A PIANO (Proper, Insufficient, Aberrant, and NO Reprogramming) Response to the Yamanaka Factors in the Initial Stages of Human iPSC Reprogramming
Abstract
:1. Introduction
2. Results
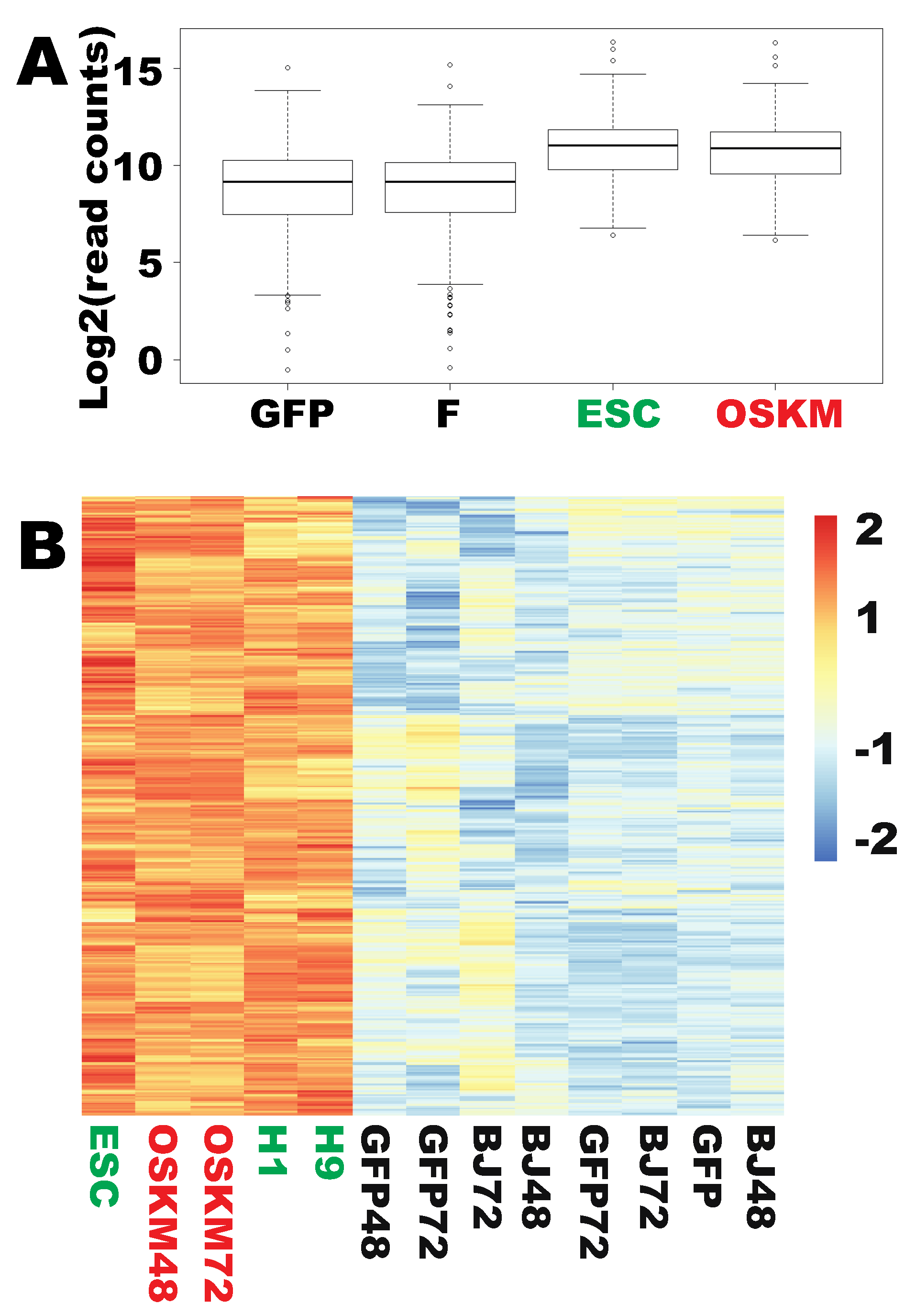
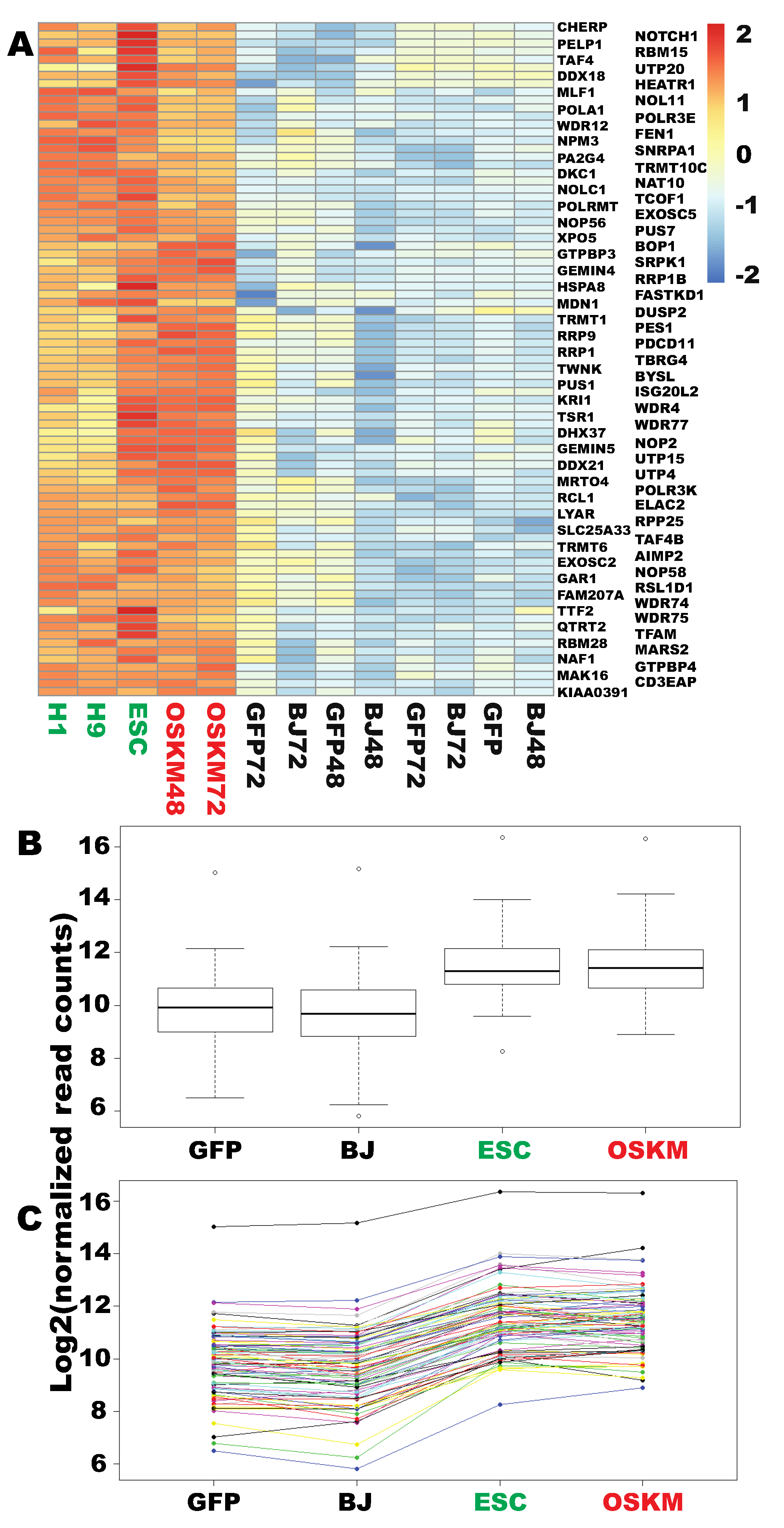
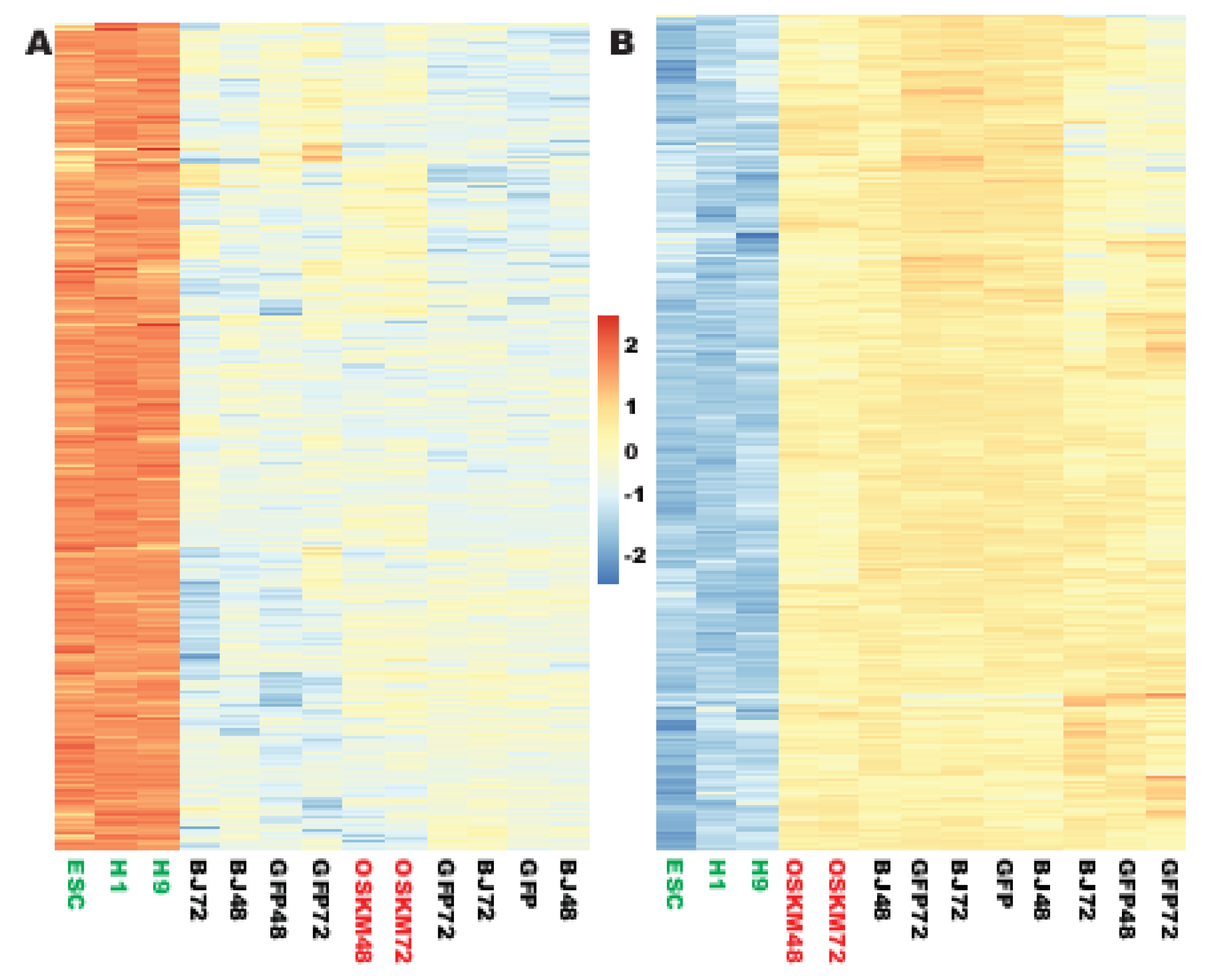
2.1. Genes in Nucleic Acid Metabolism and Ribosome Biogenesis Are Successfully Reprogrammed to the Pluripotent State within 48 h
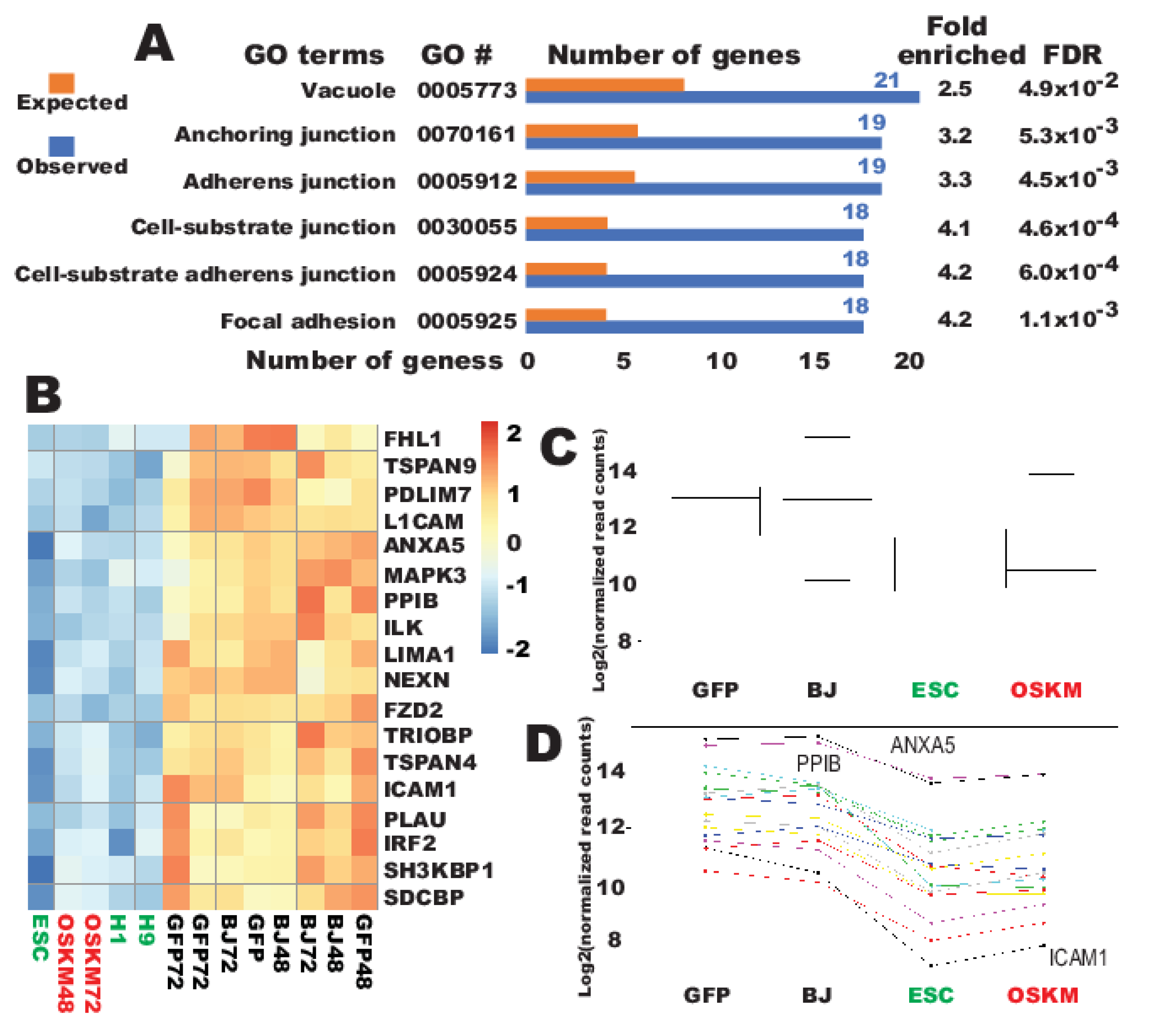
2.2. A Set of Promiscuous Fibroblast Genes and Genes for Focal Adhesion and Vacuole of Cellular Components Are Quickly Downreprogrammed to the Pluripotent State
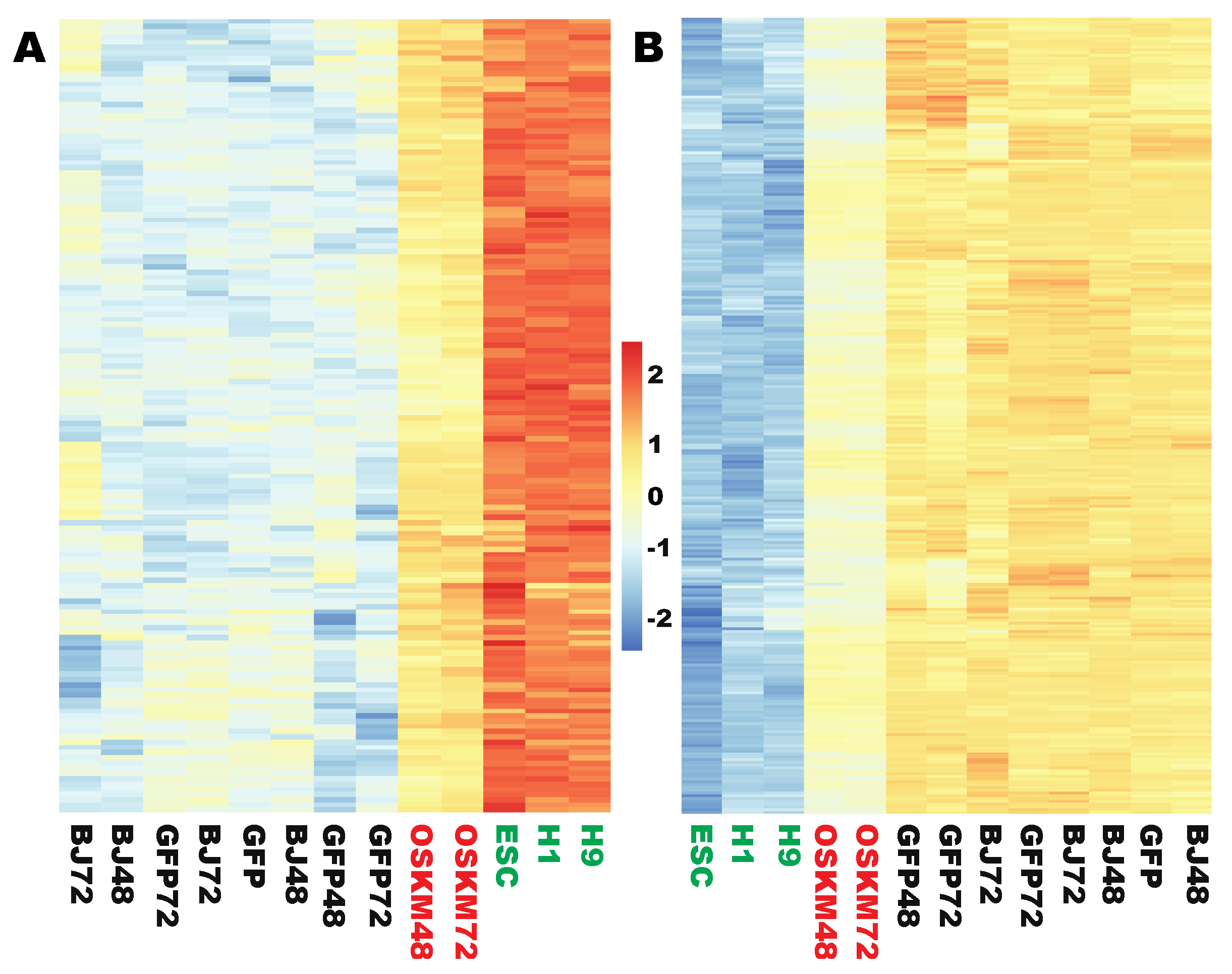
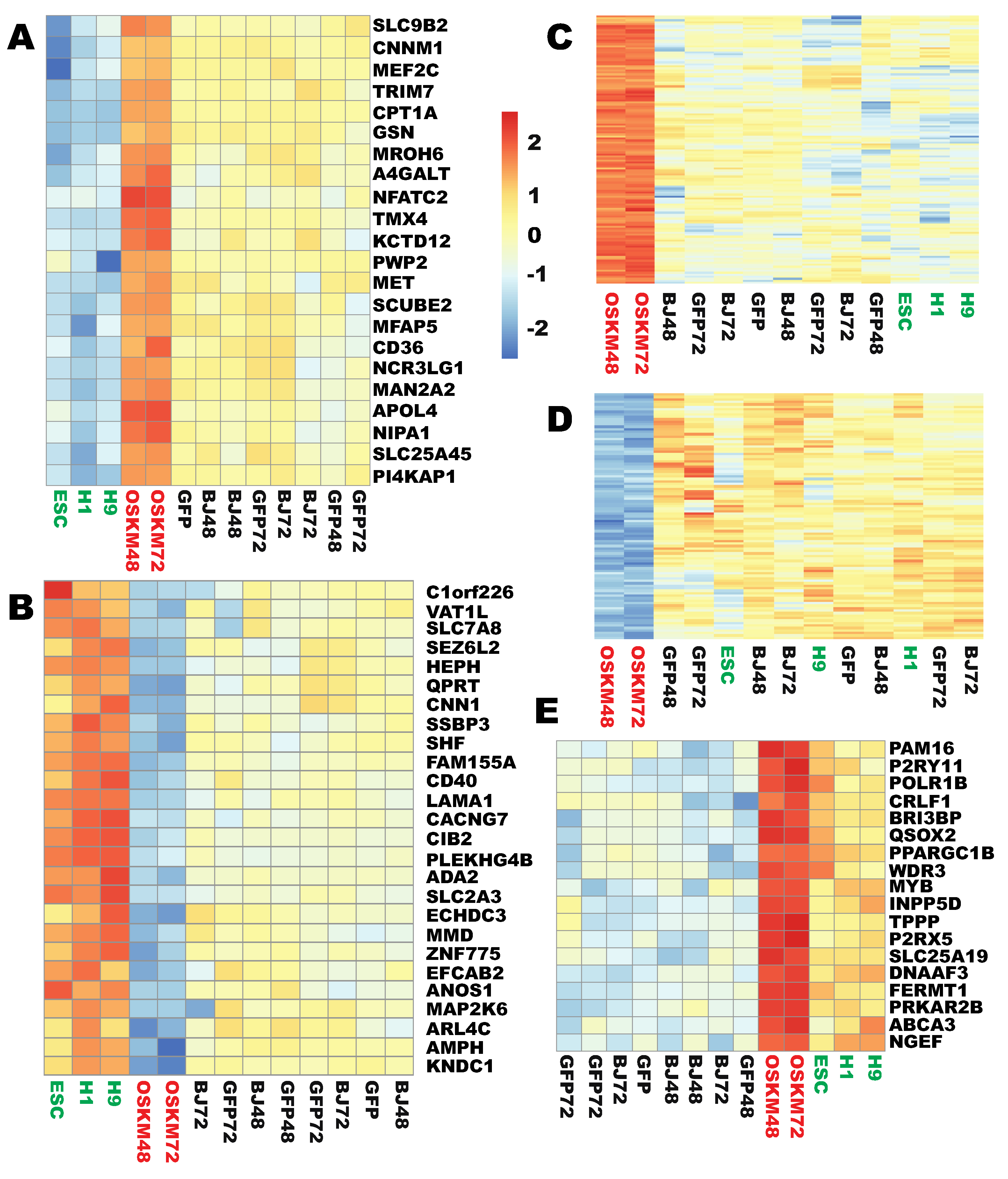
2.3. Two Sets of Genes Were Significantly but Insufficiently Reprogrammed at the Early Stages
2.4. Two Large Sets of Highly Enriched and Robustly Expressed Genes Did Not Respond to Reprogramming Factors at the Initial Stages
2.5. Six Types of Aberrant Reprogramming
3. Discussion
4. Materials and Methods
4.1. Cells and Cell Culture
4.2. Viral Transduction of Human Fibroblasts
4.3. RNA Sequencing
4.4. Bioinformatics
4.5. Visualization of a Large Set of Data
4.6. Gene Ontology Analyses
4.7. Selection Criteria
4.7.1. Reprogramome
4.7.2. Criteria for Differentially Expressed Genes
4.7.3. Defining the Significantly Regulated but Insufficiently Reprogrammed Genes by OSKM
4.7.4. Defining the Highly Enriched Gene Sets
4.7.5. Defining the Genes Irresponsive to OSKM Induction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanna, J.; Saha, K.; Pando, B.; van Zon, J.; Lengner, C.J.; Creyghton, M.P.; van Oudenaarden, A.; Jaenisch, R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 2009, 462, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Hu, K. On Mammalian Totipotency: What Is the Molecular Underpinning for the Totipotency of Zygote? Stem Cells Dev. 2019, 28, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Hu, K. Vectorology and factor delivery in induced pluripotent stem cell reprogramming. Stem Cells Dev. 2014, 23, 1301–1315. [Google Scholar] [CrossRef] [Green Version]
- Hu, K. All roads lead to induced pluripotent stem cells: The technologies of iPSC generation. Stem Cells Dev. 2014, 23, 1285–1300. [Google Scholar] [CrossRef] [Green Version]
- Shao, Z.; Yao, C.; Khodadadi-Jamayran, A.; Xu, W.; Townes, T.M.; Crowley, M.R.; Hu, K. Reprogramming by De-bookmarking the Somatic Transcriptional Program through Targeting of BET Bromodomains. Cell Rep. 2016, 16, 3138–3145. [Google Scholar] [CrossRef] [Green Version]
- Shao, Z.; Zhang, R.; Khodadadi-Jamayran, A.; Chen, B.; Crowley, M.R.; Festok, M.A.; Crossman, D.K.; Townes, T.M.; Hu, K. The acetyllysine reader BRD3R promotes human nuclear reprogramming and regulates mitosis. Nat. Commun. 2016, 7, 10869. [Google Scholar] [CrossRef] [Green Version]
- Samavarchi-Tehrani, P.; Golipour, A.; David, L.; Sung, H.K.; Beyer, T.A.; Datti, A.; Woltjen, K.; Nagy, A.; Wrana, J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010, 7, 64–77. [Google Scholar] [CrossRef] [Green Version]
- Polo, J.M.; Anderssen, E.; Walsh, R.M.; Schwarz, B.A.; Nefzger, C.M.; Lim, S.M.; Borkent, M.; Apostolou, E.; Alaei, S.; Cloutier, J.; et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012, 151, 1617–1632. [Google Scholar] [CrossRef] [Green Version]
- Sridharan, R.; Tchieu, J.; Mason, M.J.; Yachechko, R.; Kuoy, E.; Horvath, S.; Zhou, Q.; Plath, K. Role of the murine reprogramming factors in the induction of pluripotency. Cell 2009, 136, 364–377. [Google Scholar] [CrossRef] [Green Version]
- Mikkelsen, T.S.; Hanna, J.; Zhang, X.; Ku, M.; Wernig, M.; Schorderet, P.; Bernstein, B.E.; Jaenisch, R.; Lander, E.S.; Meissner, A. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008, 454, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Yu, J.; Suknuntha, K.; Tian, S.; Montgomery, K.; Choi, K.D.; Stewart, R.; Thomson, J.A.; Slukvin, I.I. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 2011, 117, e109–e119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, K.; Ianov, L.; Crossman, D.K. Profiling of the reprogramome and quantification of fibroblast reprogramming to pluripotency. BioRxiv 2020. [Google Scholar] [CrossRef]
- Kang, L.; Yao, C.; Khodadadi-Jamayran, A.; Xu, W.; Zhang, R.; Banerjee, N.S.; Chang, C.W.; Chow, L.T.; Townes, T.; Hu, K. The Universal 3D3 Antibody of Human PODXL Is Pluripotent Cytotoxic, and Identifies a Residual Population After Extended Differentiation of Pluripotent Stem Cells. Stem Cells Dev. 2016, 25, 556–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mah, N.; Wang, Y.; Liao, M.C.; Prigione, A.; Jozefczuk, J.; Lichtner, B.; Wolfrum, K.; Haltmeier, M.; Flottmann, M.; Schaefer, M.; et al. Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS ONE 2011, 6, e24351. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Cevallos, R.; Hu, K. Reprogramming Human Fibroblasts to Induced Pluripotent Stem Cells Using the GFP-marked Lentiviral Vectors in the Chemically Defined Medium. Methods Mol. Biol. 2020, in press. [Google Scholar]
- Hansson, J.; Rafiee, M.R.; Reiland, S.; Polo, J.M.; Gehring, J.; Okawa, S.; Huber, W.; Hochedlinger, K.; Krijgsveld, J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012, 2, 1579–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, K.; Slukvin, I. Induction of pluripotent stem cells from umbilical cord blood. Rev. Cell Biol. Mol. Med. 2012, (Suppl. 13). [Google Scholar] [CrossRef]
- Brambrink, T.; Foreman, R.; Welstead, G.G.; Lengner, C.J.; Wernig, M.; Suh, H.; Jaenisch, R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2008, 2, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Stadtfeld, M.; Maherali, N.; Breault, D.T.; Hochedlinger, K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2008, 2, 230–240. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [Green Version]
- Khoo, T.S.; Jamal, R.; Abdul Ghani, N.A.; Alauddin, H.; Hussin, N.H.; Abdul Murad, N.A. Retention of Somatic Memory Associated with Cell Identity, Age and Metabolism in Induced Pluripotent Stem (iPS) Cells Reprogramming. Stem Cell Rev. Rep. 2020, 16, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Koster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, K. Become competent in generating RNA-seq heat maps in one day for novices without prior R experience. Nucl. Reprogramm. Methods Mol. Biol. 2020, in press. [Google Scholar]



© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, K. A PIANO (Proper, Insufficient, Aberrant, and NO Reprogramming) Response to the Yamanaka Factors in the Initial Stages of Human iPSC Reprogramming. Int. J. Mol. Sci. 2020, 21, 3229. https://doi.org/10.3390/ijms21093229
Hu K. A PIANO (Proper, Insufficient, Aberrant, and NO Reprogramming) Response to the Yamanaka Factors in the Initial Stages of Human iPSC Reprogramming. International Journal of Molecular Sciences. 2020; 21(9):3229. https://doi.org/10.3390/ijms21093229
Chicago/Turabian StyleHu, Kejin. 2020. "A PIANO (Proper, Insufficient, Aberrant, and NO Reprogramming) Response to the Yamanaka Factors in the Initial Stages of Human iPSC Reprogramming" International Journal of Molecular Sciences 21, no. 9: 3229. https://doi.org/10.3390/ijms21093229



