Tubulin Folding Cofactor TBCB is a Target of the Salmonella Effector Protein SseK1
Abstract
:1. Introduction
2. Results
2.1. Salmonella SseK1 Interacts with Human TBCB in the Yeast Two-Hybrid System
2.2. SseK1 Copurifies and Coimmunoprecipitates with TBCB
2.3. SseK1 Glycosylates TBCB in Vitro
2.4. SseK1 Glycosylates TBCB In Vivo
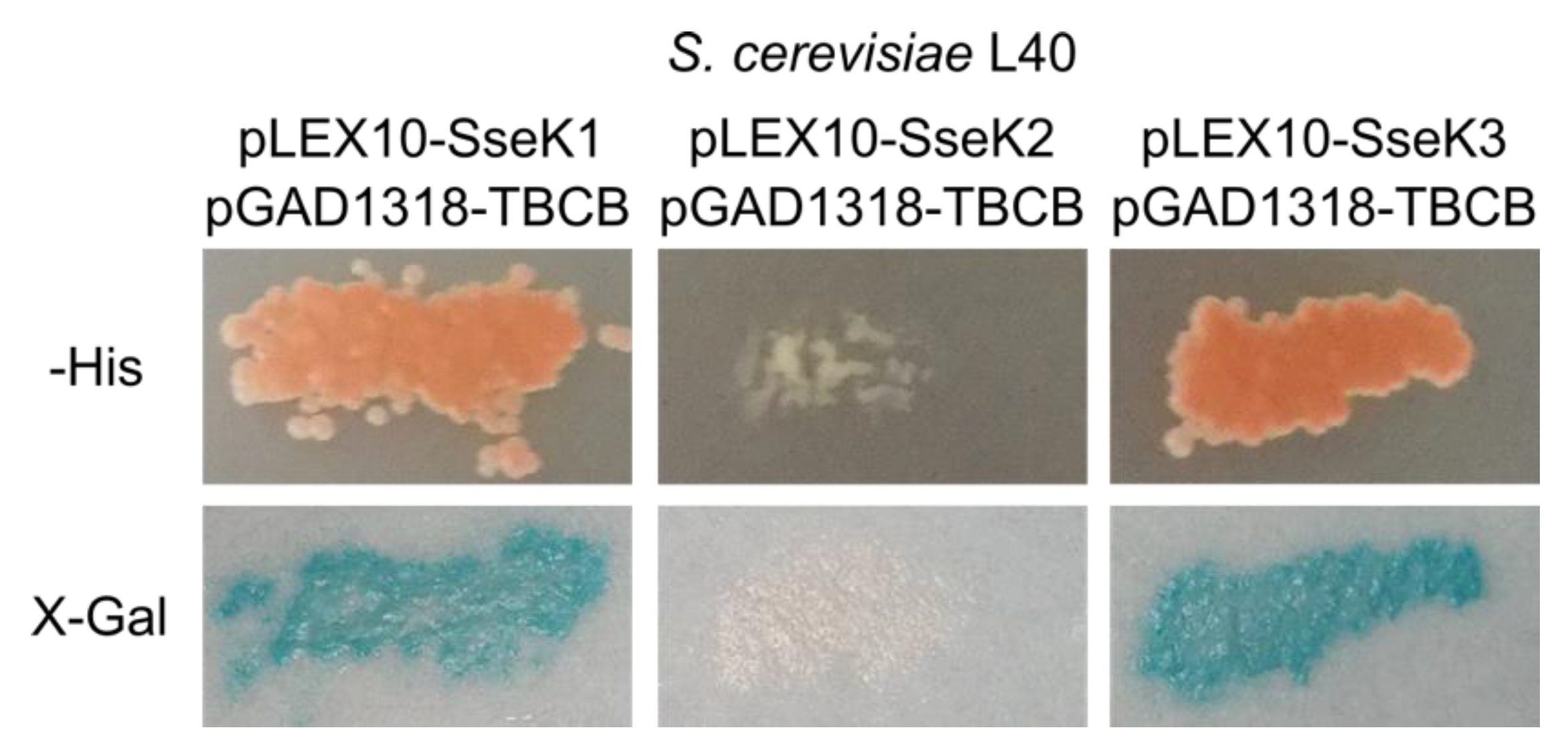
2.5. SseK3 Interacts with TBCB
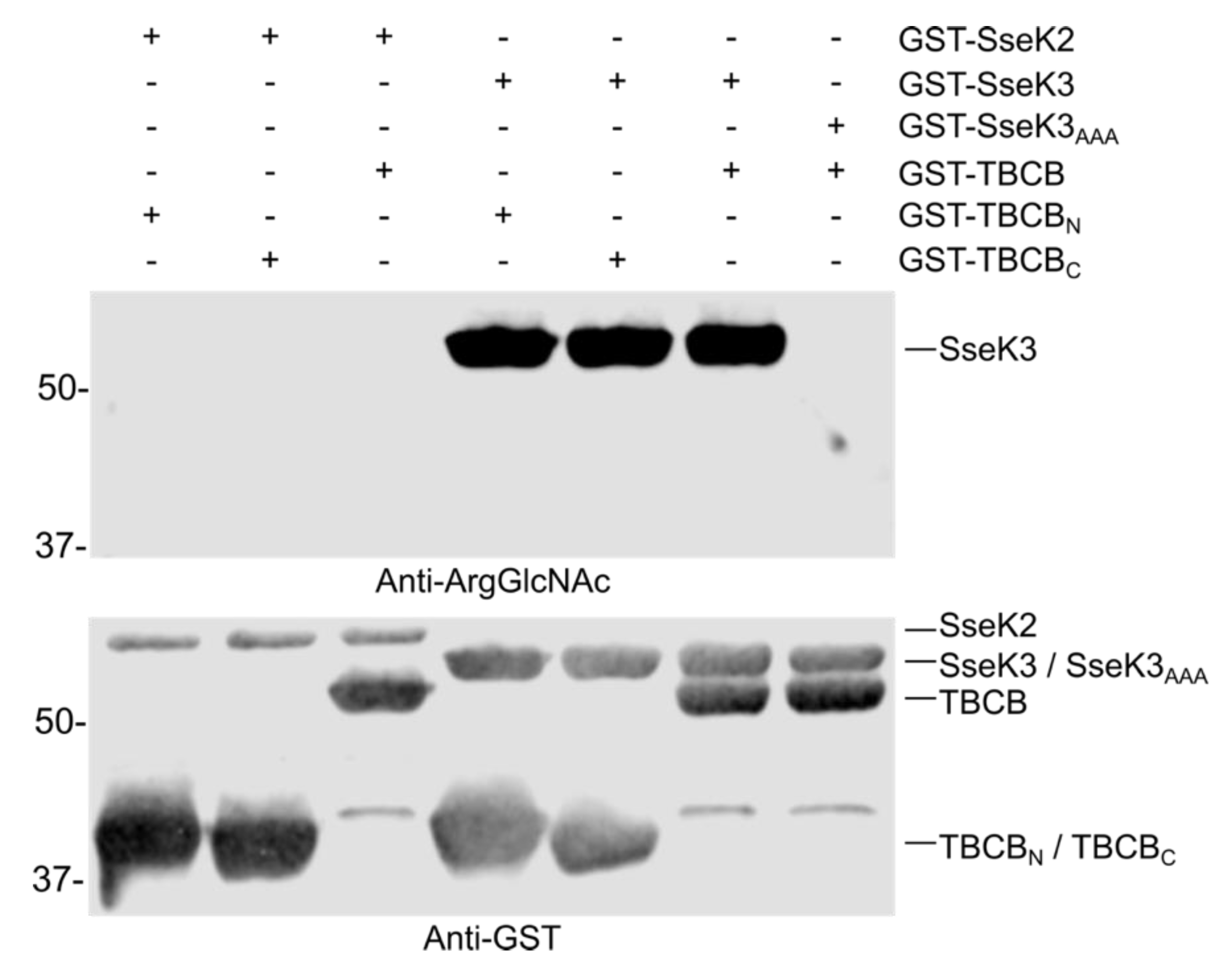
2.6. TBCB is not Glycosylated by SseK2 or SseK3
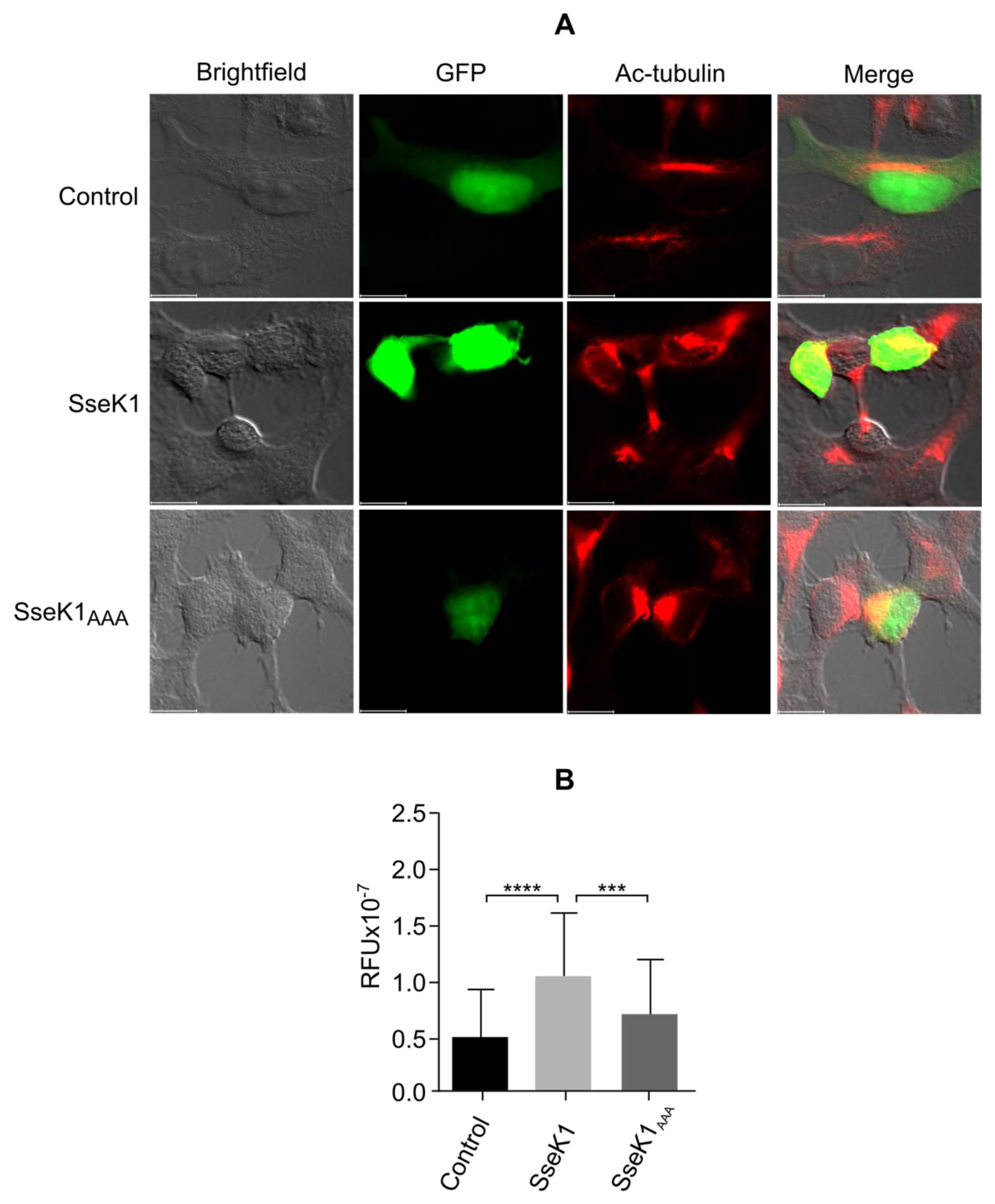
2.7. SseK1 Promotes Microtubule Stability
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Yeast Strains, and Plasmids
4.2. DNA Amplification With the Polymerase Chain Reaction and Sequencing
4.3. Bacterial Culture
4.4. Yeast Two-Hybrid Methods
4.5. Cell Culture, Lysis, and Transfection
4.6. GST Fusion Proteins, Electrophoresis, and Immunoblot
4.7. Coimmunoprecipitation Experiments
4.8. In Vitro and in Vivo Glycosylation Assays
4.9. Mutagenesis
4.10. Immunofluorescence Microscopy
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DMEM | Dulbecco’s modified Eagle’s medium |
| FADD | FAS-associated death domain protein |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| GshB | glutathione synthetase |
| GST | glutathione S-transferase |
| LB | lysogeny broth |
| NF-κB | nuclear factor kappa B |
| PMSF | phenylmethylsulfonyl fluoride |
| SPI | Salmonella pathogenicity island |
| SCV | Salmonella containing vacuole |
| T3SS | type III secretion system |
| TBCB | tubulin-folding cofactor B |
| TRADD | TNFR1-associated death domain protein |
References
- Condry, D.L.J.; Nilles, M.L. Introduction to Type III Secretion Systems. Methods Mol. Biol. (Clifton, N.J.) 2017, 1531, 1–10. [Google Scholar]
- Lara-Tejero, M.; Galán, J.E. The Injectisome, a Complex Nanomachine for Protein Injection into Mammalian Cells. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Hume, P.J.; Singh, V.; Davidson, A.C.; Koronakis, V. Swiss Army Pathogen: The Salmonella Entry Toolkit. Front. Cell. Infect. Microbiol. 2017, 7, 348. [Google Scholar] [CrossRef]
- Jennings, E.; Thurston, T.L.M.; Holden, D.W. Salmonella SPI-2 Type III Secretion System Effectors: Molecular Mechanisms and Physiological Consequences. Cell Host Microbe 2017, 22, 217–231. [Google Scholar] [CrossRef]
- Kujat Choy, S.L.; Boyle, E.C.; Gal-Mor, O.; Goode, D.L.; Valdez, Y.; Vallance, B.A.; Finlay, B.B. SseK1 and SseK2 are novel translocated proteins of Salmonella enterica serovar Typhimurium. Infect. Immun. 2004, 72, 5115–5125. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.F.; Coombes, B.K.; Bishop, J.L.; Wickham, M.E.; Lowden, M.J.; Gal-Mor, O.; Goode, D.L.; Boyle, E.C.; Sanderson, K.L.; Finlay, B.B.; et al. Salmonella Phage ST64B Encodes a Member of the SseK/NleB Effector Family. PLoS ONE 2011, 6, e17824. [Google Scholar] [CrossRef] [Green Version]
- Araujo-Garrido, J.L.; Bernal-Bayard, J.; Ramos-Morales, F. Type III Secretion Effectors with Arginine N-Glycosyltransferase Activity. Microorganisms 2020, 8, 357. [Google Scholar] [CrossRef]
- Li, S.; Zhang, L.; Yao, Q.; Li, L.; Dong, N.; Rong, J.; Gao, W.; Ding, X.; Sun, L.; Chen, X.; et al. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature 2013, 501, 242–246. [Google Scholar] [CrossRef]
- Pearson, J.S.; Giogha, C.; Ong, S.Y.; Kennedy, C.L.; Kelly, M.; Robinson, K.S.; Lung, T.W.F.; Mansell, A.; Riedmaier, P.; Oates, C.V.L.; et al. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature 2013, 501, 247–251. [Google Scholar] [CrossRef]
- Günster, R.A.; Matthews, S.A.; Holden, D.W.; Thurston, T.L.M. SseK1 and SseK3 T3SS effectors inhibit NF-κB signalling and necroptotic cell death in Salmonella-infected macrophages. Infect. Immun. 2017, 85, e00010–e00017. [Google Scholar] [PubMed] [Green Version]
- El Qaidi, S.; Chen, K.; Halim, A.; Siukstaite, L.; Rueter, C.; Hurtado-Guerrero, R.; Clausen, H.; Hardwidge, P.R. NleB/SseK effectors from Citrobacter rodentium, Escherichia coli, and Salmonella enterica display distinct differences in host substrate specificity. J. Biol. Chem. 2017, 292, 11423–11430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Soderholm, A.; Lung, T.W.F.; Giogha, C.; Hill, M.M.; Brown, N.F.; Hartland, E.; Teasdale, R.D. SseK3 Is a Salmonella Effector That Binds TRIM32 and Modulates the Host’s NF-κB Signalling Activity. PLoS ONE 2015, 10, e0138529. [Google Scholar] [CrossRef] [Green Version]
- Newson, J.P.; Scott, N.E.; Yeuk Wah Chung, I.; Wong Fok Lung, T.; Giogha, C.; Gan, J.; Wang, N.; Strugnell, R.; Brown, N.F.; Cygler, M.; et al. Salmonella effectors SseK1 and SseK3 target death domain proteins in the TNF and TRAIL signaling pathways. Mol. Cell. Proteom. 2019, 18, 1138–1156. [Google Scholar] [CrossRef]
- Baisón-Olmo, F.; Galindo-Moreno, M.; Ramos-Morales, F. Host cell type-dependent translocation and PhoP-mediated positive regulation of the effector SseK1 of Salmonella enterica. Front. Microbiol. 2015, 6, 396. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Li, S.; Li, X.; Shao, F.; Liu, L.; Hu, H.-G. Synthesis of and specific antibody generation for glycopeptides with arginine N-GlcNAcylation. Angew. Chem. Int. Ed. Engl. 2014, 53, 14517–14521. [Google Scholar] [CrossRef] [PubMed]
- Carranza, G.; Castaño, R.; Fanarraga, M.L.; Villegas, J.C.; Gonçalves, J.; Soares, H.; Avila, J.; Marenchino, M.; Campos-Olivas, R.; Montoya, G.; et al. Autoinhibition of TBCB regulates EB1-mediated microtubule dynamics. Cell. Mol. Life Sci. 2013, 70, 357–371. [Google Scholar] [CrossRef]
- Kortazar, D.; Fanarraga, M.L.; Carranza, G.; Bellido, J.; Villegas, J.C.; Avila, J.; Zabala, J.C. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp. Cell Res. 2007, 313, 425–436. [Google Scholar] [CrossRef]
- Schulze, E.; Asai, D.J.; Bulinski, J.C.; Kirschner, M. Posttranslational modification and microtubule stability. J. Cell Biol. 1987, 105, 2167–2177. [Google Scholar] [CrossRef] [Green Version]
- Webster, D.R.; Borisy, G.G. Microtubules are acetylated in domains that turn over slowly. J. Cell Sci. 1989, 92 (Pt 1), 57–65. [Google Scholar]
- Tan, H.; Liao, H.; Zhao, L.; Lu, Y.; Jiang, S.; Tao, D.; Liu, Y.; Ma, Y. HILI destabilizes microtubules by suppressing phosphorylation and Gigaxonin-mediated degradation of TBCB. Sci. Rep. 2017, 7, 46376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.S.; Hartland, E.L. A surprising sweetener from enteropathogenic Escherichia coli. Gut Microbes 2014, 5, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Scott, N.E.; Giogha, C.; Pollock, G.L.; Kennedy, C.L.; Webb, A.I.; Williamson, N.A.; Pearson, J.S.; Hartland, E.L. The bacterial arginine glycosyltransferase effector NleB preferentially modifies Fas-associated death domain protein (FADD). J. Biol. Chem. 2017, 292, 17337–17350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Wang, X.; Pham, T.H.; Feuerbacher, L.A.; Lubos, M.-L.; Huang, M.; Olsen, R.; Mushegian, A.; Slawson, C.; Hardwidge, P.R. NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-κB activation. Cell Host Microbe 2013, 13, 87–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Pham, T.H.; Feuerbacher, L.A.; Chen, K.; Hays, M.P.; Singh, G.; Rueter, C.; Hurtado-Guerrero, R.; Hardwidge, P.R. Citrobacter rodentium NleB Protein Inhibits Tumor Necrosis Factor (TNF) Receptor-associated Factor 3 (TRAF3) Ubiquitination to Reduce Host Type I Interferon Production. J. Biol. Chem. 2016, 291, 18232–18238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Liu, X.; Zha, H.; Fan, S.; Zhang, D.; Li, S.; Xiao, W. A pathogen-derived effector modulates host glucose metabolism by arginine GlcNAcylation of HIF-1α protein. PLoS Pathog. 2018, 14, e1007259. [Google Scholar] [CrossRef] [Green Version]
- Hirota, K. Basic biology of hypoxic responses mediated by the transcription factor HIFs and its implication for medicine. Biomedicines 2020, 8, 32. [Google Scholar] [CrossRef] [Green Version]
- Blasche, S.; Arens, S.; Ceol, A.; Siszler, G.; Schmid, M.A.; Häuser, R.; Schwarz, F.; Wuchty, S.; Aloy, P.; Uetz, P.; et al. The EHEC-host interactome reveals novel targets for the translocated intimin receptor. Sci. Rep. 2014, 4, 7531. [Google Scholar] [CrossRef]
- Law, R.J.; Law, H.T.; Scurll, J.M.; Scholz, R.; Santos, A.S.; Shames, S.R.; Deng, W.; Croxen, M.A.; Li, Y.; de Hoog, C.L.; et al. Quantitative Mass Spectrometry Identifies Novel Host Binding Partners for Pathogenic Escherichia coli Type III Secretion System Effectors. J. Proteome Res. 2016, 15, 1613–1622. [Google Scholar] [CrossRef]
- El Qaidi, S.; Scott, N.E.; Hays, M.P.; Geisbrecht, B.V.; Watkins, S.; Hardwidge, P.R. An intra-bacterial activity for a T3SS effector. Sci. Rep. 2020, 10, 1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Fanarraga, M.; Avila, J.; Guasch, A.; Coll, M.; Zabala, J.C. Review: Postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J. Struct. Biol. 2001, 135, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Lundin, V.F.; Leroux, M.R.; Stirling, P.C. Quality control of cytoskeletal proteins and human disease. Trends Biochem. Sci. 2010, 35, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Fontalba, A.; Paciucci, R.; Avila, J.; Zabala, J.C. Incorporation of tubulin subunits into dimers requires GTP hydrolysis. J. Cell Sci. 1993, 106, 627–632. [Google Scholar]
- Tian, G.; Huang, Y.; Rommelaere, H.; Vandekerckhove, J.; Ampe, C.; Cowan, N.J. Pathway leading to correctly folded β-tubulin. Cell 1996, 86, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Fanarraga, M.L.; Villegas, J.C.; Carranza, G.; Castaño, R.; Zabala, J.C. Tubulin cofactor B regulates microtubule densities during microglia transition to the reactive states. Exp. Cell Res. 2009, 315, 535–541. [Google Scholar] [CrossRef]
- Wang, W.; Ding, J.; Allen, E.; Zhu, P.; Zhang, L.; Vogel, H.; Yang, Y. Gigaxonin interacts with tubulin folding cofactor B and controls its degradation through the ubiquitin-proteasome pathway. Curr. Biol. 2005, 15, 2050–2055. [Google Scholar] [CrossRef] [Green Version]
- Allen, E.; Ding, J.; Wang, W.; Pramanik, S.; Chou, J.; Yau, V.; Yang, Y. Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature 2005, 438, 224–228. [Google Scholar] [CrossRef]
- Ding, J.; Liu, J.J.; Kowal, A.S.; Nardine, T.; Bhattacharya, P.; Lee, A.; Yang, Y. Microtubule-associated protein 1B: A neuronal binding partner for gigaxonin. J. Cell Biol. 2002, 158, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.D.; Lo, S.C.; Sun, Z.; Habib, G.M.; Lieberman, M.W.; Hannink, M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 2005, 280, 30091–30099. [Google Scholar] [CrossRef] [Green Version]
- Vadlamudi, R.K.; Barnes, C.J.; Rayala, S.; Li, F.; Balasenthil, S.; Marcus, S.; Goodson, H.V.; Sahin, A.A.; Kumar, R. p21-Activated Kinase 1 Regulates Microtubule Dynamics by Phosphorylating Tubulin Cofactor B. Mol. Cell. Biol. 2005, 25, 3726–3736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serna, M.; Carranza, G.; Martín-Benito, J.; Janowski, R.; Canals, A.; Coll, M.; Zabala, J.C.; Valpuesta, J.M. The structure of the complex between α-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism. J. Cell Sci. 2015, 128, 1824–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadadhar, S.; Bodakuntla, S.; Natarajan, K.; Janke, C. The tubulin code at a glance. J. Cell Sci. 2017, 130, 1347–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza Santos, M.; Orth, K. Subversion of the cytoskeleton by intracellular bacteria: Lessons from Listeria, Salmonella and Vibrio. Cell. Microbiol. 2015, 17, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Van Nhieu, G.T.; Romero, S. Common Themes in Cytoskeletal Remodeling by Intracellular Bacterial Effectors. Handb. Exp. Pharmacol. 2017, 235, 207–235. [Google Scholar]
- Ohlson, M.B.; Huang, Z.; Alto, N.M.; Blanc, M.; Dixon, J.E.; Chai, J.; Miller, S.I. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe 2008, 4, 434–446. [Google Scholar] [CrossRef] [Green Version]
- Raines, S.A.; Hodgkinson, M.R.; Dowle, A.A.; Pryor, P.R. The Salmonella effector SseJ disrupts microtubule dynamics when ectopically expressed in normal rat kidney cells. PLoS ONE 2017, 12, e0172588. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Cook, T.A.; Alberts, A.S.; Gundersen, G.G. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 2001, 3, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Schmieger, H. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 1972, 119, 75–88. [Google Scholar] [CrossRef]
- Maloy, S.R. Experimental Techniques in Bacterial Genetics; Jones and Bartlett: Burlington, MA, USA, 1990; ISBN 0867201185. [Google Scholar]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Boyer, H.W.; Roulland-Dussoix, D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 1969, 41, 459–472. [Google Scholar] [CrossRef]
- Vojtek, A.B.; Hollenberg, S.M.; Cooper, J.A. Mammalian Ras interacts directly with the serine/threonine kinase raf. Cell 1993, 74, 205–214. [Google Scholar] [CrossRef]
- Van Aelst, L. Two-hybrid analysis of Ras-Raf interactions. Methods Mol. Biol. 1998, 84, 201–222. [Google Scholar] [PubMed]
- Selig, L.; Benichou, S.; Rogel, M.E.; Wu, L.I.; Vodicka, M.A.; Sire, J.; Benarous, R.; Emerman, M. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 1997, 71, 4842–4846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman, F.; Fink, G.R.; Hicks, J.B. Laboratory Course Manual for Methods in Yeast Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1987; ISBN 0879691972. [Google Scholar]
- Fuxman Bass, J.I.; Reece-Hoyes, J.S.; Walhout, A.J.M. Colony Lift Colorimetric Assay for β-Galactosidase Activity. Cold Spring Harb. Protoc. 2016. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
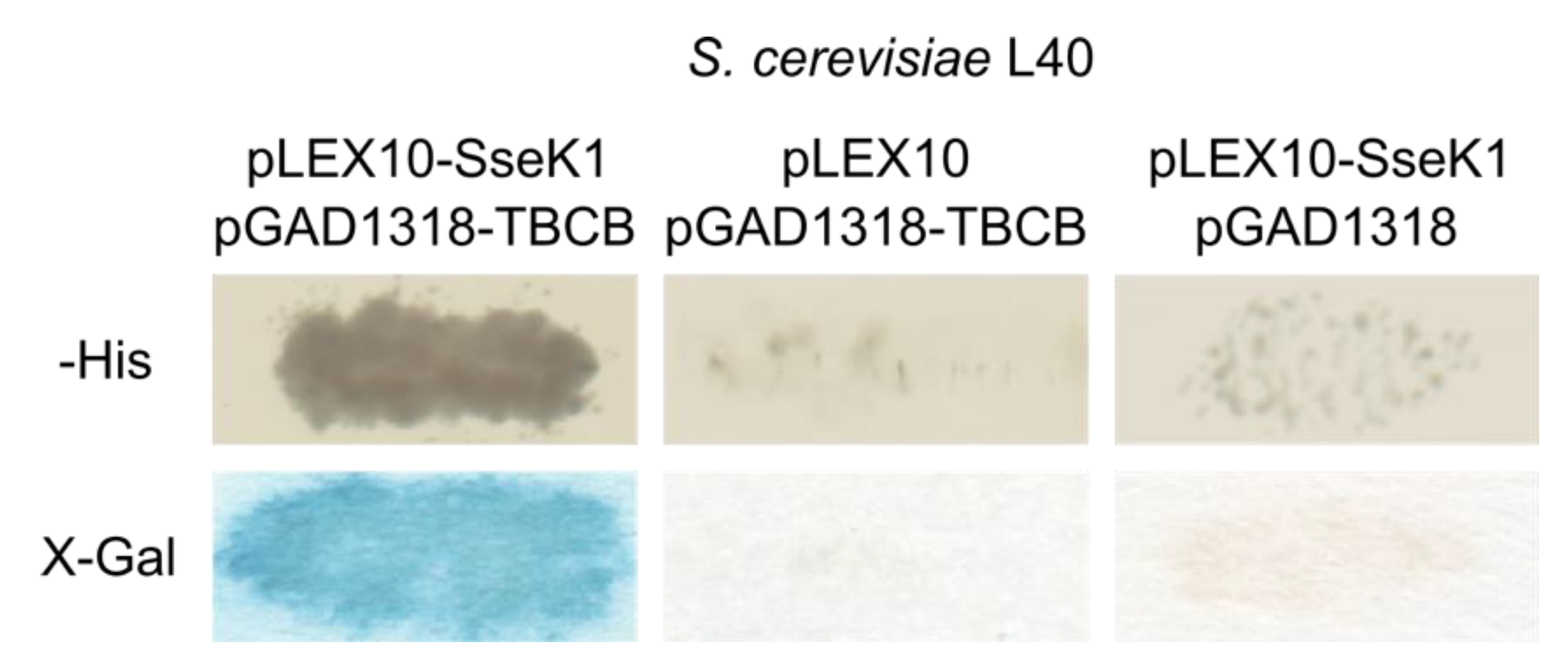
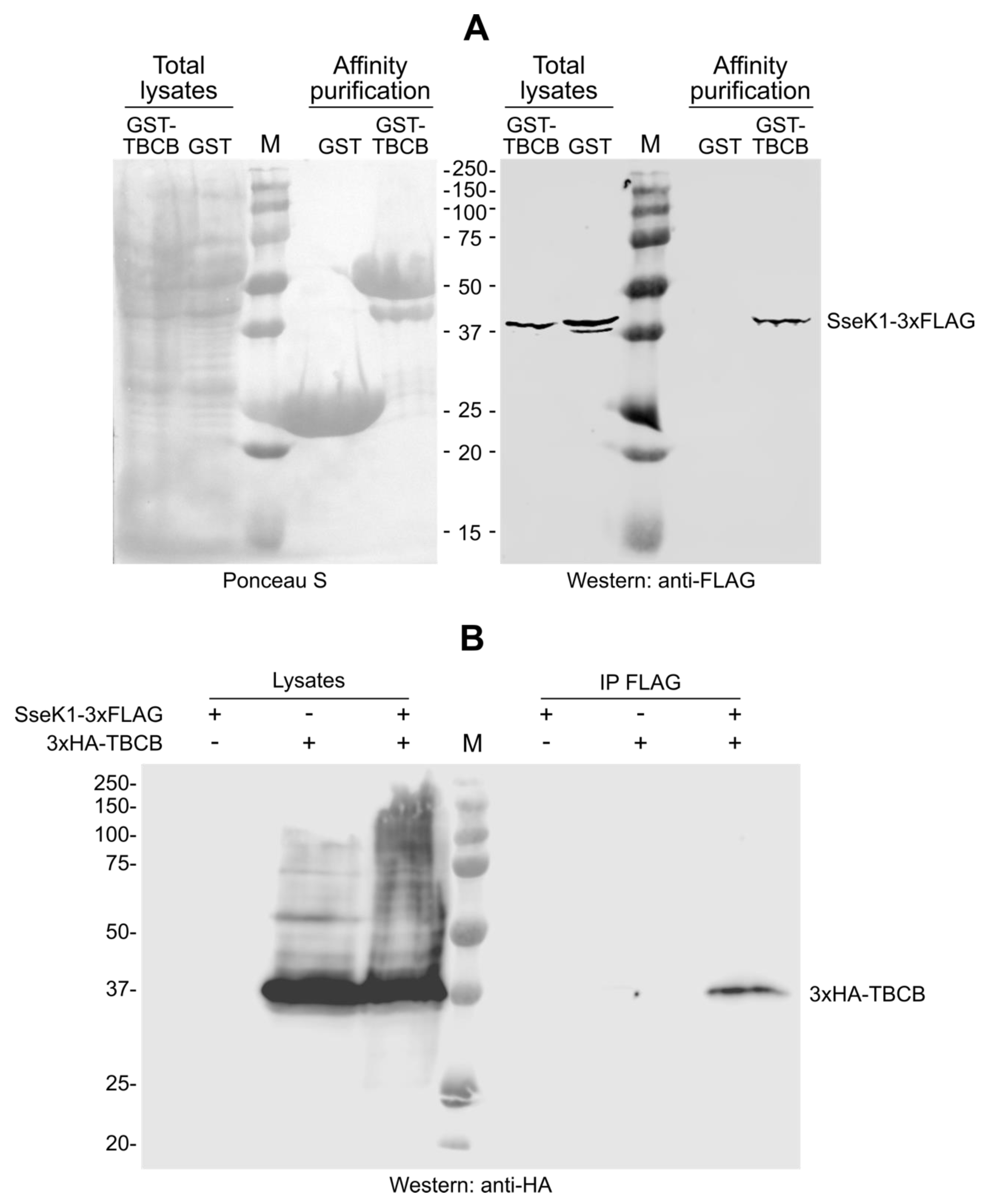
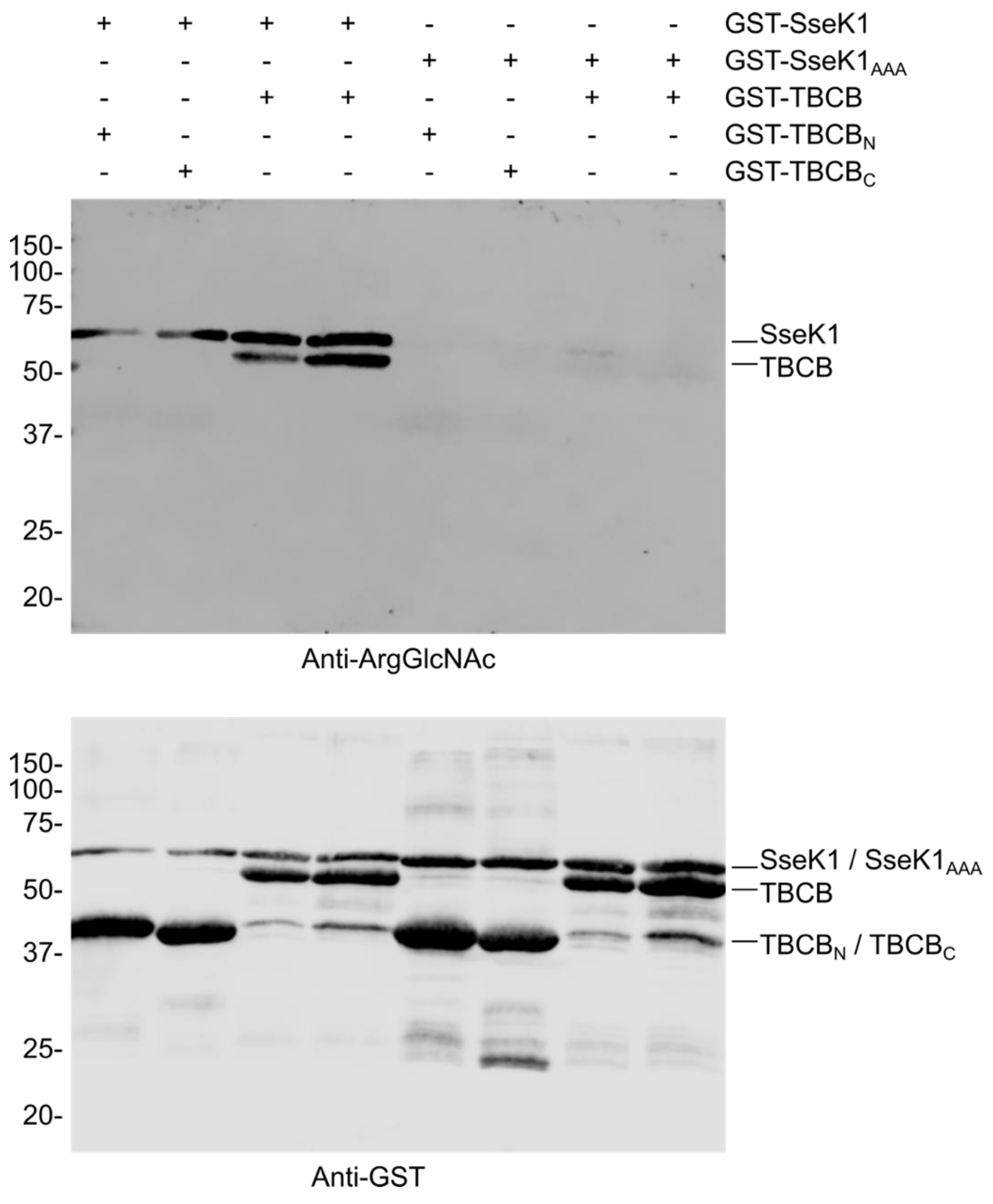
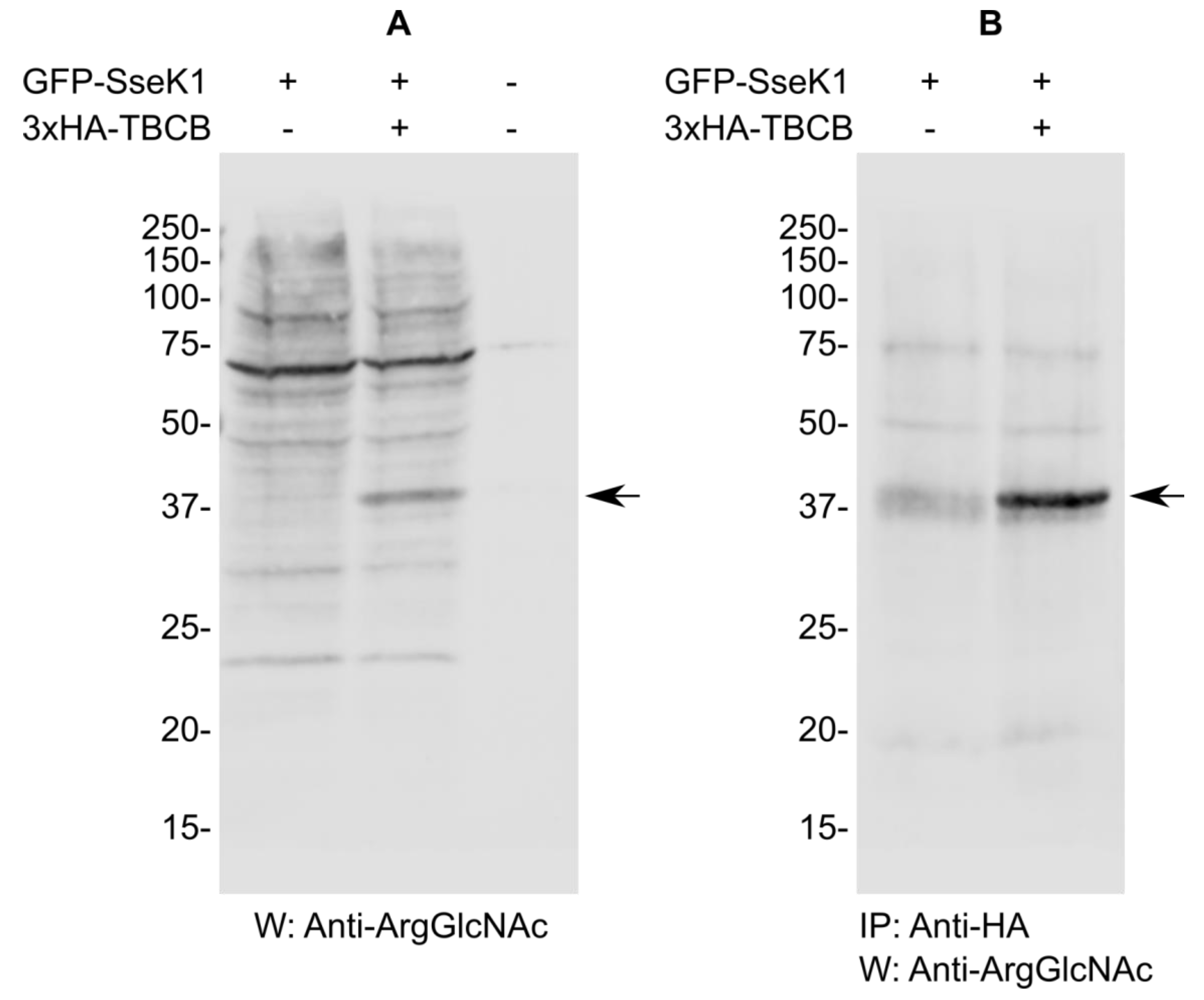
| Gene | Number of Clones | Description of the Product | Amino Acids Included in the Identified Clones |
|---|---|---|---|
| TBCB | 116 | Tubulin-binding cofactor B | 1–244 4–244 29–244 |
| ZBTB16 | 7 | Zing finger and BTB domain-containing protein 16 | 420–673 448–673 552–673 |
| H3F3B | 3 | Histone H3.3 | 1–136 40–136 |
| H3F3A | 1 | Histone H3.3 | 18–136 |
| CENPA | 1 | Histone H3-like centromeric protein A | 1–140 |
| HIRIP3 | 1 | HIRA-interacting protein 3 | 340–556 |
| XRCC6 | 1 | X-ray repair cross-complementing protein 6 | 335–609 |
| Strain/Plasmid | Relevant Characteristics | Source/Reference |
|---|---|---|
| Escherichia coli | ||
| BL21(DE3) | F− ompT gal dcm lon hsdSB (r− m−; E. coli B strain), with DE3, a λ prophage carrying the T7 RNA pol gene | Stratagene |
| DH5α | supE44 ∆lacU169 (Ø80 lacZ∆M15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | [51] |
| HB101 | F- mcrB mrr hsdS20 (rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20(SmR) glnV44 λ− | [52] |
| Salmonella enterica serovar Typhimuriuma | ||
| 14028 | Wild type | ATCC |
| SV7071 | sseK1::3xFLAG, Kmr | [15] |
| Saccharomyces cerevisiae | ||
| L40 | MATα trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ | [53] |
| Plasmids | ||
| pEGFPC1 | GFP fusion vector, Kmr | Clontech |
| pGEX-4T-1 | GST fusion vector, Apr | GE Healthcare |
| pGEX-4T-3 | GST fusion vector, Apr | GE Healthcare |
| pGAD1318 | Yeast two-hybrid vector, Apr | [54] |
| pLEX10 | Yeast two-hybrid vector, Apr | [55] |
| pIZ2047 | pBABEpuro-SseK1-3xFLAG | This work |
| pIZ2203 | pLEX10-SseK1 | This work |
| pIZ2260 | pGAD1318-TBCB | This work |
| pIZ2336 | pEGFPC1-SseK1 | This work |
| pIZ2339 | pGEX-4T-3-SseK1 | This work |
| pIZ3405 | pLEX10-SseK2 | This work |
| pIZ3406 | pLEX10-SseK3 | This work |
| pIZ3423 | pCS2-3xHA-TBCB | This work |
| pIZ3509 | pGEX-4T-1-TBCB | This work |
| pIZ3510 | pGEX-4T-1-TBCB(1–125) | This work |
| pIZ3511 | pGEX-4T-1-TBCB(126–244) | This work |
| pIZ3516 | pGEX-4T-1-SseK2 | This work |
| pIZ3517 | pGEX-4T-1-SseK3 | This work |
| pIZ3518 | pGEX-4T-3-SseK1(D223A/D225A) | This work |
| pIZ3519 | pGEX-4T-1-SseK3(D226A/D228A) | This work |
| pIZ3522 | pEGFPC1-SseK1(D223A/D225A) | This work |
| Oligonucleotide/Use | Sequence 5’-3’ |
|---|---|
| Construction of pIZ2047 | |
| SseK1pBABE_5’ | GTCAGGATCCGCCGCCACCATGATCCCACCATTAAATAG |
| 3FlagSal3’ | CTGAGTCGACTTACTATTTATCGTCGTCATC |
| Construction of pIZ2203 | |
| SseK1pLex10EcoRIfw | ATGCGAATTCATGATCCCACCATTAAATAG |
| SseK1pLex10SalIrev | ATGCGTCGACCTACTGCACATGCCTCGCCC |
| Construction of pIZ2336 | |
| SseKIGFPCEcoRIfw | ATCGGAATTCGATGATCCCACCATTAAATAGATATG |
| SseKIGFPCBamHIrev | ATCGGGATCCATTTCCGCTACTGCACATGC |
| Construction of pIZ3405 | |
| SseK2ecofw | GATCGAATTCATGGCACGTTTTAATGCCGC |
| SseK2salrv | ACGTGTCGACTTACCTCCAAGAACTGGCAG |
| Construction of pIZ3406 | |
| SseK3ecofw | ATGCGAATTCATGTTTTCTCGAGTCAGAGG |
| SseK3salrv | ATGCGTCGACTTATCTCCAGGAGCTGATAG |
| Construction of pIZ3423 | |
| TBCBecoRIfw | ATCGGAATTCGAGGTGACGGGGGTGTCGGC |
| TBCBxbaIrev | ATCGTCTAGATCATATCTCGTCCAACCCG |
| Construction of pIZ3509 | |
| TBCBEcoRIfw | ATCGGAATTCGAGGTGACGGGGGTGTCGGC |
| TBCBSTOPxhoIrev | ATCGCTCGAGGTCATATCTCGTCCAACCCG |
| Construction of pIZ3510 | |
| TBCBEcoRIfw | ATCGGAATTCGAGGTGACGGGGGTGTCGGC |
| TBCB125STOPxhoIrev | ATCGCTCGAGTCACAGGAAAGAGCGGACCGTGTC |
| Construction of pIZ3511 | |
| TBCB126EcoRIfw | ATCGGAATTCAAGCGCAGCAAGCTCGGCCG |
| TBCBSTOPxhoIrev | ATCGCTCGAGGTCATATCTCGTCCAACCCG |
| Construction of pIZ3521 | |
| Ssek3pEGFPC1ecoriFW | ATCGGAATTCTATGTTTTCTCGAGTCAGAGG |
| SseK3salrv | ATGCGTCGACTTATCTCCAGGAGCTGATAG |
| Identification of candidates carrying TBCB | |
| TBCBfw | GCTCCTACCCTGTAGATGACG |
| TBCBrev | CATATCTCGTCCAACCCGTAG |
| Mutagenesis of SseK1 | |
| SseK1DXD223AAAfw | GGTGTATATATCTTGCTGCTGCTATGATTATCACGG |
| SseK1DXD223AAArev | CCGTGATAATCATAGCAGCAGCAAGATATATACACC |
| Mutagenesis of SseK3 | |
| SseK3DXD226AAAfw | GGCTGCATATATCTTGCTGCTGCTATGTTACTTACAGG |
| SseK3DXD226AAArev | CCTGTAAGTAACATAGCAGCAGCAAGATATATGCAGCC |
| Sequencing of two-hybrid screen candidates | |
| Gal4AD | TACCACTACAATGGATG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araujo-Garrido, J.L.; Baisón-Olmo, F.; Bernal-Bayard, J.; Romero, F.; Ramos-Morales, F. Tubulin Folding Cofactor TBCB is a Target of the Salmonella Effector Protein SseK1. Int. J. Mol. Sci. 2020, 21, 3193. https://doi.org/10.3390/ijms21093193
Araujo-Garrido JL, Baisón-Olmo F, Bernal-Bayard J, Romero F, Ramos-Morales F. Tubulin Folding Cofactor TBCB is a Target of the Salmonella Effector Protein SseK1. International Journal of Molecular Sciences. 2020; 21(9):3193. https://doi.org/10.3390/ijms21093193
Chicago/Turabian StyleAraujo-Garrido, Juan Luis, Fernando Baisón-Olmo, Joaquín Bernal-Bayard, Francisco Romero, and Francisco Ramos-Morales. 2020. "Tubulin Folding Cofactor TBCB is a Target of the Salmonella Effector Protein SseK1" International Journal of Molecular Sciences 21, no. 9: 3193. https://doi.org/10.3390/ijms21093193
APA StyleAraujo-Garrido, J. L., Baisón-Olmo, F., Bernal-Bayard, J., Romero, F., & Ramos-Morales, F. (2020). Tubulin Folding Cofactor TBCB is a Target of the Salmonella Effector Protein SseK1. International Journal of Molecular Sciences, 21(9), 3193. https://doi.org/10.3390/ijms21093193





