High Expression of microRNA-143 is Associated with Favorable Tumor Immune Microenvironment and Better Survival in Estrogen Receptor Positive Breast Cancer
Abstract
1. Introduction
2. Results
2.1. Low Expression of miR-143 in Breast Cancer Compared with Normal Breast Tissue
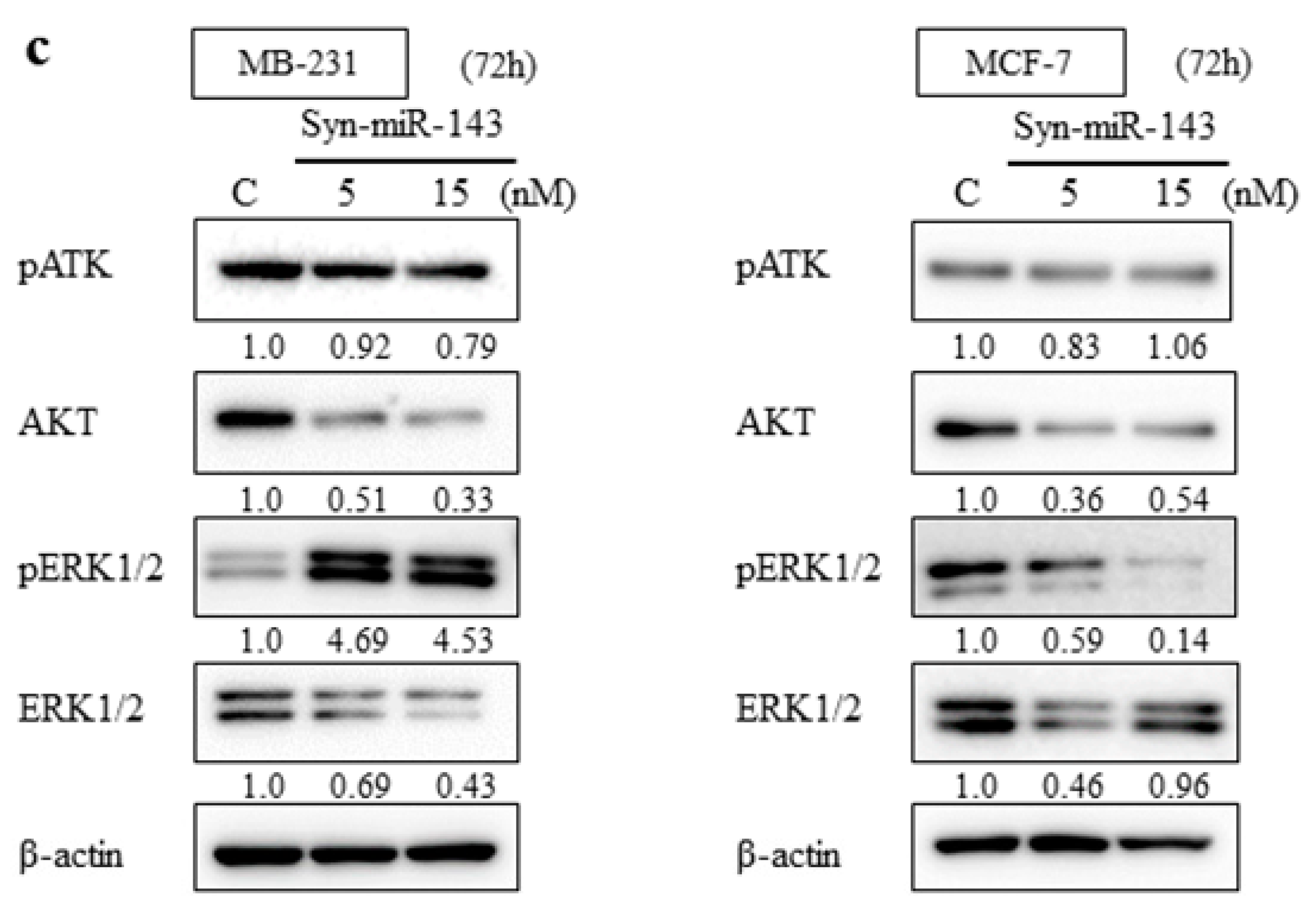
2.2. Overexpression of miR-143 Inhibited Cell Growth of MB-231 and MCF-7 Cells by Targeting KRAS and Its Effector Molecules
2.3. Introduction of syn-miR-143 Induced Apoptosis in MB-231 Cells
2.4. Anti-Tumor Effect of syn-miR-143 on Breast Cancer Xenograft Tumor In Vivo
2.5. No Significant Difference in Patient Clinicopathological Features between miR-143 High and miR-143 Low Group in Clinical Samples
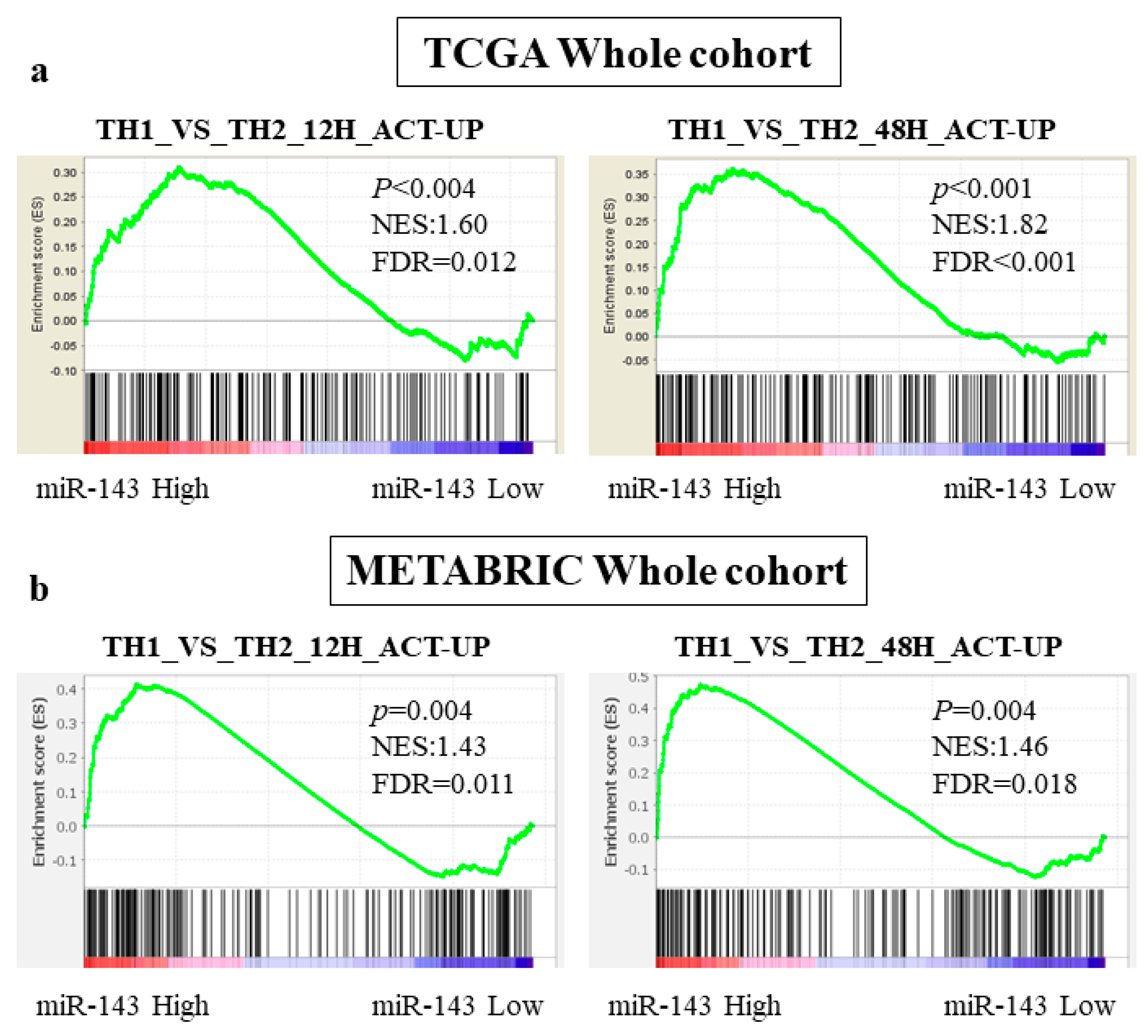
2.6. miR-143 High Expression Tumors Were Associated with Enrichment of Th1 related Gene Sets
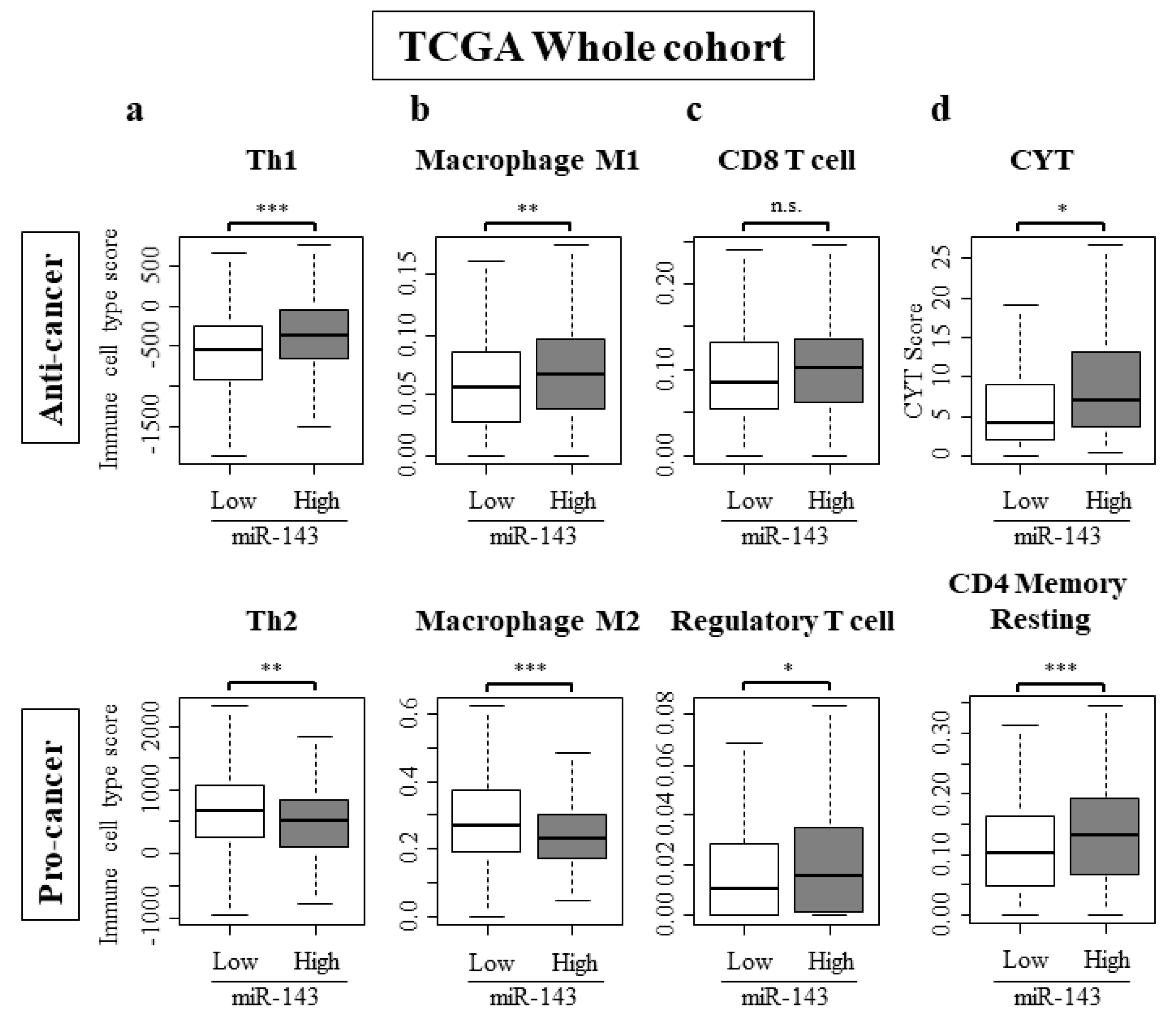
2.7. High Expression of miR-143 Was Associated with Increase in Anti-Cancer Immune Cells, Decrease in Pro-Cancer Immune Cells, and Elevated Cytolytic Activity in the Tumor Immune Microenvironment
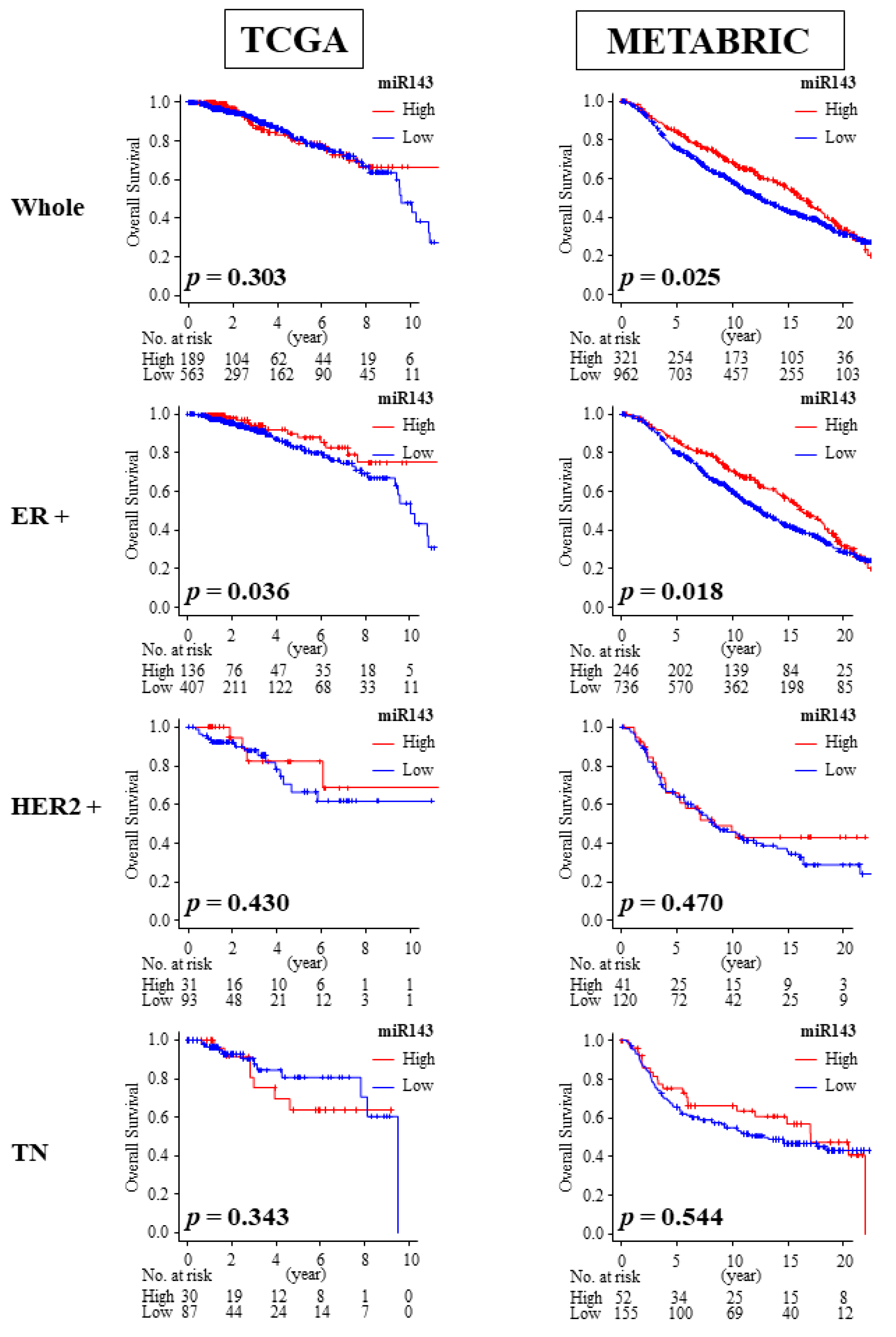
2.8. High Expression of miR-143 Was Associated with Better OS in ER Positive Patients
2.9. miR-143 Expression Did Not Correlate with ER Expression in Breast Cancer Patients
2.10. High Expression of miR-143 Was Associated with TNF-α Signaling Pathway in ER Positive Subtype but Not in TN Subtype in TCGA and METABRIC Cohorts
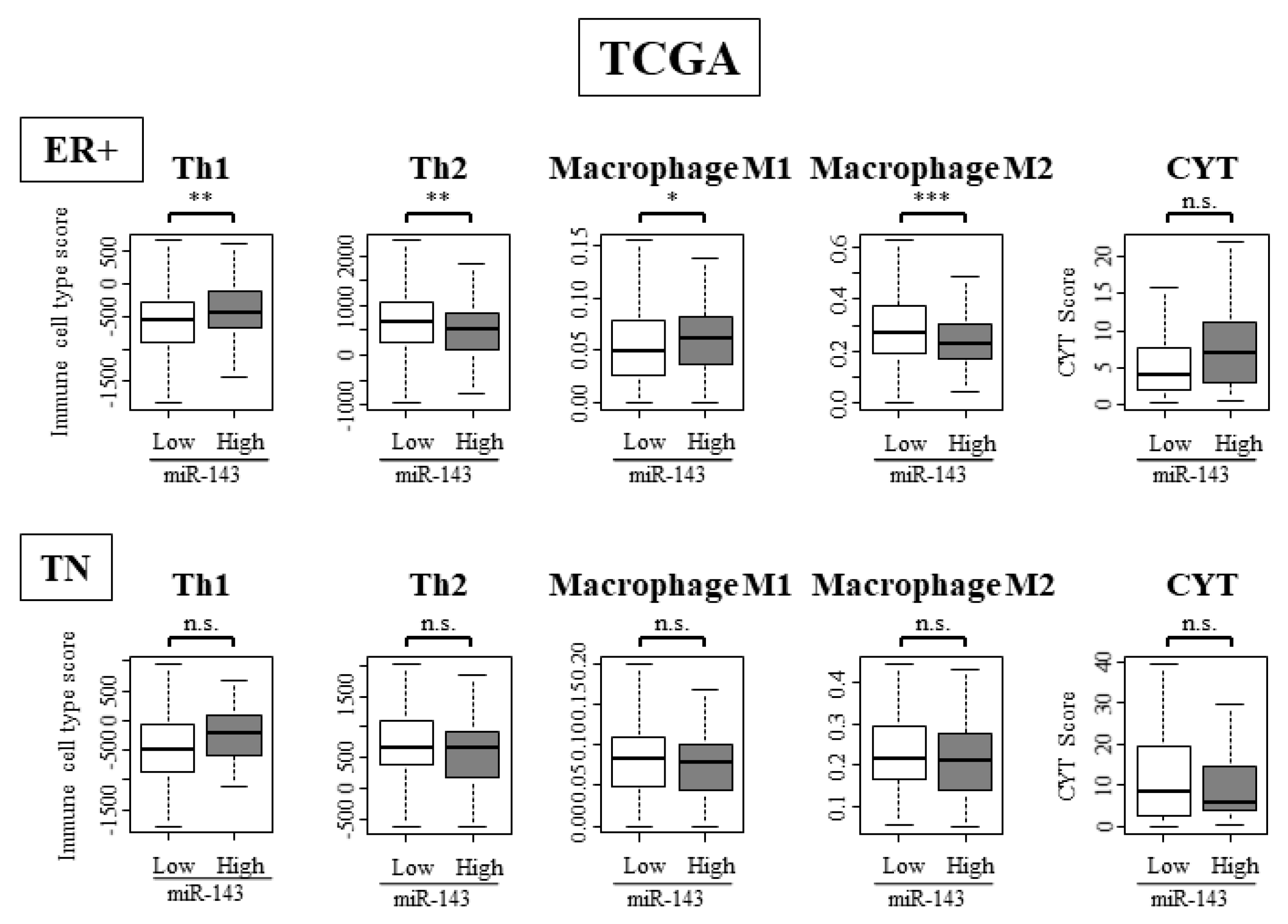
2.11. High Expression of miR-143 Was Associated with High Infiltration of Anti-Cancer Immune Cells and Anti-Cancer Activity in Tumor Immune Microenvironment Only in ER Positive Subtypes and Not in TN Subtype
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Cell Viability
4.2. Cell Transfection with miRNA
4.3. Western Blot Analysis
4.4. Quantitative RT-PCR
4.5. Hoechst 33352 Staining
4.6. Human Tumor Xenograft Model
4.7. Extraction the Clinical Data and microRNA Expression Data from TCGA and METABRIC
4.8. Gene Set Enrichment Analysis (GSEA)
4.9. CIBERSORT, Cell Compositions of Immune Cells and Cytolytic Activity Score
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ER | Estrogen receptor |
| HER2 | Human epidermal growth factor receptor 2 |
| TN | Triple negative |
| TCGA | The Cancer Genome Atlas |
| METABRIC | Molecular Taxonomy of Breast Cancer International Consortium |
| GSEA | Gene Set Enrichment Analysis |
| CYT | Cytolytic Activity |
| Th1 | Helper T cell type 1 |
| Th2 | Helper T cell type 2 |
| ESR1 | Estrogen receptor 1 |
| ESR2 | Estrogen receptor 2 |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Young, J.; Kawaguchi, T.; Yan, L.; Qi, Q.; Liu, S.; Takabe, K. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget 2017, 8, 99978–99989. [Google Scholar] [CrossRef]
- Drusco, A.; Croce, C.M. MicroRNAs and Cancer: A Long Story for Short RNAs. Adv. Cancer Res. 2017, 135, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kawaguchi, T.; Yan, L.; Young, J.; Qi, Q.; Takabe, K. Clinical Relevance of microRNA Expressions in Breast Cancer Validated Using the Cancer Genome Atlas (TCGA). Ann. Surg. Oncol. 2017, 24, 2943–2949. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Yan, L.; Qi, Q.; Peng, X.; Gabriel, E.M.; Young, J.; Liu, S.; Takabe, K. Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Sci. Rep. 2017, 7, 15945. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Takabe, K.; Yoshida, K.; Akao, Y. Effects of MIR143 on rat sarcoma signaling networks in solid tumors: A brief overview. Cancer Sci. 2020, 111, 1076. [Google Scholar] [CrossRef]
- He, Z.; Yi, J.; Liu, X.; Chen, J.; Han, S.; Jin, L.; Chen, L.; Song, H. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol. Cancer 2016, 15, 51. [Google Scholar] [CrossRef]
- Johannessen, C.; Moi, L.; Kiselev, Y.; Pedersen, M.I.; Dalen, S.M.; Braaten, T.; Busund, L.T. Expression and function of the miR-143/145 cluster in vitro and in vivo in human breast cancer. PLoS ONE 2017, 12, e0186658. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Tajirika, T.; Sugito, N.; Kuranaga, Y.; Shinohara, H.; Tsujino, T.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K. Synthetic miR-143 Inhibits Growth of HER2-Positive Gastric Cancer Cells by Suppressing KRAS Networks Including DDX6 RNA Helicase. Int. J. Mol. Sci. 2019, 20, 1697. [Google Scholar] [CrossRef]
- Akao, Y.; Kumazaki, M.; Shinohara, H.; Sugito, N.; Kuranaga, Y.; Tsujino, T.; Yoshikawa, Y.; Kitade, Y. Impairment of K-Ras signaling networks and increased efficacy of epidermal growth factor receptor inhibitors by a novel synthetic miR-143. Cancer Sci. 2018, 109, 1455–1467. [Google Scholar] [CrossRef]
- Takai, T.; Tsujino, T.; Yoshikawa, Y.; Inamoto, T.; Sugito, N.; Kuranaga, Y.; Heishima, K.; Soga, T.; Hayashi, K.; Miyata, K.; et al. Synthetic miR-143 Exhibited an Anti-Cancer Effect via the Downregulation of K-RAS Networks of Renal Cell Cancer Cells In Vitro and In Vivo. Mol. Ther. J. Am. Soc. Gene Ther. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Ma, C.; Li, W.; Yang, S.; Liu, Z. miR-143 suppresses epithelial-mesenchymal transition and inhibits tumor growth of breast cancer through down-regulation of ERK5. Mol. Carcinog. 2016, 55, 1990–2000. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Chen, X.; Liang, H.; Deng, T.; Chen, W.; Zhang, S.; Liu, M.; Gao, X.; Liu, Y.; Zhao, C.; et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol. Cancer 2014, 13, 220. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Hu, J.; Song, H.; Xu, H.; Wu, C.; Zhao, B.; Xie, D.; Wu, T.; Zhao, J.; Fang, L. miR-143-3p targeting LIM domain kinase 1 suppresses the progression of triple-negative breast cancer cells. Am. J. Transl. Res. 2017, 9, 2276–2285. [Google Scholar]
- Tokumaru, Y.; Joyce, D.; Takabe, K. Current status and limitations of immunotherapy for breast cancer. Surgery 2019. [Google Scholar] [CrossRef]
- Dieci, M.V.; Griguolo, G.; Miglietta, F.; Guarneri, V. The immune system and hormone-receptor positive breast cancer: Is it really a dead end? Cancer Treat. Rev. 2016, 46, 9–19. [Google Scholar] [CrossRef]
- McDonald, K.A.; Kawaguchi, T.; Qi, Q.; Peng, X.; Asaoka, M.; Young, J.; Opyrchal, M.; Yan, L.; Patnaik, S.; Otsuji, E.; et al. Tumor Heterogeneity Correlates with Less Immune Response and Worse Survival in Breast Cancer Patients. Ann. Surg. Oncol. 2019, 26, 2191–2199. [Google Scholar] [CrossRef]
- Woo, S.R.; Corrales, L.; Gajewski, T.F. Innate immune recognition of cancer. Annu. Rev. Immunol. 2015, 33, 445–474. [Google Scholar] [CrossRef]
- Varn, F.S.; Mullins, D.W.; Arias-Pulido, H.; Fiering, S.; Cheng, C. Adaptive immunity programmes in breast cancer. Immunology 2017, 150, 25–34. [Google Scholar] [CrossRef]
- Wu, Y.; Yan, Y.; Su, Z.; Bie, Q.; Chen, X.; Barnie, P.A.; Guo, Q.; Wang, S.; Xu, H. Enhanced circulating ILC2s and MDSCs may contribute to ensure maintenance of Th2 predominant in patients with lung cancer. Mol. Med. Rep. 2017, 15, 4374–4381. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A. Breast Cancer Immunotherapy: Facts and Hopes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 511–520. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Coussens, L.M. Inflammation and breast cancer. Balancing immune response: Crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Edechi, C.A.; Ikeogu, N.; Uzonna, J.E.; Myal, Y. Regulation of Immunity in Breast Cancer. Cancers 2019, 11, 1080. [Google Scholar] [CrossRef]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef]
- Volinia, S.; Croce, C.M. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 7413–7417. [Google Scholar] [CrossRef]
- Khodadadi-Jamayran, A.; Akgol-Oksuz, B.; Afanasyeva, Y.; Heguy, A.; Thompson, M.; Ray, K.; Giro-Perafita, A.; Sanchez, I.; Wu, X.; Tripathy, D.; et al. Prognostic role of elevated mir-24-3p in breast cancer and its association with the metastatic process. Oncotarget 2018, 9, 12868–12878. [Google Scholar] [CrossRef]
- Fisher, J.N.; Terao, M.; Fratelli, M.; Kurosaki, M.; Paroni, G.; Zanetti, A.; Gianni, M.; Bolis, M.; Lupi, M.; Tsykin, A.; et al. MicroRNA networks regulated by all-trans retinoic acid and Lapatinib control the growth, survival and motility of breast cancer cells. Oncotarget 2015, 6, 13176–13200. [Google Scholar] [CrossRef][Green Version]
- Soheilyfar, S.; Velashjerdi, Z.; Sayed Hajizadeh, Y.; Fathi Maroufi, N.; Amini, Z.; Khorrami, A.; Haj Azimian, S.; Isazadeh, A.; Taefehshokr, S.; Taefehshokr, N. In vivo and in vitro impact of miR-31 and miR-143 on the suppression of metastasis and invasion in breast cancer. J. B. U. Off. J. Balk. Union Oncol. 2018, 23, 1290–1296. [Google Scholar]
- Abbas, A.R.; Baldwin, D.; Ma, Y.; Ouyang, W.; Gurney, A.; Martin, F.; Fong, S.; van Lookeren Campagne, M.; Godowski, P.; Williams, P.M.; et al. Immune response in silico (IRIS): Immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005, 6, 319–331. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [PubMed]
- Kodahl, A.R.; Lyng, M.B.; Binder, H.; Cold, S.; Gravgaard, K.; Knoop, A.S.; Ditzel, H.J. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: A case control study. Mol. Oncol. 2014, 8, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Miyoshi, Y. Mechanism of resistance to endocrine therapy in breast cancer: The important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer 2018, 25, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Giltnane, J.M.; Balko, J.M. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov. Med. 2014, 17, 275–283. [Google Scholar] [PubMed]
- Tokumaru, Y.; Oshi, M.; Katsuta, E.; Yan, L.; Satyananda, V.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K.; Takabe, K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am. J. Cancer Res. 2020, 10, 897–907. [Google Scholar] [PubMed]
- Yoshikawa, Y.; Taniguchi, K.; Tsujino, T.; Heishima, K.; Inamoto, T.; Takai, T.; Minami, K.; Azuma, H.; Miyata, K.; Hayashi, K.; et al. Anti-cancer Effects of a Chemically Modified miR-143 on Bladder Cancer by Either Systemic or Intravesical Treatment. Mol. Ther. Methods Clin. Dev. 2019, 13, 290–302. [Google Scholar] [CrossRef]
- Nair, J.K.; Willoughby, J.L.; Chan, A.; Charisse, K.; Alam, M.R.; Wang, Q.; Hoekstra, M.; Kandasamy, P.; Kel’in, A.V.; Milstein, S.; et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. [Google Scholar] [CrossRef]
- Guo, H.; Chen, Y.; Hu, X.; Qian, G.; Ge, S.; Zhang, J. The regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol. Cancer 2013, 12, 77. [Google Scholar] [CrossRef]
- Nagahashi, M.; Ramachandran, S.; Kim, E.Y.; Allegood, J.C.; Rashid, O.M.; Yamada, A.; Zhao, R.; Milstien, S.; Zhou, H.; Spiegel, S.; et al. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 2012, 72, 726–735. [Google Scholar] [CrossRef]
- Rashid, O.M.; Nagahashi, M.; Ramachandran, S.; Dumur, C.; Schaum, J.; Yamada, A.; Terracina, K.P.; Milstien, S.; Spiegel, S.; Takabe, K. An improved syngeneic orthotopic murine model of human breast cancer progression. Breast Cancer Res. Treat. 2014, 147, 501–512. [Google Scholar] [CrossRef]
- Rashid, O.M.; Takabe, K. Animal models for exploring the pharmacokinetics of breast cancer therapies. Expert Opin. Drug Metab. Toxicol. 2015, 11, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Yan, L.; Nagahashi, M.; Raza, A.; Sturgill, J.L.; Lyon, D.E.; Rashid, O.M.; Hait, N.C.; Takabe, K. Doxorubicin effect is enhanced by sphingosine-1-phosphate signaling antagonist in breast cancer. J. Surg. Res. 2017, 219, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; DeMasi, S.C.; Terracina, K.P.; Spiegel, S.; Phan, G.Q.; Bear, H.D.; Takabe, K. Modified breast cancer model for preclinical immunotherapy studies. J. Surg. Res. 2016, 204, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Murine breast cancer mastectomy model that predicts patient outcomes for drug development. J. Surg. Res. 2017, 219, 310–318. [Google Scholar] [CrossRef]
- Nagahashi, M.; Yamada, A.; Katsuta, E.; Aoyagi, T.; Huang, W.C.; Terracina, K.P.; Hait, N.C.; Allegood, J.C.; Tsuchida, J.; Yuza, K.; et al. Targeting the SphK1/S1P/S1PR1 Axis That Links Obesity, Chronic Inflammation, and Breast Cancer Metastasis. Cancer Res. 2018, 78, 1713–1725. [Google Scholar] [CrossRef]
- Katsuta, E.; Oshi, M.; Rashid, O.M.; Takabe, K. Generating a Murine Orthotopic Metastatic Breast Cancer Model and Performing Murine Radical Mastectomy. J. Vis. Exp. 2018. [Google Scholar] [CrossRef]
- Yamada, A.; Nagahashi, M.; Aoyagi, T.; Huang, W.C.; Lima, S.; Hait, N.C.; Maiti, A.; Kida, K.; Terracina, K.P.; Miyazaki, H.; et al. ABCC1-Exported Sphingosine-1-phosphate, Produced by Sphingosine Kinase 1, Shortens Survival of Mice and Patients with Breast Cancer. Mol. Cancer Res. 2018, 16, 1059–1070. [Google Scholar] [CrossRef]
- Akao, Y.; Nakagawa, Y.; Hirata, I.; Iio, A.; Itoh, T.; Kojima, K.; Nakashima, R.; Kitade, Y.; Naoe, T. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010, 17, 398–408. [Google Scholar] [CrossRef]
- Taniguchi, K.; Sugito, N.; Kumazaki, M.; Shinohara, H.; Yamada, N.; Nakagawa, Y.; Ito, Y.; Otsuki, Y.; Uno, B.; Uchiyama, K.; et al. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015, 363, 17–27. [Google Scholar] [CrossRef]
- Taniguchi, K.; Ito, Y.; Sugito, N.; Kumazaki, M.; Shinohara, H.; Yamada, N.; Nakagawa, Y.; Sugiyama, T.; Futamura, M.; Otsuki, Y.; et al. Organ-specific PTB1-associated microRNAs determine expression of pyruvate kinase isoforms. Sci. Rep. 2015, 5, 8647. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Yan, L.; Qi, Q.; Peng, X.; Edge, S.B.; Young, J.; Yao, S.; Liu, S.; Otsuji, E.; Takabe, K. Novel MicroRNA-Based Risk Score Identified by Integrated Analyses to Predict Metastasis and Poor Prognosis in Breast Cancer. Ann. Surg. Oncol. 2018, 25, 4037–4046. [Google Scholar] [CrossRef] [PubMed]
- Sporn, J.C.; Katsuta, E.; Yan, L.; Takabe, K. Expression of MicroRNA-9 is Associated With Overall Survival in Breast Cancer Patients. J. Surg. Res. 2019, 233, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Oshi, M.; Butash, A.L.; Asaoka, M.; Katsuta, E.; Peng, X.; Qi, Q.; Yan, L.; Takabe, K. Estrogen Receptor Positive Breast Cancer with High Expression of Androgen Receptor has Less Cytolytic Activity and Worse Response to Neoadjuvant Chemotherapy but Better Survival. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Kawaguchi, T.; Peng, X.; Qi, Q.; Liu, S.; Yan, L.; Takabe, K. Tumor Infiltrating Lymphocytes and Macrophages Improve Survival in Microsatellite Unstable Colorectal Cancer. Sci. Rep. 2019, 9, 13455. [Google Scholar] [CrossRef] [PubMed]
- Hoki, T.; Katsuta, E.; Yan, L.; Takabe, K.; Ito, F. Low DMT1 Expression Associates with Increased Oxidative Phosphorylation and Early Recurrence in Hepatocellular Carcinoma. J. Surg. Res. 2019, 234, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Terakawa, T.; Katsuta, E.; Yan, L.; Turaga, N.; McDonald, K.A.; Fujisawa, M.; Guru, K.A.; Takabe, K. High expression of SLCO2B1 is associated with prostate cancer recurrence after radical prostatectomy. Oncotarget 2018, 9, 14207–14218. [Google Scholar] [CrossRef][Green Version]
- Takahashi, H.; Katsuta, E.; Yan, L.; Dasgupta, S.; Takabe, K. High expression of Annexin A2 is associated with DNA repair, metabolic alteration, and worse survival in pancreatic ductal adenocarcinoma. Surgery 2019, 166, 150–156. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Okano, M.; Oshi, M.; Butash, A.L.; Katsuta, E.; Tachibana, K.; Saito, K.; Okayama, H.; Peng, X.; Yan, L.; Kono, K.; et al. Triple-Negative Breast Cancer with High Levels of Annexin A1 Expression Is Associated with Mast Cell Infiltration, Inflammation, and Angiogenesis. Int. J. Mol. Sci. 2019, 20, 4197. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Narayanan, S.; Takabe, K. ASO Author Reflections: “From Computer to Bedside”: A New Translational Approach to Immunogenomics. Ann. Surg. Oncol. 2018, 25, 846–847. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, R.; Olex, A.L.; Dozmorov, M.; Bear, H.D.; Fernandez, L.J.; Takabe, K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res. Treat. 2017, 162, 191–198. [Google Scholar] [CrossRef] [PubMed]



| Clinicopathological Factor | Whole Cohort (n = 753) | p Value | ||
|---|---|---|---|---|
| miR-143 High n = 189 | miR-143 Low n = 564 | |||
| Age | 0.098 | |||
| <65 years | 142 | 386 | ||
| ≥65 years | 47 | 177 | ||
| Unknown | 0 | 1 | ||
| Race | 0.226 | |||
| Asian | 20 | 36 | ||
| African American | 39 | 117 | ||
| White | 128 | 407 | ||
| Other | 2 | 4 | ||
| Menopause status | 0.937 | |||
| Pre | 38 | 117 | ||
| Post | 121 | 361 | ||
| Other | 30 | 86 | ||
| Stage | 0.565 | |||
| I/II/III/IV | 32/105/49/1 | 104/324/122/8 | ||
| Unknown | 2 | 6 | ||
| pT | 0.629 | |||
| T1/T2/T3/T4 | 53/101/30/5 | 158/321/66/18 | ||
| Tx | 0 | 1 | ||
| pN | 0.683 | |||
| N0/N1/N2/N3 | 88/61/21/17 | 269/195/56/35 | ||
| Nx | 2 | 9 | ||
| M | 0.761 | |||
| M0/M1 | 150/1 | 443/8 | ||
| Mx | 38 | 113 | ||
| Grade | 0.254 | |||
| G1/G2/G3 | 19/54/38 | 47/170/153 | ||
| Gx | 78 | 194 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokumaru, Y.; Asaoka, M.; Oshi, M.; Katsuta, E.; Yan, L.; Narayanan, S.; Sugito, N.; Matsuhashi, N.; Futamura, M.; Akao, Y.; et al. High Expression of microRNA-143 is Associated with Favorable Tumor Immune Microenvironment and Better Survival in Estrogen Receptor Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 3213. https://doi.org/10.3390/ijms21093213
Tokumaru Y, Asaoka M, Oshi M, Katsuta E, Yan L, Narayanan S, Sugito N, Matsuhashi N, Futamura M, Akao Y, et al. High Expression of microRNA-143 is Associated with Favorable Tumor Immune Microenvironment and Better Survival in Estrogen Receptor Positive Breast Cancer. International Journal of Molecular Sciences. 2020; 21(9):3213. https://doi.org/10.3390/ijms21093213
Chicago/Turabian StyleTokumaru, Yoshihisa, Mariko Asaoka, Masanori Oshi, Eriko Katsuta, Li Yan, Sumana Narayanan, Nobuhiko Sugito, Nobuhisa Matsuhashi, Manabu Futamura, Yukihiro Akao, and et al. 2020. "High Expression of microRNA-143 is Associated with Favorable Tumor Immune Microenvironment and Better Survival in Estrogen Receptor Positive Breast Cancer" International Journal of Molecular Sciences 21, no. 9: 3213. https://doi.org/10.3390/ijms21093213
APA StyleTokumaru, Y., Asaoka, M., Oshi, M., Katsuta, E., Yan, L., Narayanan, S., Sugito, N., Matsuhashi, N., Futamura, M., Akao, Y., Yoshida, K., & Takabe, K. (2020). High Expression of microRNA-143 is Associated with Favorable Tumor Immune Microenvironment and Better Survival in Estrogen Receptor Positive Breast Cancer. International Journal of Molecular Sciences, 21(9), 3213. https://doi.org/10.3390/ijms21093213









