelli, M.; Boccomini, C.; Genuardi, E.; Chiappella, A.; Baldini, L.; Rossi, G.; Pulsoni, A.; et al. Persistence of minimal residual disease in bone marrow predicts outcome in follicular lymphomas treated with a rituximab-intensive program. Blood 2013, 122, 3759–3766. [Google Scholar] [CrossRef]
- Drandi, D.; Kubiczkova-Besse, L.; Ferrero, S.; Dani, N.; Passera, R.; Mantoan, B.; Gambella, M.; Monitillo, L.; Saraci, E.; Ghione, P.; et al. Minimal residual disease detection by droplet digital PCR in multiple myeloma, mantle cell lymphoma, and follicular lymphoma: A comparison with real-time PCR. J. Mol. Diagn. 2015, 17, 652–660. [Google Scholar] [CrossRef]
- Pulsoni, A.; Della Starza, I.; Cappelli, L.V.; Tosti, M.E.; Annechini, G.; Cavalli, M.; De Novi, L.A.; D’Elia, G.M.; Grapulin, L.; Guarini, A.; et al. Minimal residual disease monitoring in early stage follicular lymphoma can predict prognosis and drive treatment with rituximab after radiotherapy. Br. J. Haematol. 2020, 88, 249–258. [Google Scholar] [CrossRef]
- Della Starza, I.; Cavalli, M.; De Novi, L.A.; Genuardi, E.; Mantoan, B.; Drandi, D.; Barbero, D.; Ciabatti, E.; Grassi, S.; Gazzola, A.; et al. Minimal residual disease (MRD) in non-Hodgkin lymphomas: Interlaboratory reproducibility on marrow samples with very low levels of disease within the FIL (Fondazione Italiana Linfomi) MRD Network. Hematol. Oncol. 2019, 37, 368–374. [Google Scholar] [CrossRef]
- Cavalli, M.; De Novi, L.A.; Della Starza, I.; Cappelli, L.V.; Nunes, V.; Pulsoni, A.; Del Giudice, I.; Guarini, A.; Foà, R. Comparative analysis between RQ-PCR and digital droplet PCR of BCL2/IGH gene rearrangement in the peripheral blood and bone marrow of early stage follicular lymphoma. Br. J. Haematol. 2017, 177, 588–596. [Google Scholar] [CrossRef]
- Drandi, D.; Alcantara, M.; Benmaad, I.; Söhlbrandt, A.; Lhermitte, L.; Zaccaria, G.; Ferrante, M.; Genuardi, E.; Mantoan, B.; Villarese, P.; et al. Droplet digital PCR quantification of mantle cell lymphoma follow-up samples from four prospective trials of the European MCL network. HemaSphere 2020, 4, e347. [Google Scholar]
- Bonifacio, M.; Stagno, F.; Scaffidi, L.; Krampera, M.; Di Raimondo, F. Management of chronic myeloid leukemia in advanced phase. Front. Oncol. 2019, 9, 1132. [Google Scholar] [CrossRef]
- Rossi, A.R.; Breccia, M.; Abruzzese, E.; Castagnetti, F.; Luciano, L.; Gozzini, A.; Annunziata, M.; Martino, B.; Stagno, F.; Cavazzini, F.; et al. Outcome of 82 chronic myeloid leukemia patients treated with nilotinib or dasatinib after failure of two prior tyrosine kinase inhibitors. Haematologica 2013, 98, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Alikian, M.; Gale, R.P.; Apperley, J.F.; Foroni, L.; Alikian, M. Molecular techniques for the personalised management of patients with chronic myeloid leukaemia. Biomol. Detect. Quantif. 2017, 11, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.; Lange, T.; Cross, M.; Wildenberger, K.; Niederwieser, D.; Franke, G.N. Optimized digital droplet PCR for BCR-AB. J. Mol. Diagn. 2019, 21, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Franke, G.N.; Maier, J.; Wildenberger, K.; Cross, M.; Giles, F.J.; Müller, M.C.; Hochhaus, A.; Niederwieser, D.; Lange, T. Comparison of real-time quantitative PCR and digital droplet PCR for BCR-ABL1 monitoring in patients with chronic myeloid leukemia. J. Mol. Diagn. 2020, 22, 81–89. [Google Scholar] [CrossRef]
- Chung, H.J.; Hur, M.; Yoon, S.; Hwang, K.; Lim, H.S.; Kim, H.; Moon, H.W.; Yun, Y.M. Performance evaluation of the QXdX BCR-ABL %Is droplet digital PCR assay. Ann. Lab. Med. 2020, 40, 72–75. [Google Scholar] [CrossRef]
- Wang, W.J.; Zheng, C.F.; Liu, Z.; Tan, Y.H.; Chen, X.H.; Zhao, B.L.; Li, G.X.; Xu, Z.F.; Ren, F.G.; Zhang, Y.F.; et al. Droplet digital PCR for BCR/ABL(P210) detection of chronic myeloid leukemia: A high sensitive method of the minimal residual disease and disease progression. Eur. J. Haematol. 2018, 101, 291–296. [Google Scholar] [CrossRef]
- Park, H.; Shin, D.-Y.; Kim, I.; Sohn, S.-K.; Koh, Y.; Lee, J.-H.; Lee, K.-H.; Kim, D.-Y.; Kim, H.-J.; Ahn, J.-S.; et al. Use of droplet digital polymerase chain reaction for detecting minimal residual disease: A prospective, multi-institutional study. Ann. Oncol. 2019, 33, 2273–2280. [Google Scholar]
- Mori, S.; Vagge, E.; Le Coutre, P.; Abruzzese, E.; Martino, B.; Pungolino, E.; Elena, C.; Pierri, I.; Assouline, S.; D’Emilio, A.; et al. Age and dPCR can predict relapse in CML patients who discontinued imatinib: The ISAV study. Am. J. Hematol. 2015, 90, 910–914. [Google Scholar] [CrossRef]
- Nicolini, F.E.; Dulucq, S.; Boureau, L.; Cony-Makhoul, P.; Charbonnier, A.; Escoffre-Barbe, M.; Rigal-Huguet, F.; Coiteux, V.; Varet, B.; Dubruille, V.; et al. Evaluation of residual disease and TKI duration are critical predictive factors for molecular recurrence after stopping imatinib first-line in chronic phase CML patients. Clin. Cancer Res. 2019, 25, 6606–6613. [Google Scholar] [CrossRef]
- Colafigli, G.; Scalzulli, E.; Porrazzo, M.; Diverio, D.; Loglisci, M.G.; Latagliata, R.; Guarini, A.; Foà, R.; Breccia, M. Digital droplet PCR at the time of TKI discontinuation in chronic-phase chronic myeloid leukemia patients is predictive of treatment-free remission outcome. Hematol. Oncol. 2019, 37, 652–654. [Google Scholar] [CrossRef]
- Minervini, C.F.; Cumbo, C.; Orsini, P.; Anelli, L.; Zagaria, A.; Impera, L.; Coccaro, N.; Brunetti, C.; Minervini, A.; Casieri, P.; et al. Mutational analysis in BCR-ABL1 positive leukemia by deep sequencing based on nanopore MinION technology. Exp. Mol. Pathol. 2017, 103, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Cumbo, C.; Minervini, C.F.; Orsini, P.; Anelli, L.; Zagaria, A.; Minervini, A.; Coccaro, N.; Impera, L.; Tota, G.; Parciante, E.; et al. Nanopore targeted sequencing for rapid gene mutations detection in acute myeloid leukemia. Genes 2019, 10, 1026. [Google Scholar] [CrossRef] [PubMed]
- Minervini, C.F.; Cumbo, C.; Orsini, P.; Brunetti, C.; Anelli, L.; Zagaria, A.; Minervini, A.; Casieri, P.; Coccaro, N.; Tota, G.; et al. TP53 gene mutation analysis in chronic lymphocytic leukemia by nanopore MinION sequencing. Diagn. Pathol. 2016, 11, 96. [Google Scholar] [CrossRef]
- Minervini, C.F.; Cumbo, C.; Orsini, P.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. Nanopore sequencing in blood diseases: A wide range of opportunities. Front. Genet. 2020, 11, 76. [Google Scholar] [CrossRef]
- Orsini, P.; Minervini, C.F.; Cumbo, C.; Anelli, L.; Zagaria, A.; Minervini, A.; Coccaro, N.; Tota, G.; Casieri, P.; Impera, L.; et al. Design and MinION testing of a nanopore targeted gene sequencing panel for chronic lymphocytic leukemia. Sci. Rep. 2018, 8, 11798. [Google Scholar] [CrossRef]
- Cumbo, C.; Impera, L.; Minervini, C.F.; Orsini, P.; Anelli, L.; Zagaria, A.; Coccaro, N.; Tota, G.; Minervini, A.; Casieri, P.; et al. Genomic BCR-ABL1 breakpoint characterization by a multi-strategy approach for “personalized monitoring” of residual disease in chronic myeloid leukemia patients. Oncotarget 2018, 9, 10978–10986. [Google Scholar] [CrossRef]
- Bassan, R.; Spinelli, O.; Oldani, E.; Intermesoli, T.; Tosi, M.; Peruta, B.; Rossi, G.; Borlenghi, E.; Pogliani, E.M.; Terruzzi, E.; et al. Improved risk classification for risk-specific therapy based on the molecular study of minimal residual disease (MRD) in adult acute lymphoblastic leukemia (ALL). Blood 2009, 113, 4153–4162. [Google Scholar] [CrossRef]
- Della Starza, I.; Nunes, V.; Cavalli, M.; De Novi, L.A.; Ilari, C.; Apicella, V.; Vitale, A.; Testi, A.M.; Del Giudice, I.; Chiaretti, S.; et al. Comparative analysis between RQ-PCR and digital-droplet-PCR of immunoglobulin/T-cell receptor gene rearrangements to monitor minimal residual disease in acute lymphoblastic leukaemia. Br. J. Haematol. 2016, 174, 541–549. [Google Scholar] [CrossRef]
- Della Starza, I.; De Novi, L.A.; Santoro, A.; Salemi, D.; Tam, W.; Cavalli, M.; Menale, L.; Soscia, R.; Apicella, V.; Ilari, C.; et al. Digital droplet PCR and next-generation sequencing refine minimal residual disease monitoring in acute lymphoblastic leukemia. Leuk. Lymphoma 2019, 60, 2838–2840. [Google Scholar] [CrossRef]
- Kotrova, M.; Muzikova, K.; Mejstrikova, E.; Novakova, M.; Bakardjieva-Mihaylova, V.; Fiser, K.; Stuchly, J.; Giraud, M.; Salson, M.; Pott, C.; et al. The predictive strength of next-generation sequencing MRD detection for relapse compared with current methods in childhood ALL. Blood 2015, 126, 1045–1047. [Google Scholar] [CrossRef]
- Wetzler, M.; Dodge, R.K.; Mrózek, K.; Carroll, A.J.; Tantravahi, R.; Block, A.M.W.; Pettenati, M.J.; Le Beau, M.M.; Frankel, S.R.; Stewart, C.C.; et al. Prospective karyotype analysis in adult acute lymphoblastic leukemia: The cancer and leukemia group B experience. Blood 1999, 93, 3983–3993. [Google Scholar]
- Faderl, S.; Jeha, S.; Kantarjian, H.M. The biology and therapy of adult acute lymphoblastic leukemia. Cancer 2003, 98, 1337–1354. [Google Scholar] [CrossRef]
- Burmeister, T.; Schwartz, S.; Bartram, C.R.; Gökbuget, N.; Hoelzer, D.; Thiel, E. Patients’ age and BCR-ABL frequency in adult B-precursor ALL: A retrospective analysis from the GMALL study group. Blood 2008, 112, 918–919. [Google Scholar] [CrossRef]
- Iacobucci, I.; Lonetti, A.; Venturi, C.; Ferrari, A.; Papayannidis, C.; Ottaviani, E.; Abbenante, M.C.; Paolini, S.; Bresciani, P.; Potenza, L.; et al. Use of a high sensitive nanofluidic array for the detection of rare copies of BCR-ABL1 transcript in patients with Philadelphia-positive acute lymphoblastic leukemia in complete response. Leuk. Res. 2014, 38, 581–585. [Google Scholar] [CrossRef]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Coccaro, N.; Anelli, L.; Zagaria, A.; Casieri, P.; Tota, G.; Orsini, P.; Impera, L.; Minervini, A.; Minervini, C.F.; Cumbo, C.; et al. Droplet digital PCR is a robust tool for monitoring minimal residual disease in adult philadelphia-positive acute lymphoblastic leukemia. J. Mol. Diagn. 2018, 20, 474–482. [Google Scholar] [CrossRef]
- Fielding, A.K. Treatment of philadelphia chromosome–positive acute lymphoblastic leukemia in adults: A broader range of options, improved outcomes, and more therapeutic dilemmas. Am. Soc. Clin. Oncol. Educ. B. 2015, e352–e359. [Google Scholar] [CrossRef]
- Foà, R.; Vitale, A.; Vignetti, M.; Meloni, G.; Guarini, A.; De Propris, M.S.; Elia, L.; Paoloni, F.; Fazi, P.; Cimino, G.; et al. Dasatinib as first-line treatment for adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 2011, 118, 6521–6528. [Google Scholar] [CrossRef]
- Ravandi, F.; Jorgensen, J.L.; Thomas, D.A.; O’Brien, S.; Garris, R.; Faderl, S.; Huang, X.; Wen, S.; Burger, J.A.; Ferrajoli, A.; et al. Detection of MRD may predict the outcome of patients with Philadelphia chromosome-positive ALL treated with tyrosine kinase inhibitors plus chemotherapy. Blood 2013, 122, 1214–1221. [Google Scholar] [CrossRef]
- Short, N.J.; Jabbour, E.; Sasaki, K.; Patel, K.; O’Brien, S.M.; Cortes, J.E.; Garris, R.; Issa, G.C.; Garcia-Manero, G.; Luthra, R.; et al. Impact of complete molecular response on survival in patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 2016, 128, 504–507. [Google Scholar] [CrossRef]
- Yoon, J.H.; Yhim, H.Y.; Kwak, J.Y.; Ahn, J.S.; Yang, D.H.; Lee, J.J.; Kim, S.J.; Kim, J.S.; Park, S.J.; Choi, C.W.; et al. Minimal residual disease-based effect and long-term outcome of first-line dasatinib combined with chemotherapy for adult Philadelphia chromosomepositive acute lymphoblastic leukemia. Ann. Oncol. 2016, 27, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Chiaretti, S.; Vitale, A.; Vignetti, M.; Piciocchi, A.; Fazi, P.; Elia, L.; Falini, B.; Ronco, F.; Ferrara, F.; De Fabritiis, P.; et al. A sequential approach with imatinib, chemotherapy and transplant for adult Ph+ acute lymphoblastic leukemia: Final results of the GIMEMA LAL 0904 study. Haematologica 2016, 101, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- George, D.; Czech, J.; John, B.; Yu, M.; Jennings, L.J. Detection and quantification of chimerism by droplet digital PCR. Chimerism 2013, 4, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Stahl, T.; Böhme, M.U.; Kröger, N.; Fehse, B. Digital PCR to assess hematopoietic chimerism after allogeneic stem cell transplantation. Exp. Hematol. 2015, 43, 462–468.e1. [Google Scholar] [CrossRef] [PubMed]
- Santurtún, A.; Riancho, J.A.; Arozamena, J.; López-Duarte, M.; Zarrabeitia, M.T. Indel analysis by droplet digital PCR: A sensitive method for DNA mixture detection and chimerism analysis. Int. J. Legal Med. 2017, 131, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Kliman, D.; Castellano-Gonzalez, G.; Withers, B.; Street, J.; Tegg, E.; Mirochnik, O.; Lai, J.; Clancy, L.; Gottlieb, D.; Blyth, E. Ultra-sensitive droplet digital PCR for the assessment of microchimerism in cellular therapies. Biol. Blood Marrow Transplant. 2018, 24, 1069–1078. [Google Scholar] [CrossRef]
- Mika, T.; Baraniskin, A.; Ladigan, S.; Wulf, G.; Dierks, S.; Haase, D.; Schork, K.; Turewicz, M.; Eisenacher, M.; Schmiegel, W.; et al. Digital droplet PCR-based chimerism analysis for monitoring of hematopoietic engraftment after allogeneic stem cell transplantation. Int. J. Lab. Hematol. 2019, 41, 615–621. [Google Scholar] [CrossRef]
- Waterhouse, M.; Pfeifer, D.; Duque-Afonso, J.; Follo, M.; Duyster, J.; Depner, M.; Bertz, H.; Finke, J. Droplet digital PCR for the simultaneous analysis of minimal residual disease and hematopoietic chimerism after allogeneic cell transplantation. Clin. Chem. Lab. Med. 2019, 57, 641–647. [Google Scholar] [CrossRef]
- Doescher, A.; Casper, J.; Kraemer, D.; Kapels, H.H.; Petershofen, E.K.; Müller, T.H. Platelet engraftment after allogenic stem cell transplantation is monitored by digital polymerase chain reaction without interference by platelet support. Exp. Hematol. 2018, 68, 21–29. [Google Scholar] [CrossRef]
- Research CCfibamt. Instructions for Post-Transplant Essential Data (Post-TED) Form (Revision 2), National Marrow Donor Program and the Medical College of Wisconsin. Available online: https://www.cibmtr.org/ (accessed on 15 March 2020).
- EBMT Home|EBMT. Available online: https://www.ebmt.org/ (accessed on 15 March 2020).
- Bill, M.; Grimm, J.; Jentzsch, M.; Kloss, L.; Goldmann, K.; Schulz, J.; Beinicke, S.; Häntschel, J.; Cross, M.; Vucinic, V.; et al. Digital droplet PCR-based absolute quantification of pre-transplant NPM1 mutation burden predicts relapse in acute myeloid leukemia patients. Ann. Hematol. 2018, 97, 1757–1765. [Google Scholar] [CrossRef]
- Welch, J.S.; Ley, T.J.; Link, D.C.; Miller, C.A.; Larson, D.E.; Koboldt, D.C.; Wartman, L.D.; Lamprecht, T.L.; Liu, F.; Xia, J.; et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012, 150, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, G.; Hoadley, K.; Triche, T.J.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [PubMed]
- Im, A.P.; Sehgal, A.R.; Carroll, M.P.; Smith, B.D.; Tefferi, A.; Johnson, D.E.; Boyiadzis, M. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: Associations with prognosis and potential treatment strategies. Leukemia 2014, 28, 1774–1783. [Google Scholar] [CrossRef] [PubMed]
- Krönke, J.; Bullinger, L.; Teleanu, V.; Tschürtz, F.; Gaidzik, V.I.; Kühn, M.W.M.; Rücker, F.G.; Holzmann, K.; Paschka, P.; Kapp-Schwörer, S.; et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood 2013, 122, 100–108. [Google Scholar] [CrossRef]
- Brambati, C.; Galbiati, S.; Xue, E.; Toffalori, C.; Crucitti, L.; Greco, R.; Sala, E.; Crippa, A.; Chiesa, L.; Soriani, N.; et al. Droplet digital polymerase chain reaction for DNMT3A and IDH1/2 mutations to improve early detection of acute myeloid leukemia relapse after allogeneic hematopoietic stem cell transplantation. Haematologica 2016, 101, e157–e161. [Google Scholar] [CrossRef]
- Jentzsch, M.; Bill, M.; Grimm, J.; Schulz, J.; Goldmann, K.; Beinicke, S.; Häntschel, J.; Pönisch, W.; Franke, G.N.; Vucinic, V.; et al. High BAALC copy numbers in peripheral blood prior to allogeneic transplantation predict early relapse in acute myeloid leukemia patients. Oncotarget 2017, 8, 87944–87954. [Google Scholar] [CrossRef]
- Jentzsch, M.; Bill, M.; Grimm, J.; Schulz, J.; Beinicke, S.; Häntschel, J.; Goldmann, K.; Pönisch, W.; Franke, G.-N.; Vucinic, V.; et al. Prognostic impact of blood MN1 copy numbers before allogeneic stem cell transplantation in patients with acute myeloid leukemia. HemaSphere 2019, 3, e167. [Google Scholar] [CrossRef]
- Weber, S.; Alpermann, T.; Dicker, F.; Jeromin, S.; Nadarajah, N.; Eder, C.; Fasan, A.; Kohlmann, A.; Meggendorfer, M.; Haferlach, C.; et al. BAALC expression: A suitable marker for prognostic risk stratification and detection of residual disease in cytogenetically normal acute myeloid leukemia. Blood Cancer J. 2014, 4, e173. [Google Scholar] [CrossRef]
- Najima, Y.; Ohashi, K.; Kawamura, M.; Onozuka, Y.; Yamaguchi, T.; Akiyama, H.; Sakamaki, H. Molecular monitoring of BAALC expression in patients with CD34-positive acute leukemia. Int. J. Hematol. 2010, 91, 636–645. [Google Scholar] [CrossRef]
- Schwind, S.; Marcucci, G.; Kohlschmidt, J.; Radmacher, M.D.; Mrózek, K.; Maharry, K.; Becker, H.; Metzeler, K.H.; Whitman, S.P.; Wu, Y.Z.; et al. Low expression of MN1 associates with better treatment response in older patients with de novo cytogenetically normal acute myeloid leukemia. Blood 2011, 118, 4188–4198. [Google Scholar] [CrossRef]
- Bienz, M.; Ludwig, M.; Mueller, B.U.; Leibundgut, E.O.; Ratschiller, D.; Solenthaler, M.; Fey, M.F.; Pabst, T. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin. Cancer Res. 2005, 11, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Langer, C.; Radmacher, M.D.; Ruppert, A.S.; Whitman, S.P.; Paschka, P.; Mrózek, K.; Baldus, C.D.; Vukosavljevic, T.; Liu, C.G.; Ross, M.E.; et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: A Cancer and Leukemia Group B (CALGB) study. Blood 2008, 111, 5371–5379. [Google Scholar] [PubMed]
- Metzeler, K.H.; Dufour, A.; Benthaus, T.; Hummel, M.; Sauerland, M.C.; Heinecke, A.; Berdel, W.E.; Büchner, T.; Wörmann, B.; Mansmann, U.; et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: A comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J. Clin. Oncol. 2009, 27, 5031–5038. [Google Scholar] [CrossRef]
- Schwind, S.; Marcucci, G.; Maharry, K.; Radmacher, M.D.; Mrózek, K.; Holland, K.B.; Margeson, D.; Becker, H.; Whitman, S.P.; Wu, Y.Z.; et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: A cancer and leukemia group B study. Blood 2010, 116, 5660–5669. [Google Scholar] [CrossRef]
- Weber, S.; Haferlach, T.; Alpermann, T.; Perglerová, K.; Schnittger, S.; Haferlach, C.; Kern, W. Feasibility of BAALC gene expression for detection of minimal residual disease and risk stratification in normal karyotype acute myeloid leukaemia. Br. J. Haematol. 2016, 175, 904–916. [Google Scholar] [CrossRef]
- Yoon, J.H.; Kim, H.J.; Shin, S.H.; Yahng, S.A.; Lee, S.E.; Cho, B.S.; Eom, K.S.; Kim, Y.J.; Lee, S.; Min, C.K.; et al. BAALC and WT1 expressions from diagnosis to hematopoietic stem cell transplantation: Consecutive monitoring in adult patients with core-binding-factor-positive AML. Eur. J. Haematol. 2013, 91, 112–121. [Google Scholar] [CrossRef]
- Heuser, M.; Beutel, G.; Krauter, J.; Döhner, K.; Von Neuhoff, N.; Schlegelberger, B.; Ganser, A. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood 2006, 108, 3898–3905. [Google Scholar] [CrossRef]
- Langer, C.; Marcucci, G.; Holland, K.B.; Radmacher, M.D.; Maharry, K.; Paschka, P.; Whitman, S.P.; Mrózek, K.; Baldus, C.D.; Vij, R.; et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: A cancer and leukemia group B study. J. Clin. Oncol. 2009, 27, 3198–3204. [Google Scholar] [CrossRef]
- Fehse, B.; Badbaran, A.; Berger, C.; Sonntag, T.; Riecken, K.; Geffken, M.; Kröger, N.; Ayuk, F.A. Digital PCR assays for precise quantification of CD19-CAR-T cells after treatment with axicabtagene ciloleucel. Mol. Ther. Methods Clin. Dev. 2020, 16, 172–178. [Google Scholar] [CrossRef]
- Jespersen, D.S.; Schönherz, A.A.; Due, H.; Bøgsted, M.; Sondergaard, T.E.; Dybkær, K. Expression of NOTCH3 exon 16 differentiates Diffuse Large B-cell Lymphoma into molecular subtypes and is associated with prognosis. Sci. Rep. 2019, 9, 335. [Google Scholar] [CrossRef]
- Beheshti, A.; Vanderburg, C.; McDonald, J.T.; Ramkumar, C.; Kadungure, T.; Zhang, H.; Gartenhaus, R.B.; Evens, A.M. A circulating microRNA signature predicts age-based development of lymphoma. PLoS ONE 2017, 12, e0170521. [Google Scholar] [CrossRef] [PubMed]
- Kotrova, M.; van der Velden, V.H.J.; van Dongen, J.J.M.; Formankova, R.; Sedlacek, P.; Brüggemann, M.; Zuna, J.; Stary, J.; Trka, J.; Fronkova, E. Next-generation sequencing indicates false-positive MRD results and better predicts prognosis after SCT in patients with childhood ALL. Bone Marrow Transpl. 2017, 52, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Postel, M.; Roosen, A.; Laurent-Puig, P.; Taly, V.; Wang-Renault, S.F. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: A cancer diagnostic perspective. Expert Rev. Mol. Diagn. 2018, 18, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Taly, V.; Pekin, D.; Benhaim, L.; Kotsopoulos, S.K.; Le Corre, D.; Li, X.; Atochin, I.; Link, D.R.; Griffiths, A.D.; Pallier, K.; et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 2013, 59, 1722–1731. [Google Scholar] [CrossRef]
- Zhong, Q.; Bhattacharya, S.; Kotsopoulos, S.; Olson, J.; Taly, V.; Griffiths, A.D.; Link, D.R.; Larson, J.W. Multiplex digital PCR: Breaking the one target per color barrier of quantitative PCR. Lab Chip 2011, 11, 2167–2174. [Google Scholar] [CrossRef]
- Whale, A.S.; Huggett, J.F.; Tzonev, S. Fundamentals of multiplexing with digital PCR. Biomol. Detect. Quantif. 2016, 10, 15–23. [Google Scholar] [CrossRef]
- Pécuchet, N.; Zonta, E.; Didelot, A.; Combe, P.; Thibault, C.; Gibault, L.; Lours, C.; Rozenholc, Y.; Taly, V.; Laurent-Puig, P.; et al. Base-position error rate analysis of next-generation sequencing applied to circulating tumor DNA in non-small cell lung cancer: A prospective study. PLoS Med. 2016, 13, e1002192. [Google Scholar] [CrossRef] [PubMed]
- Pietrasz, D.; Pécuchet, N.; Garlan, F.; Didelot, A.; Dubreuil, O.; Doat, S.; Imbert-Bismut, F.; Karoui, M.; Vaillant, J.C.; Taly, V.; et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin. Cancer Res. 2017, 23, 116–123. [Google Scholar] [CrossRef]
- Garlan, F.; Laurent-Puig, P.; Sefrioui, D.; Siauve, N.; Didelot, A.; Sarafan-Vasseur, N.; Michel, P.; Perkins, G.; Mulot, C.; Blons, H.; et al. Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL study). Clin. Cancer Res. 2017, 23, 5416–5425. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Khong, T.; Ramachandran, M.; Chow, A.; Klarica, D.; Mai, L.; Walsh, S.; Broemeling, D.; Marziali, A.; Wiggin, M.; et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia 2017, 31, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Coccaro, N.; Tota, G.; Zagaria, A.; Anelli, L.; Casieri, P.; Impera, L.; Minervini, A.; Minervini, C.F.; Orsini, P.; Cumbo, C.; et al. Monitoring minimal residual disease by ddPCR in acute lymphoblastic leukemia associated with the FGFR1 gene rearrangement. Int. J. Lab. Hematol. 2018, 40, e117–e120. [Google Scholar] [CrossRef]
- Coccaro, N.; Tota, G.; Anelli, L.; Zagaria, A.; Casieri, P.; Cellamare, A.; Minervini, C.F.; Minervini, A.; Cumbo, C.; Impera, L.; et al. MYEOV gene overexpression in primary plasma cell leukemia with t(11;14)(q13;q32). Oncol. Lett. 2016, 12, 1460–1464. [Google Scholar] [CrossRef]
- Zagaria, A.; Anelli, L.; Coccaro, N.; Tota, G.; Casieri, P.; Cellamare, A.; Impera, L.; Brunetti, C.; Minervini, A.; Minervini, C.F.; et al. BCR–ABL1 e6a2 transcript in chronic myeloid leukemia: Biological features and molecular monitoring by droplet digital PCR. Virchows Arch. 2015, 467, 357–363. [Google Scholar] [CrossRef]
- Coccaro, N.; Brunetti, C.; Tota, G.; Pierri, C.L.; Anelli, L.; Zagaria, A.; Casieri, P.; Impera, L.; Minervini, C.F.; Minervini, A.; et al. A novel t(3;9)(q21.2; p24.3) associated with SMARCA2 and ZNF148 genes rearrangement in myelodysplastic syndrome. Leuk. Lymphoma 2018, 59, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Coccaro, N.; Zagaria, A.; Orsini, P.; Anelli, L.; Tota, G.; Casieri, P.; Impera, L.; Minervini, A.; Minervini, C.F.; Cumbo, C.; et al. RARA and RARG gene downregulation associated with EZH2 mutation in acute promyelocytic-like morphology leukemia. Hum. Pathol. 2018, 80, 82–86. [Google Scholar] [CrossRef]
- Handschuh, L.; Wojciechowski, P.; Kazmierczak, M.; Marcinkowska-Swojak, M.; Luczak, M.; Lewandowski, K.; Komarnicki, M.; Blazewicz, J.; Figlerowicz, M.; Kozlowski, P. NPM1 alternative transcripts are upregulated in acute myeloid and lymphoblastic leukemia and their expression level affects patient outcome. J. Transl. Med. 2018, 16, 232. [Google Scholar] [CrossRef]
- Storlazzi, C.T.; Albano, F.; Lo Cunsolo, C.; Doglioni, C.; Guastadisegni, M.C.; Impera, L.; Lonoce, A.; Funes, S.; Macrí, E.; Iuzzolino, P.; et al. Upregulation of the SOX5 by promoter swapping with the P2RY8 gene in primary splenic follicular lymphoma. Leukemia 2007, 21, 2221–2225. [Google Scholar] [CrossRef][Green Version]
- Kjaer, L.; Skov, V.; Andersen, M.T.; Aggerholm, A.; Clair, P.; Gniot, M.; Soeby, K.; Udby, L.; Dorff, M.H.; Hasselbalch, H.; et al. Variant-specific discrepancy when quantitating BCR-ABL1 e13a2 and e14a2 transcripts using the Europe Against Cancer qPCR assay. Eur. J. Haematol. 2019, 103, 26–34. [Google Scholar] [CrossRef]
- Bernardi, S.; Bonifacio, M.; Iurlo, A.; Zanaglio, C.; Tiribelli, M.; Binotto, G.; Abruzzese, E.; Russo, D. “Variant-specific discrepancy when quantitating BCR-ABL1 e13a2 and e14a2 transcripts using the Europe Against Cancer qPCR assay.” Is dPCR the key? Eur. J. Haematol. 2019, 103, 272–273. [Google Scholar] [CrossRef] [PubMed]
- Lund, H.L.; Hughesman, C.B.; McNeil, K.; Clemens, S.; Hocken, K.; Pettersson, R.; Karsan, A.; Foster, L.J.; Haynes, C. Initial diagnosis of chronic myelogenous leukemia based on quantification of M-BCR status using droplet digital PCR. Anal. Bioanal. Chem. 2016, 408, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Orsini, P.; Impera, L.; Parciante, E.; Cumbo, C.; Minervini, C.F.; Minervini, A.; Zagaria, A.; Anelli, L.; Coccaro, N.; Casieri, P.; et al. Droplet digital PCR for the quantification of Alu methylation status in hematological malignancies. Diagn. Pathol. 2018, 13, 98. [Google Scholar] [CrossRef] [PubMed]
- Findlay, S.D.; Vincent, K.M.; Berman, J.R.; Postovit, L.M. A digital pcr-based method for efficient and highly specific screening of genome edited cells. PLoS ONE 2016, 11, e0153901. [Google Scholar] [CrossRef] [PubMed]
- Decraene, C.; Silveira, A.B.; Bidard, F.C.; Vallée, A.; Michel, M.; Melaabi, S.; Vincent-Salomon, A.; Saliou, A.; Houy, A.; Milder, M.; et al. Multiple hotspot mutations scanning by single droplet digital PCR. Clin. Chem. 2018, 64, 317–328. [Google Scholar] [CrossRef]
- Decraene, C.; Bortolini Silveira, A.; Michel, M.; Bidard, F.C.; Pierga, J.Y.; Stern, M.H.; Proudhon, C. Single droplet digital polymerase chain reaction for comprehensive and simultaneous detection of mutations in hotspot regions. J. Vis. Exp. 2018, 139, e58051. [Google Scholar] [CrossRef]
- Mock, U.; MacHowicz, R.; Hauber, I.; Horn, S.; Abramowski, P.; Berdien, B.; Hauber, J.; Fehse, B. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 2015, 43, 5560–5571. [Google Scholar] [CrossRef]
- Berman, J.R.; Cooper, S.; Zhang, B.; Karlin-Neumann, G.; Litterst, C.; Jouvenot, Y.; Hefner, E.; Miyaoka, Y.; Conklin, B.R. Ultra-sensitive quantification of genome editing events using Droplet-Digital PCR. Bio-Rad Bullettin 2015, 6712, 1–6. [Google Scholar]
- Alcaide, M.; Yu, S.; Bushell, K.; Fornika, D.; Nielsen, J.S.; Nelson, B.H.; Mann, K.K.; Assouline, S.; Johnson, N.A.; Morin, R.D. Multiplex droplet digital PCR quantification of recurrent somatic mutations in diffuse large b-cell and follicular lymphoma. Clin. Chem. 2016, 62, 1238–1247. [Google Scholar] [CrossRef]
- De Smith, A.J.; Walsh, K.M.; Hansen, H.M.; Endicott, A.A.; Wiencke, J.K.; Metayer, C.; Wiemels, J.L. Somatic mutation allelic ratio test using ddPCR (SMART-ddPCR): An accurate method for assessment of preferential allelic imbalance in tumor DNA. PLoS ONE 2015, 10, e0143343. [Google Scholar] [CrossRef]
- Lee, S.; Kim, D.W.; Cho, B.S.; Yoon, J.H.; Shin, S.H.; Yahng, S.A.; Lee, S.E.; Eom, K.S.; Kim, Y.J.; Chung, N.G.; et al. Impact of minimal residual disease kinetics during imatinib-based treatment on transplantation outcome in Philadelphia chromosome-positive acute lymphoblastic leukemia. Leukemia 2012, 26, 2367–2374. [Google Scholar] [CrossRef]
- Kim, D.Y.; Joo, Y.D.; Lim, S.N.; Kim, S.D.; Lee, J.H.; Lee, J.H.; Kim, D.H.; Kim, K.; Jung, C.W.; Kim, I.; et al. Nilotinib combined with multiagent chemotherapy for newly diagnosed Philadelphia-positive acute lymphoblastic leukemia. Blood 2015, 126, 746–756. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coccaro, N.; Tota, G.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. Digital PCR: A Reliable Tool for Analyzing and Monitoring Hematologic Malignancies. Int. J. Mol. Sci. 2020, 21, 3141. https://doi.org/10.3390/ijms21093141
Coccaro N, Tota G, Anelli L, Zagaria A, Specchia G, Albano F. Digital PCR: A Reliable Tool for Analyzing and Monitoring Hematologic Malignancies. International Journal of Molecular Sciences. 2020; 21(9):3141. https://doi.org/10.3390/ijms21093141
Chicago/Turabian StyleCoccaro, Nicoletta, Giuseppina Tota, Luisa Anelli, Antonella Zagaria, Giorgina Specchia, and Francesco Albano. 2020. "Digital PCR: A Reliable Tool for Analyzing and Monitoring Hematologic Malignancies" International Journal of Molecular Sciences 21, no. 9: 3141. https://doi.org/10.3390/ijms21093141
APA StyleCoccaro, N., Tota, G., Anelli, L., Zagaria, A., Specchia, G., & Albano, F. (2020). Digital PCR: A Reliable Tool for Analyzing and Monitoring Hematologic Malignancies. International Journal of Molecular Sciences, 21(9), 3141. https://doi.org/10.3390/ijms21093141







