Regulation of Sulfur Homeostasis in Mycorrhizal Maize Plants Grown in a Fe-Limited Environment
Abstract
:1. Introduction
2. Results
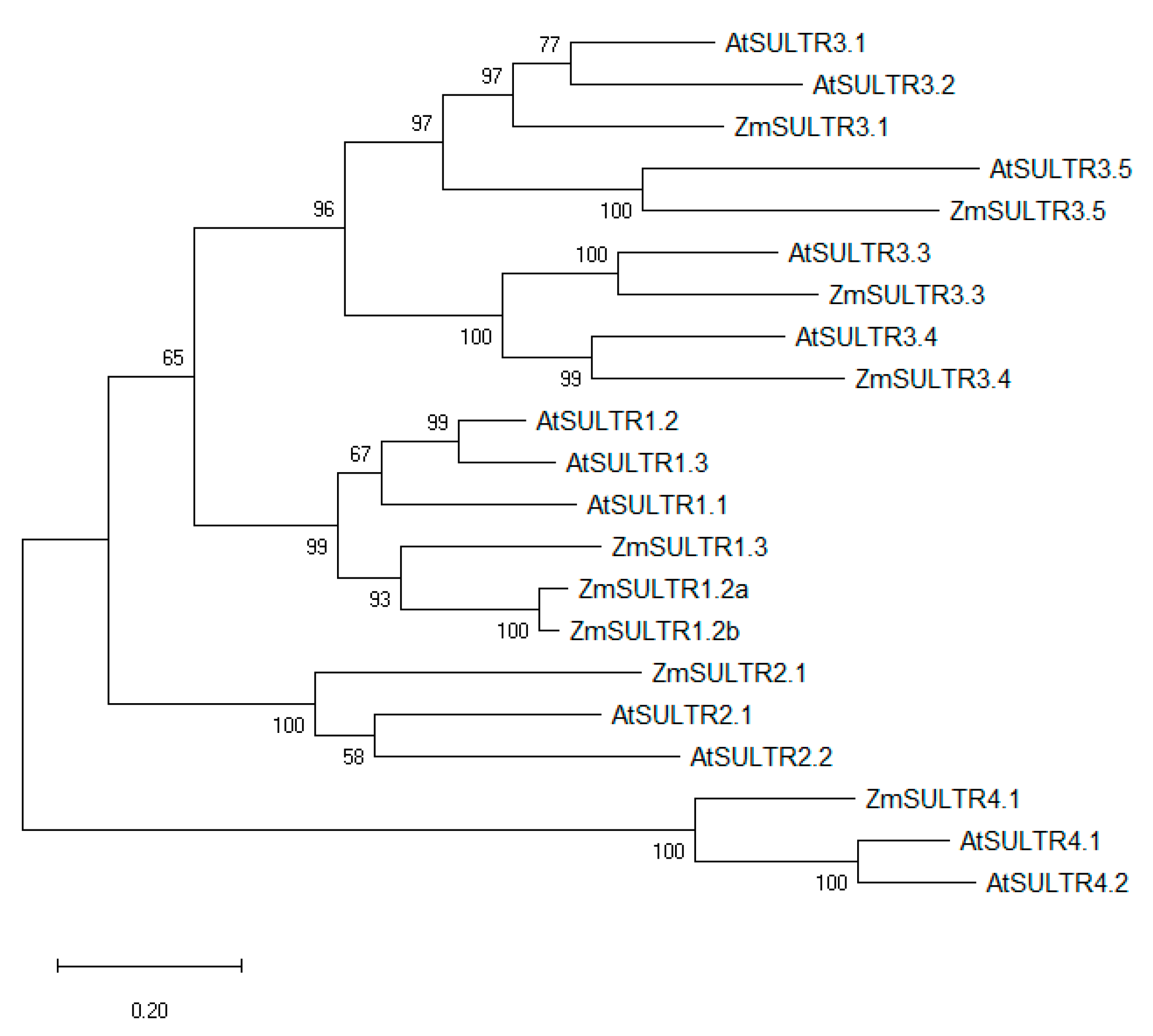
2.1. In Silico Analysis
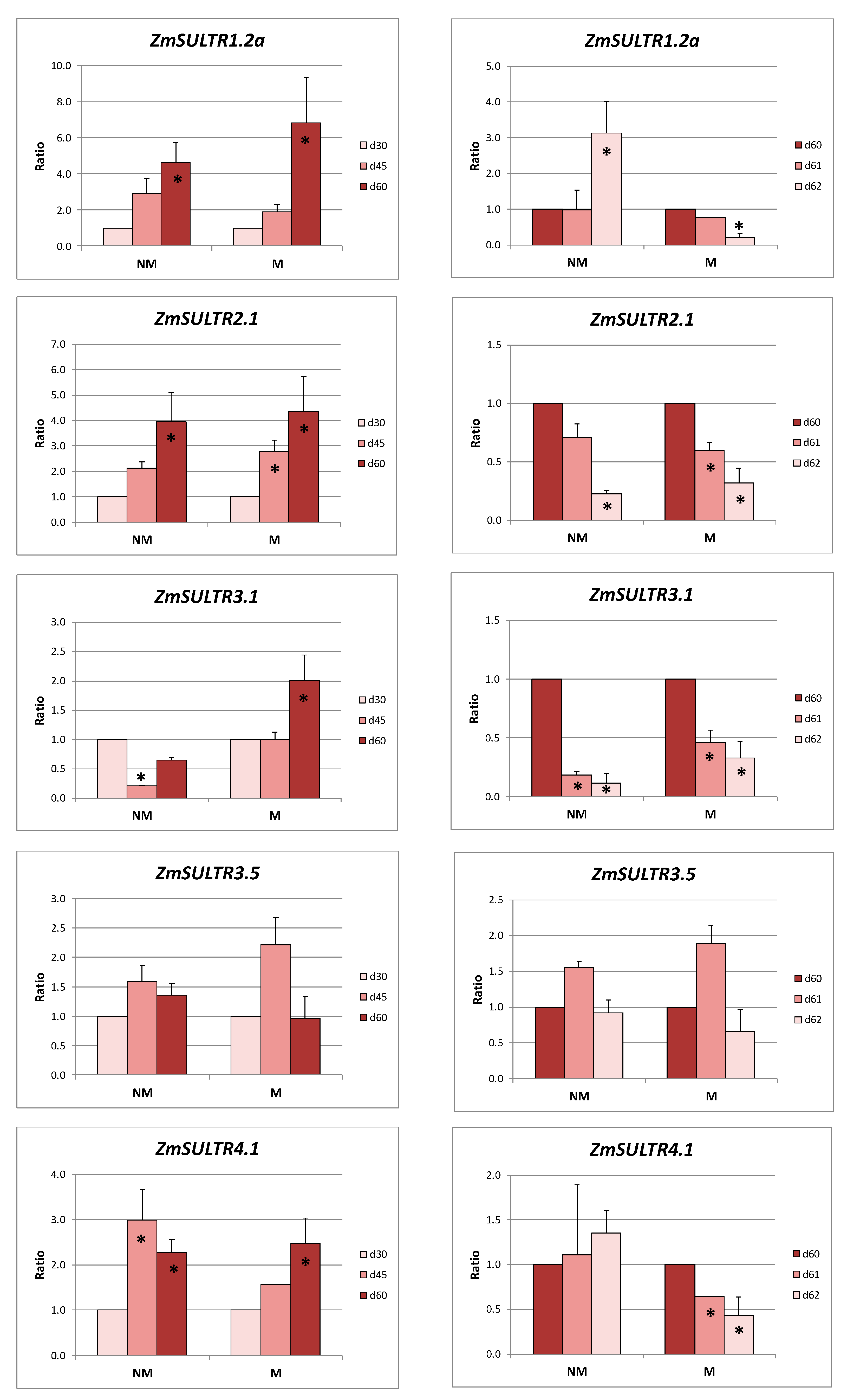
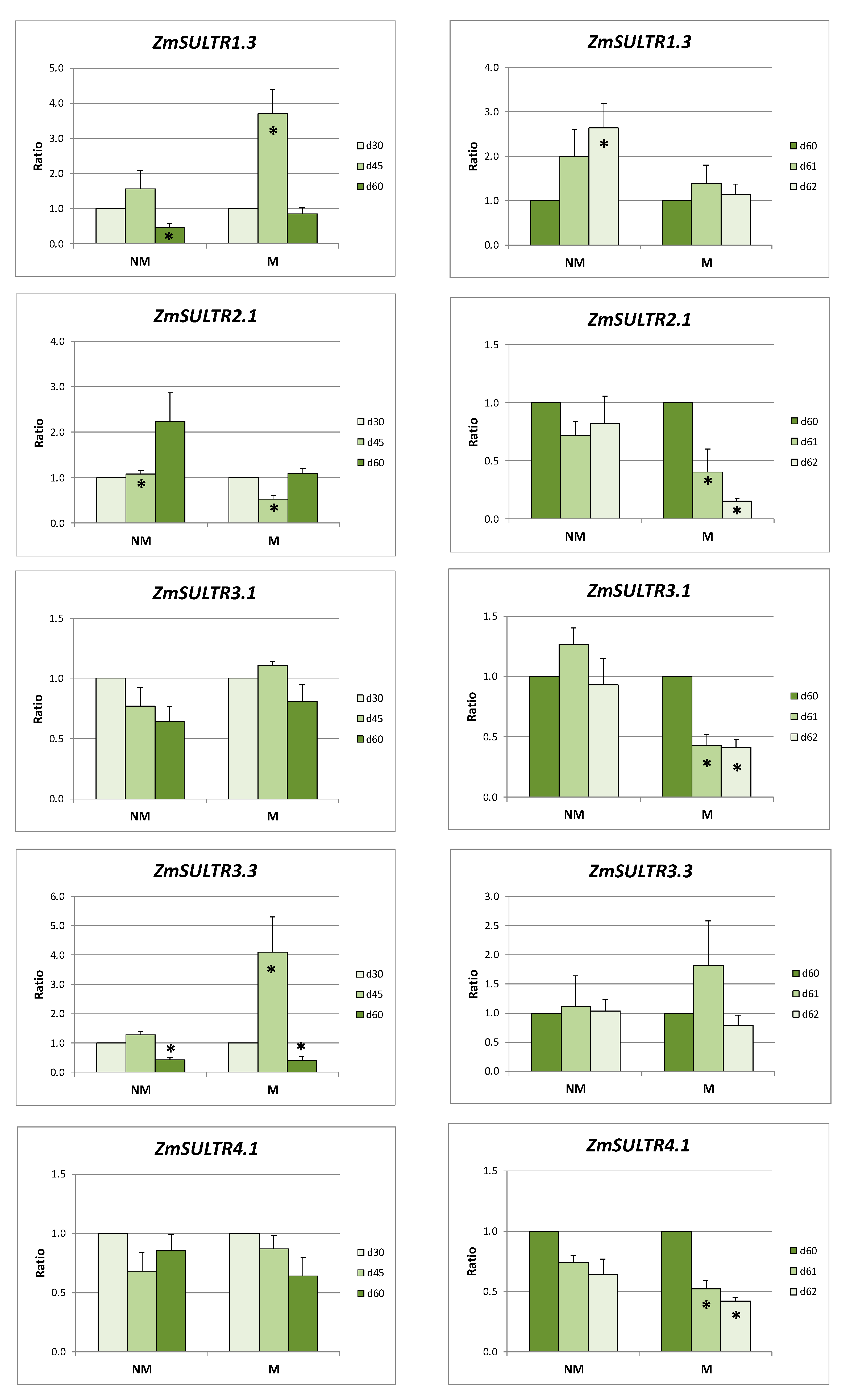
2.2. Sulfate Uptake and Transport in Roots
2.3. Sulfate Transport in Leaves
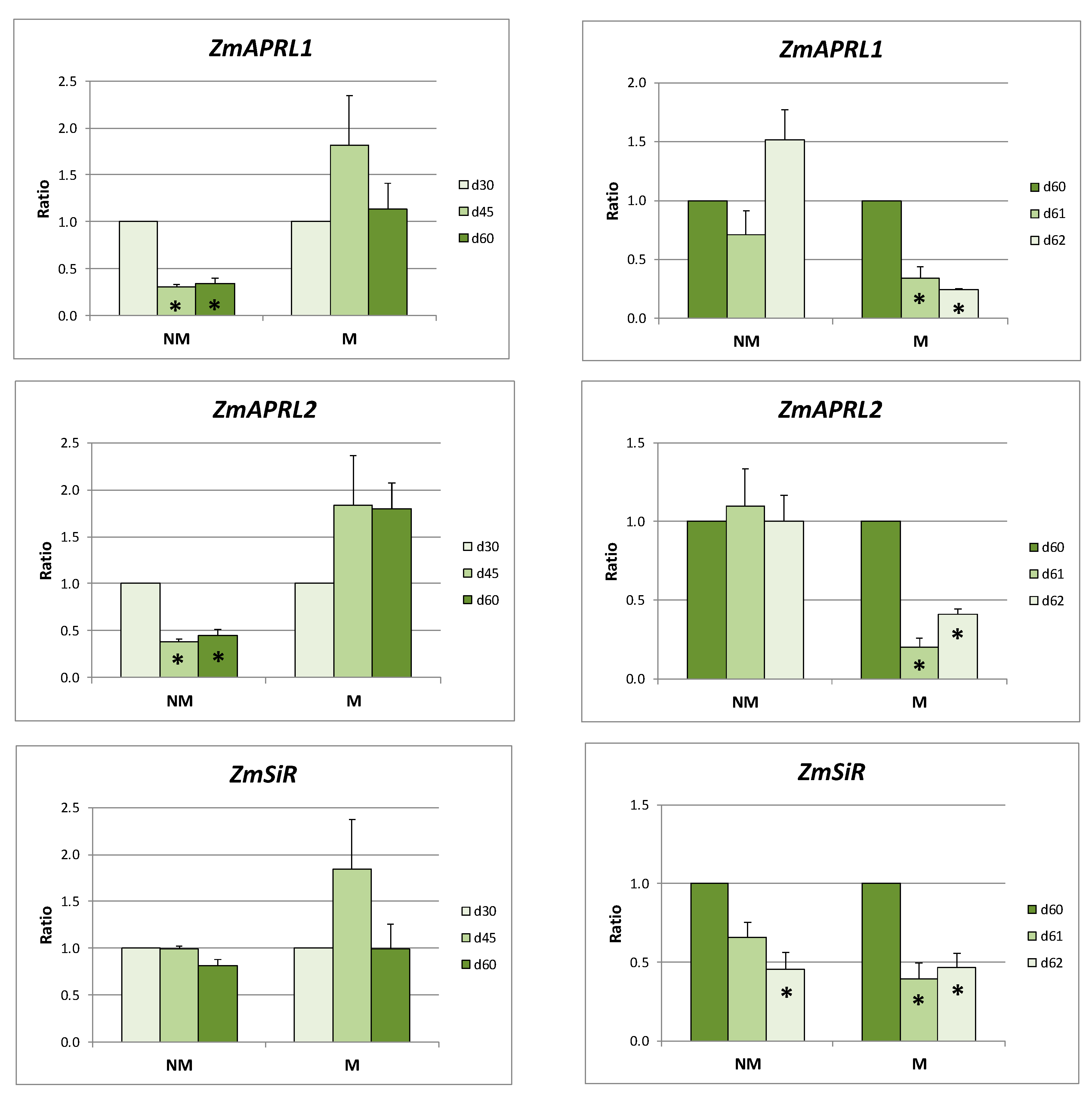
2.4. Sulfate Assimilation in Leaves and Roots
3. Discussion
3.1. Sulfate Uptake from the Rhizosphere
3.2. Sulfate Distribution in Tissues and Organs
3.3. Intracellular Transport of Sulfate
3.4. Sulfate Assimilation
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Plant Samplings
4.3. In Silico and Gene Expression Analysis
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NM | Non-mycorrhizal |
| M | Mycorrhizal |
| SULTR | Sulfate Transporter |
| APS | Adenosine 5’-phosphosulfate |
| APR | APS reductase |
| SiR | Sulfite reductase |
| DMA | Deoxymugineic acid |
References
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef] [PubMed]
- Bohrer, A.S.; Takahashi, H. Compartmentalization and regulation of sulfate assimilation pathways in plants. Int. Rev. Cell Mol. Biol. 2016, 326, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Buchner, P.; Takahashi, H.; Hawkesford, M.J. Plant sulfate transporters: Co-ordination of uptake intracellular and long distance transport. J. Exp. Bot. 2004, 55, 1765–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, H.; Watanabe-Takahashi, A.; Smith, F.W.; Blake-Kalff, M.; Hawkesford, M.J.; Saito, K. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 2000, 23, 171–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidmar, J.J.; Tagmount, A.; Cathala, N.; Touraine, B.; Davidian, J.-C.E. Cloning and characterization of a root specific high-affinity sulfate transporter from Arabidopsis thaliana. FEBS Lett. 2000, 475, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, N.; Takahashi, H.; Smith, F.W.; Yamaya, T.; Saito, K. Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 2002, 29, 465–473. [Google Scholar] [CrossRef]
- Yoshimoto, N.; Inoue, E.; Watanabe-Takahashi, A.; Saito, K.; Takahashi, H. Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiol. 2007, 145, 378–388. [Google Scholar] [CrossRef] [Green Version]
- Cao, M.J.; Wang, Z.; Wirtz, M.; Hell, R.; Oliver, D.J.; Xiang, C.B. SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J. 2013, 73, 607–616. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, P.X.; Miao, Z.Q.; Qi, G.F.; Wang, Z.; Yuan, Y.; Ahmad, N.; Cao, M.J.; Hell, R.; Wirtz, M.; et al. SULTR3s function in chloroplast sulfate uptake and affect ABA biosynthesis and the stress response. Plant Physiol. 2019, 180, 593–604. [Google Scholar] [CrossRef]
- Kataoka, T.; Watanabe-Takahashi, A.; Hayashi, N.; Ohnishi, M.; Mimura, T.; Buchner, P.; Hawkesford, M.J.; Yamaya, T.; Takahashi, H. Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 2004, 16, 2693–2704. [Google Scholar] [CrossRef] [Green Version]
- Kopriva, S.; Calderwood, A.; Weckopp, S.C.; Koprivova, A. Plant sulfur and Big Data. Plant Sci. 2015, 241, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopriva, S.; Mugford, S.G.; Baraniecka, P.; Lee, B.; Matthewman, C.A.; Koprivova, A. Control of sulfur partitioning between primary and secondary metabolism in Arabidopsis. Front. Plant Sci. 2012, 3, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidian, J.C.; Kopriva, S. Regulation of sulfate uptake and assimilation—The same or not the same? Mol. Plant 2010, 3, 314–325. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Kopriva, S. Transporters in plant sulfur metabolism. Front. Plant Sci. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vauclare, P.; Kopriva, S.; Fell, D.; Suter, M.; Sticher, L.; von Ballmoos, P.; Krähenbühl, U.; den Camp, R.O.; Brunold, C. Flux control of sulphate assimilation in Arabidopsis thaliana: Adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphur-ylase to negative control by thiols. Plant J. 2002, 31, 729–740. [Google Scholar] [CrossRef]
- Lee, B.R.; Koprivova, A.; Kopriva, S. The key enzyme of sulfate assimilation, adenosine 5′-phosphosulfate reductase, is regulated by HY5 in Arabidopsis. Plant J. 2011, 67, 1042–1054. [Google Scholar] [CrossRef]
- Chorianopoulou, S.N.; Saridis, Y.I.; Dimou, M.; Katinakis, P.; Bouranis, D.L. Arbuscular mycorrhizal symbiosis alters the expression patterns of three key iron homeostasis genes, ZmNAS1, ZmNAS3, and ZmYS1, in S deprived maize plants. Front. Plant Sci. 2015, 6, 257. [Google Scholar] [CrossRef] [Green Version]
- Portwood, J.L., II; Woodhouse, M.R.; Cannon, E.K.; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Walsh, J.R.; Sen, T.Z.; Cho, K.T.; Schott, D.A.; et al. MaizeGDB 2018: The maize multi-genome genetics and genomics database. Nucleic Acids Res. 2019, 47, D1146–D1154. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Nakamura, Y.; Watanabe-Takahashi, A.; Inoue, E.; Yamaya, T.; Takahashi, H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005, 42, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Rouached, H.; Wirtz, M.; Alary, R.; Hell, R.; Arpat, A.B.; Davidian, J.-C.; Fourcroy, P.; Berthomieu, P. Differential regulation of the expression of two high-affinity sulfate transporters, SULTR1.1 and SULTR1.2, in Arabidopsis. Plant Physiol. 2008, 147, 897–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saridis, G.; Chorianopoulou, S.N.; Ventouris, Y.E.; Sigalas, P.P.; Bouranis, D.L. An exploration of the roles of ferric iron chelation-strategy components in the leaves and roots of maize plants. Plants 2019, 8, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimoto, N.; Inoue, E.; Saito, K.; Yamaya, T.; Takahashi, H. Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis plant physiology. Plant Physiol. 2003, 131, 1511–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]

| Gene | Maize Genome Database Gene ID | Chromosome | |
|---|---|---|---|
| B73 RefGen_v3 | Zm-B73-REFERENCE-GRAMENE-4.0 | ||
| ZmSULTR1.2a | GRMZM2G159632 | Zm00001d028162 | 1 |
| ZmSULTR1.2b | GRMZM2G342907 | Zm00001d048189 | 9 |
| ZmSULTR1.3 | GRMZM2G080178 | Zm00001d050283 | 4 |
| ZmSULTR2.1 | GRMZM2G042171 | Zm00001d028164 | 1 |
| ZmSULTR3.1 | GRMZM2G154211 | Zm00001d027749 | 1 |
| ZmSULTR3.3 | GRMZM2G395114 | Zm00001d002038 | 2 |
| ZmSULTR3.4 | GRMZM2G444801 | Zm00001d000204 | 9 |
| ZmSULTR3.5 | GRMZM2G158013 | Zm00001d043614 | 3 |
| ZmSULTR4.1 | GRMZM2G068212 | Zm00001d005257 | 2 |
| ZmAPRL1 | AC189750.4_FG004 | Zm00001d021596 | 7 |
| ZmAPRL2 | GRMZM2G087254 | Zm00001d006467 | 2 |
| ZmSiR | GRMZM2G090338 | Zm00001d038625 | 6 |
| Days after Sowing | ||||||
|---|---|---|---|---|---|---|
| Gene | 30 | 45 | 60 | 61 | 62 | |
| Leaves | ZmSiR | 0.89 ± 0.15 | 1.49 ± 0.31 | 1.35 ± 0.26 | 0.85 ± 0.11 | 1.52 ± 0.30 |
| ZmAPRL2 | 0.70 ± 0.10 | 1.87 ± 0.18 | 2.48 ± 0.58 | 1.04 ± 0.21 | 1.07 ± 0.17 | |
| ZmAPRL1 | 0.82 ± 0.24 | 2.30 ± 0.42 | 2.22 ± 0.39 | 1.33 ± 0.28 | 0.38 ± 0.09 | |
| ZmSULTR3.3 | 1.01 ± 0.17 | 3.02 ± 0.33 | 0.91 ± 0.13 | 1.27 ± 0.35 | 0.44 ± 0.11 | |
| ZmSULTR3.1 | 1.19 ± 0.31 | 1.81 ± 0.14 | 1.62 ± 0.57 | 0.90 ± 0.31 | 1.07 ± 0.19 | |
| ZmSULTR4.1 | 1.89 ± 0.22 | 2.28 ± 0.38 | 1.34 ± 0.27 | 1.13 ± 0.14 | 1.00 ± 0.13 | |
| ZmSULTR2.1 | 2.84 ± 0.47 | 1.19 ± 0.29 | 2.24 ± 0.36 | 2.27 ± 0.76 | 0.64 ± 0.08 | |
| ZmSULTR1.3 | 1.30 ± 0.25 | 2.74 ± 0.16 | 1.97 ± 0.24 | 1.22 ± 0.20 | 0.68 ± 0.15 | |
| Roots | ZmSiR | 1.39 ± 0.29 | 1.79 ± 0.14 | 1.89 ± 0.22 | 3.21 ± 0.84 | 0.33 ± 0.07 |
| ZmAPRL2 | 0.65 ± 0.11 | 1.18 ± 0.30 | 2.97 ± 0.28 | 1.21 ± 0.23 | 0.56 ± 0.14 | |
| ZmAPRL1 | 0.70 ± 0.19 | 1.54 ± 0.41 | 3.40 ± 0.49 | 0.80 ± 0.25 | 0.54 ± 0.19 | |
| ZmSULTR3.6 | 1.54 ± 0.32 | 1.00 ± 0.16 | 0.53 ± 0.18 | 0.64 ± 0.15 | 0.43 ± 0.12 | |
| ZmSULTR3.1 | 0.45 ± 0.21 | 2.49 ± 0.67 | 1.85 ± 0.32 | 1.82 ± 0.17 | 4.07 ± 1.24 | |
| ZmSULTR4.1 | 1.97 ± 0.58 | 1.35 ± 0.37 | 1.68 ± 0.13 | 34.80 ± 4.90 | 0.40 ± 0.08 | |
| ZmSULTR2.1 | 1.00 ± 0.16 | 3.51 ± 0.96 | 1.94 ± 0.33 | 0.66 ± 0.11 | 1.26 ± 0.14 | |
| ZmSULTR1.2a | 1.00 ± 0.26 | 1.12 ± 0.21 | 3.17 ± 0.74 | 1.21 ± 0.30 | 0.09 ± 0.02 | |
| Primer Sequences Used in RT-PCR (5’ to 3’) | ||
|---|---|---|
| Gene | Forward | Reverse |
| ZmSULTR1.2a | AAGATATTCCTCACGGTCGGC | AGACTTCTGGCTCGTACTG |
| ZmSULTR1.3 | CTCCAAAAGCGAGGCATTCAG | GCGACCGTGAGGAAGATATGAC |
| ZmSULTR2.1 | CGGGATGGAAGGCAGTTCA | TCCACCGCTTCACCTACTGT |
| ZmSULTR3.1 | CCGGTCTGACGTAACCAGTC | AATTCGTTTTTCCCGCGACG |
| ZmSULTR3.3 | ATGCAGATCACAAGGCCGAA | TGGCGAAGTTTATCGGTGCT |
| ZmSULTR3.5 | CAGGCAGGAAAGCAAGCATC | GGAGAGACCAGATGCCACAC |
| ZmSULTR4.1 | CGTGTTGACCGATTCTGTTTCG | AAGGTTTTGCCGGAGCAGT |
| ZmAPRL1 | GACACAGGAAGGAACGAGGG | CTCAGCTACCAAGCACAGGG |
| ZmAPRL2 | CCATCTAGGTGCGCTGTACG | AAATCTCTCCCTCAGCTGCC |
| ZmSiR | GCTGAACGGGGGATCTTACC | CTAGCGCATCCATTAGGGCA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chorianopoulou, S.N.; Sigalas, P.P.; Tsoutsoura, N.; Apodiakou, A.; Saridis, G.; Ventouris, Y.E.; Bouranis, D.L. Regulation of Sulfur Homeostasis in Mycorrhizal Maize Plants Grown in a Fe-Limited Environment. Int. J. Mol. Sci. 2020, 21, 3249. https://doi.org/10.3390/ijms21093249
Chorianopoulou SN, Sigalas PP, Tsoutsoura N, Apodiakou A, Saridis G, Ventouris YE, Bouranis DL. Regulation of Sulfur Homeostasis in Mycorrhizal Maize Plants Grown in a Fe-Limited Environment. International Journal of Molecular Sciences. 2020; 21(9):3249. https://doi.org/10.3390/ijms21093249
Chicago/Turabian StyleChorianopoulou, Styliani N., Petros P. Sigalas, Niki Tsoutsoura, Anastasia Apodiakou, Georgios Saridis, Yannis E. Ventouris, and Dimitris L. Bouranis. 2020. "Regulation of Sulfur Homeostasis in Mycorrhizal Maize Plants Grown in a Fe-Limited Environment" International Journal of Molecular Sciences 21, no. 9: 3249. https://doi.org/10.3390/ijms21093249
APA StyleChorianopoulou, S. N., Sigalas, P. P., Tsoutsoura, N., Apodiakou, A., Saridis, G., Ventouris, Y. E., & Bouranis, D. L. (2020). Regulation of Sulfur Homeostasis in Mycorrhizal Maize Plants Grown in a Fe-Limited Environment. International Journal of Molecular Sciences, 21(9), 3249. https://doi.org/10.3390/ijms21093249







