Abstract
Colorectal cancer (CRC) is a malignant disease with an incidence of over 1.8 million new cases per year worldwide. CRC outcome is closely related to the respective stage of CRC and is more favorable at less advanced stages. Detection of early colorectal adenomas is the key to survival. In spite of implemented screening programs showing efficiency in the detection of early precancerous lesions and CRC in asymptomatic patients, a significant number of patients are still diagnosed in advanced stages. Research on CRC accomplished during the last decade has improved our understanding of the etiology and development of colorectal adenomas and revealed weaknesses in the general approach to their detection and elimination. Recent studies seek to find a reliable non-invasive biomarker detectable even in the blood. New candidate biomarkers could be selected on the basis of so-called liquid biopsy, such as long non-coding RNA, microRNA, circulating cell-free DNA, circulating tumor cells, and inflammatory factors released from the adenoma into circulation. In this work, we focused on both genetic and epigenetic changes associated with the development of colorectal adenomas into colorectal carcinoma and we also discuss new possible biomarkers that are detectable even in adenomas prior to cancer development.
1. Introduction
Colorectal cancer (CRC) is a serious heterogeneous disease that stands in third place in cancer incidence and represents the second cause of death in the world (nearly1.8 million patients newly diagnosed and 1 million patients who die every year) [1]. CRC has become predominant cancer in Western countries, which could be partially explained by the aged population and adverse lifestyle habits such as smoking, increased consumption of red meat and alcohol, lack of physical activity related to obesity, and diabetes, connected usually with low diversity of intestine microflora. Risk factors also include positive family history reflecting individual genetic equipment [2]. The screening programs aim to identify patients with precancerous lesions or those with resectable CRC stages 0, I, and II, who have a generally better prognosis than symptomatic patients with the pre-existing disease [3]. Despite screening programs, many patients are diagnosed in the stages III and IV of CRC that lead to a worse overall prognosis [4]. According to data from National Cancer Institute from the United States of America (USA) [5], five years survival rates for stage IV account only for 12% at colon cancer (CC) and 13% at rectal cancer (RC), while detection at an early stage I can increase the chance to survive up to 92% at CC and 88% at RC [6].
Several screening methods such as stool testing, blood testing, and endoscopic and radiological examination are currently available [7,8]. Polyps or tumors can manifest by microscopic bleeding (so-called occult bleeding). First-line test detecting occult bleeding is a fecal occult blood test (FOBT). Guaiac fecal occult blood test (gFOBT) detects hemoglobin (Hb) by peroxidase activity. Nevertheless, gFOBT is insufficiently specific to human hemoglobin and connected with a risk of false-positive results and omission of small polyps or non-bleeding polyps [7]. Despite its disadvantages, it has been able to contribute to a 33% reduction in CRC mortality [9]. Nowadays, gFOBT has predominantly been replaced by a fecal immunochemical test (FIT) based on antibody assay, which provides qualitative and quantitate results on Hb concentration per gram feces. Positive results of gFOBT or FIT are followed by endoscopic examination [8].
Endoscopic methods include colonoscopy examination, sigmoidoscopy, or capsule endoscopy. Colonoscopy is the main investigative method of the large bowel, this technique provides visualization of the entire large intestine, precise localization, biopsy, or complete removal of a potential precancerous lesion in a single session [10]. Early polypectomy leads to a 76–90% reduction in CRC incidence [11]. The weakness of this method is its invasiveness, it is an unpleasant procedure requiring several days of diet restriction and bowel preparation. These could pose an obstacle for many people, and among other things, it is expensive with the necessary presence of a very well-trained examiner [12].
Sigmoidoscopy compared to colonoscopy reduces time-consuming examination and patient discomfort and provides a lower risk of complications without the need for sedation, but allows investigation of only the rectum and the sigmoid. The study of sigmoidoscopy screening of individuals between 55 and 64 years in the United Kingdom (UK), indicated subsequent CRC incidence reduction by 33% and mortality by 43% [13].
Colon capsule endoscopy (CCE) is a non-invasive method suitable for individuals who are unwilling to undergo colonoscopy because of discomfort or any other obstacles. Meta-analysis showed that CCE for any polyp has a specificity of 89% and sensitivity of 73%. Though CCE is not as accurate as colonoscopy, it could decrease the need for its application [14].
CRC screening by radiology using computed tomographic (CT) colonography is able to visualize the entire colorectum and with no need for sedation. Even though it still requires bowel preparation, it is a relatively non-invasive method. This technique can detect only large adenomas and tumors with size ≥10 mm, nevertheless with sensitivity of 90% [15].
Screening methods based on blood testing were enriched by a highly promising biomarker, methylated gene septin9 (mSEPT9) in the last few years. mSEPT9 is released from CRC cells into circulation and is detectable in peripheral blood. A recent study showed that mSEPT9 assay, approved by Food and Drug Administration (FDA) in the USA, has a higher specificity (94.5%) than FOBT at advanced stages of CRC, but not at asymptomatic patients with early neoplasia [16,17].
Several types of a lesion can be histologically described from the colonoscopy biopsy. A colon polyp is a small clump of cells that forms on the lining of the colon epithelium. There are two main classes of polyps, non-neoplastic and neoplastic (Table 1) [18]. In general, the larger the neoplastic adenoma the greater the risk of cancer. Table 2 shows the recommended follow up after patient polypectomy [19,20,21]. Although the recommended surveillance guideline has been widely accepted, clinicians still detect the incidence of CRC (<10%) developed during the initial colonoscopy and the subsequent follow-up examination. This subgroup of CRCs is referred to as interval CRC (I-CRC) and represents one of the problems that screening programs face [22].

Table 1.
Classification of non-neoplastic and neoplastic polyps and polyposis [18].

Table 2.
Current surveillance recommendation [20,30].
Around 5–10% of CRC cases are related to heredity including most common syndromes such as hereditary non-polyposis colorectal cancer (HNPCC), familial adenomatous polyposis (FAP) and attenuated familial adenomatous polyposis (aFAP), MUTYH-associated polyposis (MAP), Juvenile polyposis syndrome (JPS), Peutz-Jeghers syndrome (PJS), Polymerase proofreading-associated polyposis (PPAP), PTEN hamartoma tumors syndrome (PHTS), Cowden syndrome, and Familial colorectal cancer type X, while more than 90% of CRC cases are of sporadic origin [6,7]. Syndromes are usually detected at an early age. However, sporadic CRC correlates with increasing age due to the accumulation of mutations in intestine cells [23,24].
In the study by Brenner et al. [25], 10 years of cumulative risk of CRC among both sex with advanced adenomas increases from 25.4–25.2% at age 55 years to 42.9–39.7% at age 80 years. The development of carcinoma from adenoma tissue can last 5 to 20 years, and it is not influenced purely by one pathway [26,27]. This transition is a complex, multifactorial process that has been characterized by chromosomal instability (CIN), microsatellite instability (MSI), and DNA methylation in CpG islands areas (CIMP). All these pathways may overlap with each other and are responsible for genetic instability in adenoma that could undergo malignant transformation [28] (Figure 1). The events contributing to these processes are constantly subject to intensive investigations [27].
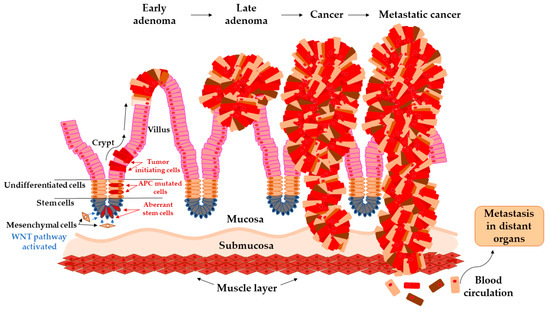
Figure 1.
Arise of tumor-initiating cells from aberrant colon crypt and subsequent transition of early adenoma to metastatic cancer.
Considering the current knowledge about the CRC development and with an application of screening programs, we are still missing identification of patients with asymptomatic disease progression in early stages, where detection plays a key role in cancer survival. Recent studies seek to find new non-invasive biomarkers measurable even in early stages of CRC from an area of non-coding RNA, inflammatory biomarkers, or cell-free DNA [29].
Transition of Adenoma to Carcinoma in Colon
The colon epithelium is constantly and rapidly renewing tissue. Old cells on the top of the villus are released into the lumen and replaced with new cells raised from colonic crypts. On the bottom of colonic crypts are stem cells that proliferate and differentiate into the cellular compartment of colon epithelium [31]. Vogelstein et al. [32] proposed the classical model of tumor evolution in the large bowel (Figure 1). Cells with high WNT signaling activity arise from aberrant crypts and evolve into a tubular or tubule-villous polyp. The subsequent proliferation of polyp may lead to the development of early adenoma with a low grade of dysplasia. Early adenoma expanses into advanced adenoma with a high grade of dysplasia and with increasing accumulation of mutations in daughter cells progressing ultimately further into carcinoma [2,32,33].
Each mutation that provides tumor cell-selective growth advantage is called driver mutation. This advantage slightly increases the growth rate of clonal expansion around 0.4% and is increasing with every new driving mutation [34]. Driver mutations enhance the accumulation of a large number of somatic mutations due to altering the cell condition and reduce the population fitness landscape. The predominant mutations, so-called passenger mutations, are mutations without selective growth advantage. With each clonal expansion of cancer cells, heterogeneous passenger mutations are generated that constitute the enormous variations of unique tumors [35].
Thanks to the next-generation sequencing (NGS) technique, thousands of mutations in the human genome were identified and some of them contribute to malignant evolution [36]. The driver mutations in the APC gene, predominantly frameshift at codon 1,554 [37], provide cell-selective growth advantage [32], and cause loss of cell ability to control the concentration level of protein β-catenin in the cytoplasm. β-catenin implements in the WNT signaling pathway and its concentration imbalances lead to uncontrolled growth and cell division [38]. Following mutations in TP53 or SMAD4 genes induce transformation into a malignant tumor, which overgrows into basal tissue and has an ability to metastasize into lymph nodes and distant organs [27].
2. Genetic Changes in Adenoma
The evolution of adenoma to carcinoma contains a wide range of genetic and epigenetic alterations. Here, we described the most relevant genetic changes associated with precancerous stages of colorectal adenoma.
2.1. Chromosomal Instability (CIN)
Chromosomal instability is associated with about 70% of sporadic CRC cases and is caused by aberrant segregation during mitoses, breaks in DNA due to nucleotide excision repair genes (NER) deficiency, or fusion of telomeres.
Chromosomal rearrangement could be classified as a numerical CIN, involving gains or losses of whole chromosomes, or it could be described as a structural CIN involving translocations, inversions, amplifications, or deletions certain parts of chromosomes [39]. CIN acts as a cancer driver by changing the copy number of large gene cohorts within tumor suppressor genes, oncogenes, DNA repair genes, and apoptotic genes [40]. Besides, the loss of one of the parental alleles during mitosis has a consequence in the loss of heterozygosity (LOH) [41].
In the study by Hermsen et al. chromosomal aberrations of 66 non-progressed colorectal adenomas, 46 progressed adenomas, and 36 colorectal carcinomas were analyzed by comparative genomic hybridization (CGH) method [42]. Authors observed that even in small adenomas a certain degree of CIN was found, independent of the degree of dysplasia. In particular, losses of chromosomal regions were observed in small non-progressed adenomas while in progressed adenomas predominantly gains of chromosomal regions and increased CIN were detected. The higher accumulation of losses at 8p21-pter, 15q11-q21, 17p12-13,18q12-21 and gains at 8q23-qter, 13q14-3, 20q13 chromosomes correlated with tumor progression [42]. Further, the most common losses were found at 1p, 4, 8p, 14, 15, 17p, 18, and most common gains at chromosomes 7, 8q, 13, 20 [43].
2.2. Microsatellite Instability (MSI)
MSI is defined as the change in microsatellite length, caused by the insertion or deletion of repetitive sequences in a tumor compared to the length of microsatellite in non-malignant tissue in the same individual. MSI is caused by a deficiency of the DNA repair mechanism, particularly the mismatch repair pathway (MMR). Under normal physiological conditions, the role of MMR is to correct DNA errors formed during the replication process, however deactivating some MMR genes (e.g., MLH1, MSH2, MSH6, or PMS1, and PMS2) results in MSI [44].
Originally, MSI has been reported to be associated with germline mutation of MMR genes in hereditary non-polyposis colorectal syndromes (HNPCC) known as a Lynch syndrome (LS). Sporadic tumors with MSI, commonly caused by biallelic promoter hypermethylation of the MHL1 gene, has a tendency to arise in the proximal colon. The frequency of the MSI is 80% to 95% in HNPCC cancers and 10% to 15% in sporadic CRC [45,46].
HNPCC tumors demonstrate high MSI, especially those located in the proximal colon, and the presence of tumor-infiltrating lymphocytes, nevertheless associated with better prognosis [47].
2.3. DNA Methylation in CpG Islands
DNA methylation, an important epigenetic modification, is closely related to the occurrence and development of tumors [48] and takes place at the 5-position of the pyrimidine ring of the cytosine residues within CpG sites to form 5-methylcytosines. CpG islands, 0.5- to 2-kb regions rich in cytosine-guanine dinucleotides, are present in approximately half of all human genes; comprising about 30,000 CpG islands in the human genome. The presence of multiple methylated CpG sites in CpG islands leads to the stable silencing of gene expression [49,50,51]. The CpG island methylator phenotype (CIMP) was firstly introduced in CRC by Toyota et al. [52] as a mechanism of CRC development. CIMP-positive CRC is characterized by a high degree of methylation in multiple CpG islands of genes associated with CRC, such as tumor suppressor genes MLH1, MGMT, and p16 [52,53].
CIMP level corresponds to the histological stage of dysplasia. In the study of Rashid [54], the methylation status of p16, MINT2, and MINT31 was determined and the methylation of these loci was present at 41% tubular adenomas and at 73% of tubulo-villous or villous adenomas. Interestingly, tubulo-villous and villous types of adenomas were more frequently found to evolve into invasive carcinomas. K-ras mutation was also observed in larger adenomas associated with higher CIMP [53,54], and this mutation also occurred at sessile serrated adenomas with significantly frequent methylation of MINT1 and MINT2 genes [55,56].
Although it is well known that hypermethylation of MHL1 gene leads to MSI, these two pathways, CIMP and MSI, are shown to be independent ways. It has been suggested that hypermethylation of MHL1 gene is a consequence in late stages, which was supported by the presence of previous mutations or allelic loss [54,57].
3. Insight into Novel Candidate Biomarkers of CRC
Research on CRC reported during the last decade has improved our understanding of the etiology and development of colorectal neoplasia and revealed weaknesses in the general approach to their detection and elimination.
The aims of recent studies on early CRC detection are oriented on the identification of new biomarkers and a more comprehensive understanding of existing biomarkers. A significant role in this field is played by non-coding RNA (long and small non-coding RNAs), cell-free DNA, circulating tumor cells, and inflammatory agents or length of telomeres. Here, we present the most promising biomarkers, which could serve as a diagnostic tool.
3.1. Long Non-Coding RNAs (lncRNAs)
Only 1–2% of the human genome encodes proteins whereas 70–90% is transcribed into non-coding RNA (ncRNA) [58,59]. The role of non-coding RNAs in organisms comprises numerous biological functions such as RNA splicing, regulation of transcription and translation, epigenetic modification, cell metabolism, interaction with RNA, DNA, and proteins [60]. Recently, it has been documented their involvement in a several diseases, especially in cancer. LncRNAs, transcripts longer than 200 nucleotides, can act as oncogenes or tumor suppressor genes in CRC and are involved in all phases of cancer evolution, tumor progression including migration of cancer cells, proliferation, tumor invasion, and metastasis formation [61]. LncRNAs can be detected not only in target tissue but also in peripheral blood. Therefore, these transcripts could represent promising diagnostic biomarkers for CRC and even in precancerous stages. A number of lncRNA e.g., CCAT1, CAHM, CRNDE, CRCAL1-4, H19, HOTAIR, MALAT1 was found significantly differentially expressed in carcinomas compared to adjacent colon tissue [62,63].
Colon-cancer associated transcript 1 (CCAT1) was recently found significantly up-regulated in all stages of adenoma-carcinoma cascades, in adenomatous polyps (100 - fold), in tumors (5 - fold) or in metastases (over - 100 fold) when compared to adjacent mucosa [64]. CCAT1 locus is located nearby of family MYC regulator genes, a well-known transcription factor. Observations of Xiang et al. [65] suggest that CCAT1-L lncRNA is involved in MYC regulation by intra-chromosome looping between the MYC gene promoter and distal upstream enhancer elements that regulate MYC transcription. The up-regulation of lncRNA-CCAT1 is highly abundant in the premalignant stages of CRC [60,66].
The H19, an oncofetal gene for lncRNA, is located in an imprinted region of chromosome 11, close to the telomeric region. H19 is overexpressed during the early stages of embryogenesis, downregulated after birth, and re-expressed during tumor genesis [67]. H19-lncRNA regulates gene expression of CDK4, CCND, and certain cancer-related proteins, such as RB1, and indirectly the activity of β-catenin via reduction of CDK8 expression by interacting with macroH2A [68]. H19 lncRNA is highly abundant in many tumors, including CRC [69]. Yoshimizu et al. [70] demonstrated that lack of H19-lncRNA expression may be considered as an initiating step in increasing the number of polyp appearance in APC mutated carcinogenesis mice model.
A gene locus (Chr16: hCG_1815491), named colorectal neoplasia differentially expressed (CRNDE), encodes lncRNA that is activated in early stages in colorectal neoplasia [71]. Elevated expression was detected in more than 90% of colorectal adenomas and adenocarcinomas in comparison to adjacent tissue. Moreover, transcripts of CRNDE were also found in the plasma of 13 out of 15 CRC patients [71]. A significant up-regulation of lncRNA, CRNDE-h variant transcript was found in serum exosomes of individuals with adenoma and CRC patients compared to control healthy subjects [72]. In addition, the level of CRNDE-lncRNA correlated with tumor size and advanced CRC stages and patient survival. CRNDE knockout suppressed CRC cell proliferation and supported apoptosis both in vitro and in vivo in the mouse model [73]. CRNDE-lncRNA also plays a significant role in CRC development by enhancing an activity of Ras/MAPK and WNT/β-catenin signaling pathways [74].
3.2. MicroRNAs (miRNAs)
MicroRNAs (miRNAs), class of small non-coding RNAs (with an average of 22 nucleotides in length), regulate the gene expression through RNA interference. MiRNAs may act as oncogenes or tumor suppressors, and their differential expression has been involved in many cancers, including CRC [75]. Their role in the silencing or triggering several pathways has been observed, thus contributing to the transition from normal epithelial colonic mucosa to adenoma and carcinoma [76]. To date, numerous studies have been focused on the differences in the miRNA’s expression between patients with CRC and healthy individuals. However, the investigation of precancerous adenomas is scarce 2 [77].
Differential miRNAs expression levels between several types of adenomas were described by studies shown in (Table 3).

Table 3.
Summary of studies focusing on miRNA profiles in colorectal cancer (CRC) adenomas (in chronological order).
The study by Tsikitis et al. focused on several types of adenomas characterized by histology and malignant potential. MiR-145, -143, -107a, -194, and -26a exhibited higher expression in low risk adenomas than in high risk adenomas, whereas miR-663b, -1268, -320a, -320b, and -1275 were highly expressed in high risk adenomas. Authors suggested the potential value of comprehensive miRNAs profiling to identify patients with high-risk malignant potential adenomas [78]. Kanth et al. analyzed miRNAs isolated from in formalin-fixed paraffin-embedded (FFPE) samples from 6 patients with sessile serrated polyps (SSA/Ps), hyperplastic polyps (HPs), and paired adjacent colon mucosa by small noncoding RNA sequencing. Several differentially expressed miRNAs (miR-135b, -378a, -548, -31, and -196b) were observed in SSA/Ps in comparison with HPs. The authors suggested that these miRNAs might serve as a good diagnostic biomarker of serrated polyps [79]. Aslam et al. also analyzed miRNAs isolated from FFPE samples and the expression level of miR-135b was progressively increased with the sequential progression of non-affected tissue to adenoma and carcinoma [80]. Oberg et al. identified several miRNAs (including miR-31 and -135) that evinced a significant difference in their expression profiles between adenomas and adjacent mucosa [81]. The involvement of miR-31 in transition from adenoma to carcinoma was also proposed by [82].
A study by Wang et al. focused on miRNA as biomarkers for prediction of adenoma recurrence. Patients with advanced colorectal adenomas were monitored for 22–24 months and almost 50% of them experienced adenoma recurrence. Authors identified that low expression of miR-194 can serve as a potential independent factor for adenoma recurrence. Moreover, this parameter was a better predictor than number of adenomas and adenoma size [83].
In the last decade, many studies focused on the concept of liquid biopsy with the goal to identify diagnostic and prognostic biomarkers from body fluids. As miRNAs can be secreted into the circulation, they might represent promising diagnostic candidates [51]. It has been observed that circulating miRNA can also aid to distinguish between CRC and adenoma patients (Table 3).
Ardila et al. analyzed circulating miRNAs in the serum of advanced adenomas, hyperplastic polyps and controls and found that miR-141, -143, and -200c were overexpressed in the serum of patients with adenomas compared to all the others [84].
Analysis of miRNAs in plasma of patients with adenomas was the task of the study conducted by Nagy et al. The authors identified three miRNAs (miR-31, -4506 and -452) differentially expressed in adenomas when compared with adjacent mucosa and the similar trend was also noticed in their plasma samples [85]. Four other miRNAs (miR-21, -29a, -92a and -135b) displayed significantly higher expression levels in adenomas when compared with non-affected adjacent mucosa. Alongside, patients with adenomas also evinced higher expression levels of miR-21 and -29a in their serum and exosomes than healthy individuals [86].
Besides plasma and serum, miRNAs can be also detected in the stool specimens [87]. In the study of Wu et al., authors identified miR-31 and -135b to be the most upregulated miRNAs in both CRC tissue and advanced adenomas tissue. These miRNAs were validated in stool specimens. The expression level of stool miR-135b was significantly higher in subjects with CRC compared with control individuals. However, there was no significant difference in the stool levels of miR-31. Authors repeated this analysis in patients upon removal of colorectal tumors and advanced adenomas and observed significant drop in miR-135b expression level in comparison with their level before removal [88].
3.3. Circulating Cell-Free DNA
As mentioned before, multiple genetic aberrations gradually accumulate over time, first in normal cells that develop into precursor lesions with the potential to develop into cancer. Thus, it may theoretically be possible to detect these genetic changes in plasma DNA taken from individuals with precursor lesions and monitor them over time to detect the progression. However, there are very few published studies on this issue.
One attractive way to improve adenoma detection and compliance is an analysis using cell-free DNA (cfDNA). Dying cells release their fragmented DNA into the circulation. In cancer patients, the cfDNA fraction that originates from tumor cells (circulating tumor DNA (ctDNA)) carries tumor-related alterations that can be detected using next-generation sequencing and PCR-based methodologies [102]. The cfDNA analysis, known as the liquid biopsy approach, is cost-effective, minimally invasive, and its specificity can be increased by tailoring the assay to detect tumor-specific mutations [51,103,104]. Recently, liquid biopsies have been used to detect minimal residual disease and monitor relapse after surgical resection of a localized disease [105]. The use of liquid biopsies for the detection of benign tumors has proved to be challenging [106,107]. The probability of detecting cfDNA is low in early-stage CRC [107] and many groups showed different results in terms of diagnostic value of total ctDNA levels or analysis of KRAS mutations in the plasma of patients with adenomas [105,108,109,110,111]. Encouraging studies reported an increase in total cfDNA or even detected tumor-related mutations in patients with benign adenomas [108,110].
Several parameters can influence the detection rate of benign lesions. Adenomas are typically small and do not manifest the persisted apoptosis or necrosis that is usually observed in advanced cancers. However, heterogeneity has been described in adenomas, which might affect KRAS mutation detection [112,113] suggesting that this oncogene might be subclonal and therefore inadequate for targeted cfDNA testing. Similar conclusions have been recently emerged from the study by Myint et al. [105] as authors argued that benign lesions do not release significant quantities of DNA in the circulation and are therefore unlikely to be diagnosed by liquid biopsies, at least using current technologies.
Further studies have shown even lower sensitivity. An analysis of 96 mutations in nine cancer driver genes (BRAF, CTNNB1, EGFR, FOXL2, GNAS, KRAS, NRAS, PIK3CA, and TP53) detected mutations in plasma cfDNA in 6% (12/200) of individuals undergoing colonoscopy; 42% of these individuals had polyps, and the rest had negative finding on colonoscopy [114]. KRAS mutations were detected in 33% (9/27) of individuals with CRC, 10% (3/30) of individuals with neoplastic polyps, and in 6% (2/35) of healthy individuals with no identified polyps during a colonoscopy. The same study also analyzed BRAF mutations in plasma cfDNA, and the results were similar in all three groups: 15% in those with CRC, 20% in individuals with neoplastic polyps, and 11% in healthy controls suggesting technical or biological issues. From a biological point of view, benign diseases, especially those with inflammatory background, may be associated with elevated levels of cfDNA [115]. In addition, somatic DNA mutations associated with cancer have been identified in histologically normal skin and colonic mucosa [116,117]. KRAS and APC mutations have also been identified in aberrant crypt foci in the colon which may be precursors of adenomas and CRC [118]. It further emphasizes that apparently unaffected colon mucosa may harbor cancer gene mutations and indeed KRAS mutations have been found in colonic effluent samples of patients at increased risk of CRC, however with normal finding on colonoscopy [119]. It was assumed that the source of the ctDNA could be from a neoplasm outside of the colorectal area, from apoptotic cells or destruction of precancerous cells, benign inflammatory lesions such as endometriosis, and small neoplasms with somatic DNA mutations during the normal process of immune surveillance [119,120].
In addition to KRAS and BRAF mutations, Galanopoulos et al. [109] recently studied blood samples and colonic biopsy specimens from healthy individuals with no polyps undergoing screening colonoscopy, patients with CRC, and patients with neoplastic intestinal polyps. Based on the mutation analysis for codon 12 of the KRAS, authors were able to discriminate patients with CRC compared to healthy individuals. However, with no success in predicting the presence of colonic polyps.
Kopreski et al. [110] found KRAS mutations in plasma cfDNA in 22 of the 62 patients with adenomas and in 9 out of 65 of those with hyperplastic or other non-neoplastic lesions. In prospective colonoscopy study, Perrone et al. [111] found 22 instances of high-grade intraepithelial neoplasia in adenomas (12.9%), 54 adenomas (31.8%), and 19 hyperplastic lesions (11.2%) in the 170 investigated individuals. KRAS mutations were found in the plasma of 3/19 patients with high-grade intraepithelial neoplasia (15.8%), 1/54 patients with adenomas (1.8%), and none of the patients with hyperplasia.
Gocke et al. [121] demonstrated that either of two hotspot mutations (codons 175 and 248) in TP53 was detectable in cfDNA in 1.3% (3/240) of the individuals. However, only one of these three individuals had a polyp that carried the same TP53 mutation, and thus, the origin of the other two plasma TP53 mutations could not be determined.
Mead et al. [108] analyzed diagnostic markers utilizing cfDNA isolated from samples obtained from 35 individuals without endoscopic abnormality, a group of 26 individuals with benign colorectal adenomas, and 24 patients with CRC. The best model to discriminate physiological from neoplasia populations was based on four DNA markers (Line1 79 bp, Alu 247 bp, mitochondrial DNA, and Alu 115 bp), with ROC curve of 0.810. The final test had a positive predictive value (PPV) of 81.1% for polyps and a negative predictive value (NPV) of 73.5% (sensitivity 83% and specificity 72%) for early cancer diagnosis.
CRC screening with a multitarget stool DNA test was approved by the Food and Drug Administration in 2014. This simple, noninvasive, multitarget stool DNA (mt-sDNA)-based screening test (Cologuard; Exact Sciences, Madison, WI) has much greater sensitivity for the detection of both CRC and advanced precancerous lesions than FIT. Thus was developed to improve both non in-vasive screening performance and screening compliance [122,123]. Screening study data have similarly supported a Cologuard multiyear interval with a negative predictive value of a single test even of 99.94% for CRC and 95% for advanced adenoma [124]. Colo-guard consists of quantitative molecular assays to detect aberrantly methylated DNA (NDRG4 and BMP3) and DNA mutations (KRAS) in stool plus a fecal hemoglobin immunoassay. Berger et al. suggested [122] that screening every 3 years using a multitarget mt-sDNA test provides reasonable performance at an acceptable cost.
Taken together, these results suggest that the detection of pathogenic mutations in plasma is not synonymous with precancerous lesions or cancer. Whilst many precancerous colorectal lesions are not detectable at all, some small polyps can shed detectable amounts of ctDNA in plasma. Since adenomas are potentially premalignant and should be excised, their detection through measurement of ctDNA should be useful and the finding of a positive test might increase the rate of screening colonoscopies, which suffers from poor patient compliance.
3.4. Circulating Tumor Cells (CTCs)
The process of tumor metastasis involves the release of epithelial cancer cells, called circulating tumor cells (CTCs) into the bloodstream. It has been observed that CTCs can access the circulatory system not only in metastatic stages but even at preinvasive lesions [125]. However, even in metastatic stages, the blood concentration of CTCs is extremely low and therefore the CTCs detection is difficult and much more challenging for colorectal adenomas or carcinomas in situ. Nevertheless, in terms of diagnostic value, CTCs provide an opportunity to monitor the development of cancer at all stages with a deeper understanding of tumor biology and better treatment efficiency [51].
The analysis of CTCs in the sense of liquid biopsy requires cell enrichment and CTC detection. The gold standard is represented by the CellSearch system [126], the only FDA approved method for CTC-detection today is based on immunomagnetic CTC enrichment using an antibody against the epithelial cell adhesion molecule (EpCAM) and combined with flow cytometry. EpCAM adhesion molecule is specific for epithelial cells and most carcinomas are characterized by its overexpression. Besides CTC detection by EpCAM, different assays based on physical characteristics (e.g., size, density, deformability, and electrical charge) or on more specific biological properties such as certain tumor epithelial protein (e.g., CK20, CD45) also exist [127].
In CRC, CTCs may originate not only from epithelial tumor cells, but also from tumor cells undergoing epithelial-mesenchymal transition, and tumor stem cells [128]. As mentioned before, extremely low concentration of CTCs in peripheral blood (e.g., 1-5 CTCs per 7.5 mL blood at CRC stage III) is the main obstacle in cancer progress investigation [129]. However, the presence of CTCs could serve as an indicator of metastatic spread of the disease. In a recent meta-analysis by Tan and Wu [130] that included 15 studies with 3129 CRC patients, the association between CTCs detection and poor survival outcomes for patients with CRC was proved. Another meta-analysis by Huang et al. [131], included 13 studies with eligible 2388 CRC patients observed CTCs level in peripheral blood before initiating chemotherapy and during the chemotherapy. They confirmed CTCs high level was significantly associated with poor progression-free survival and poor overall survival. Moreover, CTCs are suitable for evaluation chemosensitivity and gene expression to evaluate the current mutational status of the tumor [132]. Interesting results were shown by Guadagni et al. [133], where CTCs from 47% of CRC patients exhibited high sensitivity to mitomycin when compared to recommended chemotherapeutic for CRC.
Considering the low occurrence of CTCs in the bloodstream during CRC metastatic process, the utilization of CTCs for a screening of CRC early stages or precancerous lesions represents a very ambitious aim. The recent study of Tsai et al. [134] has accepted this challenging topic. On the day of the colonoscopy, 8 ml of peripheral blood was collected from 667 Taiwanese subjects (in detail the study consisted of 235 healthy controls, 107 subjects with adenomatous polyps, and 325 patients with CRC across all stages I-IV). All individuals were tested for CTCs presence by using the CellMax Platform (EpCAM(+), CK20(+), CD45(−) epithelial cells). Results of the study showed high specificity 86% and sensitivity 79% for adenomatous lesion and for CRC across all stages specificity 82% and sensitivity 95%. While the CTCs presence in metastatic CRC is widely accepted, CTCs detection at adenomatous polyps or in precancerous stages appears to be rather difficult. Nevertheless, CTCs detection has the potential to serve as a diagnostic tool in CRC screening.
3.5. Circulating Inflammation Markers
Inflammation belongs between one of the colorectal neoplasia drivers; however, particular inflammatory processes that play a role in early carcinogenesis are still unknown. Recently, Huang et al. [135] compared serum levels of 78 inflammation markers between 171 pathologically confirmed colorectal adenoma cases and 344 controls within the frame of Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. Their results provided important new evidence implicating C-chemokine cysteine motif chemokine ligand 20 (CCL20)—and growth-related gene oncogene products (GRO) –related pathways in early CRC, and further supported a role for insulin. The CCL20/CCR6 system also appeared to play a role in organ-selective liver metastasis of CRC. A recent meta-analysis of 4 cohorts and 10 case-control studies found no associations between adenoma and 3 most studied circulating inflammation markers CRP, IL-6, and TNF-α [136]. Similarly, no significant associations were found for C-peptide, GM-CSF, interferon-α (IFN-α), IL-1β, IL-2, IL-4, IL-6, IL-7, IL-10, IL-12, IL-17A, MIP-1β, and vascular endothelial growth factor and adenoma [137,138,139]. Although associations were reported for IFN-α2, IL-7, IL-8, MCP-3, and SIL-4R in cross-sectional analyses [137,140] they have not been prospectively confirmed.
3.6. Telomere Length
Telomeres are terminal repeated sequences at the ends of chromosomes and their shortening is associated with aging. During the cell division are telomeres shortened due to the lack of enzyme telomerase that synthesizes the ends of linear nucleic acid. Nevertheless, this enzyme is active in germ cells, stem cells, and in most of the cancer cells. In the advanced stage of CRC epithelial-mesenchymal transition occurs and telomere lengthening is maintenance by the alternative lengthening of telomeres (ALT) [141,142,143].
The latest studies reported that both short and long telomeres have been involved in carcinogenesis. The relationship between the length of telomeres in colorectal adenomas and the risk of cancer development is still subject to discussion [144]. Results of comparing the telomere length between adenomas, tumors, and adjacent mucosa are inconsistent and vary among several studies. In the study by Peacock et al. [141], telomere length of colon tissue from 40 adenomas and 45 controls identified during colonoscopy was exanimated. The result suggested that long telomeres in non-affected colon tissue are related to increased risk of CRC. Bautista et al. [145] observed higher telomerase activity in adenomas compared to their adjacent control mucosa, however shorter telomere length in adenomas in comparison to control tissue. Similar results were also obtained in the other studies [146,147], where telomere shortening in adenomas compared to adjacent mucosa was detected. The largest analysis of telomere length was done in the study of Suraweera et al. [148] where relative telomere length (RLT) was measured in 90 adenomas and adjacent normal mucosa. Adenomas showed a shortening of telomeres by 79% and lengthening only in 7% of cases.
4. Conclusions
The development of colorectal cancer is a comprehensive process of genetic, epigenetic, and structural modifications from benign adenoma to invasive cancer. Early detection and complete endoscopic removal of adenomas in their early stages is the key to survival with almost zero chance for cancer development. Nowadays, the most used method for the investigation of the large bowel is a colonoscopy. Although it is a very sensitive and reliable method, it brings considerable difficulties as a dietary restriction, invasive examination, risk of omission of some adenomas, high price, and requires a well-trained examiner. Non-invasive approaches for early adenoma detection are still evolving. The future perspectives in this area are moving towards liquid biopsy as a potential minimally invasive tool for clinical use. CfDNA, CTCs, inflammatory markers or specific RNA transcripts, such as a miR-31, miR-135, lncRNAs, released from adenoma lesions into circulation are extensively studied and have been shown as promising candidate biomarkers for early CRC. Although the concentration of cfDNA in plasma is very low, it still can provide useful information about mutations in crucial genes as a KRAS or BRAF that are involved in carcinogenesis. The early appearance of KRAS or BARF mutations in circulation even in healthy individuals warrants further investigation as a potential prognostic marker. Recently acquired knowledge about new possible biomarkers can help to better understand colorectal cancer evaluation and design its future detection strategy.
Author Contributions
Conceptualization A.S., P.V., V.V., J.K.; writing—review and editing A.S., K.C., V.V., J.K., T.H., P.V.; writing—original draft preparation A.S., K.C., V.V.; data curation and investigation A.S., K.C., V.V., J.K., T.H.; funding acquisition P.V., T.H., V.V., K.C.; project administration A.S., P.V., V.V.; supervision P.V., V.V., T.H. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the Grant Agency of the Ministry of Health of the Czech Republic (AZVNV18-03-00199), the Grant Agency of the Czech Republic (GACR18-09709S) and the Charles University Grant Agency (GAUK 302119). We are thankful to Charles University Research Centre program UNCE/MED/006 “University Centre of Clinical and Experimental Liver Surgery” and National Sustainability Program I (NPU I) Nr. LO1503 provided by the Ministry of Education Youth and Sports of the Czech Republic. This article is based upon work from COST Action CA17118, supported by COST (European Cooperation in Science and Technology). www.cost.eu.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ALT | alternative lengthening of telomeres |
| aFAP | attenuated familial adenomatous polyposis |
| CC | colon cancer |
| CCE | colon capsule endoscopy |
| CCL20 | C-chemokine cysteine motif chemokine ligand 20 |
| cfDNA | cell-free DNA |
| CGH | comparative genomic hybridization |
| CIMP | CpG island methylator phenotype |
| CIN | chromosomal instability |
| CRC | colorectal cancer |
| CTCs | circulating tumor cells |
| CT | computed tomographic |
| ctDNA | circulating tumor DNA |
| EpCAM | epithelial cell adhesion molecule |
| FAP | familial adenomatous polyposis |
| FDA | Food and Drug Administration |
| FFPE | formalin-fixed paraffin-embedded |
| FIT | fecal immunochemical test |
| FOBT | fecal occult blood test |
| gFOBT | guaiac fecal occult blood test |
| Hb | hemoglobin |
| HNPCC | hereditary non-polyposis colorectal cancer |
| HPs | hyperplastic polyps |
| JPS | Juvenile polyposis syndrome |
| LOH | loss of heterozygosity |
| LS | Lynch syndrome |
| MAP | MUTYH-associated polyposis |
| MMR | mismatch repair pathway |
| mt-sDNA | multitarget stool DNA |
| ncRNA | non-coding RNA |
| NER | nucleotide excision repair genes |
| NGS | next-generation sequencing |
| NPV | negative predictive value |
| PHTS | hamartoma tumors syndrome |
| PJS | Peutz-Jeghers syndrome |
| PPAP | Polymerase proofreading-associated polyposis |
| PPV | positive predictive value |
| RC | rectal cancer |
| RLT | relative telomere length |
| SSA/Ps | sessile serrated polyps |
| UK | United Kingdom |
| USA | United States of America |
| WNT | Wingless/Int-1 pathway |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis Primers 2015, 1, 15065. [Google Scholar] [CrossRef]
- Pande, R.; Froggatt, P.; Baragwanath, P.; Harmston, C. Survival outcome of patients with screening versus symptomatically detected colorectal cancers. Colorectal Dis. 2013, 15, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Altobelli, E.; Rapacchietta, L.; Marziliano, C.; Campagna, G.; Profeta, V.F.; Fagnano, R. Differences in colorectal cancer surveillance epidemiology and screening in the WHO European Region. Oncol. Lett. 2019, 17, 2531–2542. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute, Surveillance Research Program, Surveillance Systems Branch, Surveillance, Epidemiology, and End Results (SEER) Program. SEER*Stat Database: Incidence – SEER 18 Regs Research Data + Hurricane Katrina Impacted Louisiana Cases, Nov 2015 Sub (1973–2013 varying) – Linked To County Attributes – Total U.S., 1969–2014 Counties. 2016. Available online: https://seer.cancer.gov/data-software/documentation/seerstat/nov2016/ (accessed on 4 May 2020).
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Binefa, G.; Rodriguez-Moranta, F.; Teule, A.; Medina-Hayas, M. Colorectal cancer: From prevention to personalized medicine. World J. Gastroenterol. 2014, 20, 6786–6808. [Google Scholar] [CrossRef] [PubMed]
- Schreuders, E.H.; Grobbee, E.J.; Spaander, M.C.W.; Kuipers, E.J. Advances in Fecal Tests for Colorectal Cancer Screening. Curr. Treat. Options. Gastroenterol. 2016, 14, 52–162. [Google Scholar] [CrossRef]
- Mandel, J.S.; Bond, J.H.; Church, T.R.; Snover, D.C.; Bradley, G.M.; Schuman, L.M.; Ederer, F. Reducing Mortality from Colorectal Cancer by Screening for Fecal Occult Blood. N. Engl. J. Med. 1993, 328, 1365–1371. [Google Scholar] [CrossRef]
- Ladabaum, U.; Dominitz, J.A.; Kahi, C.; Schoen, R.E. Strategies for Colorectal Cancer Screening. Gastroenterology 2020, 158, 418–432. [Google Scholar] [CrossRef]
- Winawer, S.J.; Zauber, A.G.; Ho, M.N.; O’Brien, M.J.; Gottlieb, L.S.; Sternberg, S.S.; Waye, J.D.; Schapiro, M.; Bond, J.H.; Panish, J.F.; et al. Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N. Engl. J. Med. 1993, 329, 1977–1981. [Google Scholar] [CrossRef]
- Lhewa, D.Y.; Strate, L.L. Pros and cons of colonoscopy in management of acute lower gastrointestinal bleeding. World J. Gastroenterol. 2012, 18, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Atkin, W.S.; Edwards, R.; Kralj-Hans, I.; Wooldrage, K.; Hart, A.R.; Northover, J.M.; Parkin, D.M.; Wardle, J.; Duffy, S.W.; Cuzick, J. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: A multicentre randomised controlled trial. Lancet 2010, 375, 1624–1633. [Google Scholar] [CrossRef]
- Rokkas, T.; Papaxoinis, K.; Triantafyllou, K.; Ladas, S.D. A meta-analysis evaluating the accuracy of colon capsule endoscopy in detecting colon polyps. Gastrointest. Endosc. 2010, 71, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.D.; Chen, M.-H.; Toledano, A.Y.; Heiken, J.P.; Dachman, A.; Kuo, M.D.; Menias, C.O.; Siewert, B.; Cheema, J.I.; Obregon, R.G.; et al. Accuracy of CT Colonography for Detection of Large Adenomas and Cancers. N. Engl. J. Med. 2008, 359, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Fei, F.; Zhang, M.; Li, Y.; Zhang, X.; Zhu, S.; Zhang, S. The role of (m)SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer 2019, 19, 450. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Jia, J.; Peng, X.; Xiao, W.; Li, Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis. Sci. Rep. 2017, 7, 3032. [Google Scholar] [CrossRef]
- Rubio, C.A.; Jaramillo, E.; Lindblom, A.; Fogt, F. Classification of Colorectal Polyps: Guidelines for the Endoscopist. Endoscopy 2002, 34, 226–236. [Google Scholar] [CrossRef]
- Hassan, C.; Quintero, E.; Dumonceau, J.M.; Regula, J.; Brandao, C.; Chaussade, S.; Dekker, E.; Dinis-Ribeiro, M.; Ferlitsch, M.; Gimeno-Garcia, A.; et al. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy 2013, 45, 842–851. [Google Scholar] [CrossRef]
- Levin, B.; Lieberman, D.A.; McFarland, B.; Smith, R.A.; Brooks, D.; Andrews, K.S.; Dash, C.; Giardiello, F.M.; Glick, S.; Levin, T.R.; et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: A joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J. Clin. 2008, 58, 130–160. [Google Scholar] [CrossRef]
- Shussman, N.; Wexner, S.D. Colorectal polyps and polyposis syndromes. Gastroenterol. Rep. (Oxf) 2014, 2, 1–15. [Google Scholar] [CrossRef]
- Dong, S.-H.; Huang, J.-Q.; Chen, J.-S. Interval colorectal cancer: A challenging field in colorectal cancer. Future Oncol. 2018, 14, 1307–1316. [Google Scholar] [CrossRef]
- Whiffin, N.; Hosking, F.J.; Farrington, S.M.; Palles, C.; Dobbins, S.E.; Zgaga, L.; Lloyd, A.; Kinnersley, B.; Gorman, M.; Tenesa, A.; et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum. Mol. Genet. 2014, 23, 4729–4737. [Google Scholar] [CrossRef] [PubMed]
- Archambault, A.N.; Su, Y.R.; Jeon, J.; Thomas, M.; Lin, Y.; Conti, D.V.; Win, A.K.; Sakoda, L.C.; Lansdorp-Vogelaar, I.; Peterse, E.F.P.; et al. Cumulative Burden of Colorectal Cancer-Associated Genetic Variants Is More Strongly Associated With Early-Onset vs. Late-Onset Cancer. Gastroenterology 2020, 158, 1274–1286.e12. [Google Scholar] [CrossRef]
- Brenner, H.; Hoffmeister, M.; Stegmaier, C.; Brenner, G.; Altenhofen, L.; Haug, U. Risk of progression of advanced adenomas to colorectal cancer by age and sex: Estimates based on 840,149 screening colonoscopies. Gut 2007, 56, 1585–1589. [Google Scholar] [CrossRef] [PubMed]
- Loeve, F.; Boer, R.; Zauber, A.G.; Van Ballegooijen, M.; Van Oortmarssen, G.J.; Winawer, S.J.; Habbema, J.D. National Polyp Study data: Evidence for regression of adenomas. Int. J. Cancer 2004, 111, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Cross, W.; Kovac, M.; Mustonen, V.; Temko, D.; Davis, H.; Baker, A.M.; Biswas, S.; Arnold, R.; Chegwidden, L.; Gatenbee, C.; et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol. 2018, 2, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.F.; Ibrahim, A.E.; Arends, M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016, 469, 125–134. [Google Scholar] [CrossRef]
- Pellino, G.; Gallo, G.; Pallante, P.; Capasso, R.; De Stefano, A.; Maretto, I.; Malapelle, U.; Qiu, S.; Nikolaou, S.; Barina, A.; et al. Noninvasive Biomarkers of Colorectal Cancer: Role in Diagnosis and Personalised Treatment Perspectives. Gastroenterol. Res. Pract. 2018, 2018, 2397863. [Google Scholar] [CrossRef]
- Winawer, S.J.; Zauber, A.G.; Fletcher, R.H.; Stillman, J.S.; O’Brien, M.J.; Levin, B.; Smith, R.A.; Lieberman, D.A.; Burt, R.W.; Levin, T.R.; et al. Guidelines for colonoscopy surveillance after polypectomy: A consensus update by the US Multi-Society Task Force on Colorectal Cancer and the American Cancer Society. CA Cancer J. Clin. 2006, 56, 143–159, quiz 184-5. [Google Scholar] [CrossRef]
- Cernat, L.; Blaj, C.; Jackstadt, R.; Brandl, L.; Engel, J.; Hermeking, H.; Jung, A.; Kirchner, T.; Horst, D. Colorectal Cancers Mimic Structural Organization of Normal Colonic Crypts. PLoS ONE 2014, 9, e104284. [Google Scholar] [CrossRef]
- Kinzler, K.W.; Vogelstein, B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 1997, 386, 761–763. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, C.; Vogelstein, B.; Parmigiani, G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc. Natl. Acad. Sci. USA 2013, 110, 1999–2004. [Google Scholar] [CrossRef] [PubMed]
- Khurana, E.; Fu, Y.; Colonna, V.; Mu, X.J.; Kang, H.M.; Lappalainen, T.; Sboner, A.; Lochovsky, L.; Chen, J.; Harmanci, A.; et al. Integrative annotation of variants from 1092 humans: Application to cancer genomics. Science 2013, 342, 1235587. [Google Scholar] [CrossRef] [PubMed]
- Bozic, I.; Antal, T.; Ohtsuki, H.; Carter, H.; Kim, D.; Chen, S.; Karchin, R.; Kinzler, K.W.; Vogelstein, B.; Nowak, M.A. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 18545–18550. [Google Scholar] [CrossRef] [PubMed]
- Ciriello, G.; Miller, M.L.; Aksoy, B.A.; Senbabaoglu, Y.; Schultz, N.; Sander, C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013, 45, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Rowan, A.J.; Lamlum, H.; Ilyas, M.; Wheeler, J.; Straub, J.; Papadopoulou, A.; Bicknell, D.; Bodmer, W.F.; Tomlinson, I.P.M. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc. Natl. Acad. Sci. USA 2000, 97, 3352–3357. [Google Scholar] [CrossRef] [PubMed]
- Kwong, L.N.; Dove, W.F. APC and its modifiers in colon cancer. Adv. Exp. Med. Biol. 2009, 656, 85–106. [Google Scholar]
- Burrell, R.A.; McClelland, S.E.; Endesfelder, D.; Groth, P.; Weller, M.C.; Shaikh, N.; Domingo, E.; Kanu, N.; Dewhurst, S.M.; Gronroos, E.; et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013, 494, 492–496. [Google Scholar] [CrossRef]
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 139–150. [Google Scholar] [CrossRef]
- Tariq, K.; Ghias, K. Colorectal cancer carcinogenesis: A review of mechanisms. Cancer Bio. Med. 2016, 13, 120–135. [Google Scholar] [CrossRef]
- Hermsen, M.; Postma, C.; Baak, J.; Weiss, M.; Rapallo, A.; Sciutto, A.; Roemen, G.; Arends, J.W.; Williams, R.; Giaretti, W.; et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology 2002, 123, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Haan, J.C.; Labots, M.; Rausch, C.; Koopman, M.; Tol, J.; Mekenkamp, L.J.; van de Wiel, M.A.; Israeli, D.; van Essen, H.F.; van Grieken, N.C.; et al. Genomic landscape of metastatic colorectal cancer. Nat. Commun. 2014, 5, 5457. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef] [PubMed]
- Peltomäki, P. Deficient DNA mismatch repair: A common etiologic factor for colon cancer. Hum. Mol. Gen. 2001, 10, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Loukola, A.; Salovaara, R.; Kristo, P.; Moisio, A.L.; Kääriäinen, H.; Ahtola, H.; Eskelinen, M.; Härkönen, N.; Julkunen, R.; Kangas, E.; et al. Microsatellite instability in adenomas as a marker for hereditary nonpolyposis colorectal cancer. Am. J. Pathol. 1999, 155, 1849–1853. [Google Scholar] [CrossRef]
- Steinke, V.; Engel, C.; Buttner, R.; Schackert, H.K.; Schmiegel, W.H.; Propping, P. Hereditary nonpolyposis colorectal cancer (HNPCC)/Lynch syndrome. Dtsch. Arztebl. Int. 2013, 110, 32–38. [Google Scholar] [CrossRef]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Paabo, S.; Rebhan, M.; Schubeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef]
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of Modified DNA Bases: 5-Methylcytosine and Beyond. Front. Genet. 2018, 9, 640. [Google Scholar] [CrossRef]
- Marcuello, M.; Vymetalkova, V.; Neves, R.P.L.; Duran-Sanchon, S.; Vedeld, H.M.; Tham, E.; van Dalum, G.; Flugen, G.; Garcia-Barberan, V.; Fijneman, R.J.; et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Aspects Med. 2019, 69, 107–122. [Google Scholar] [CrossRef]
- Toyota, M.; Ahuja, N.; Ohe-Toyota, M.; Herman, J.G.; Baylin, S.B.; Issa, J.-P.J. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA 1999, 96, 8681–8686. [Google Scholar] [CrossRef] [PubMed]
- Psofaki, V.; Kalogera, C.; Tzambouras, N.; Stephanou, D.; Tsianos, E.; Seferiadis, K.; Kolios, G. Promoter methylation status of hMLH1, MGMT, and CDKN2A/p16 in colorectal adenomas. World J. Gastroenterol. 2010, 16, 3553–3560. [Google Scholar] [CrossRef] [PubMed]
- Rashid, A.; Shen, L.; Morris, J.S.; Issa, J.-P.J.; Hamilton, S.R. CpG Island Methylation in Colorectal Adenomas. Am. J. Pathol. 2001, 159, 1129–1135. [Google Scholar] [CrossRef]
- Park, S.-J.; Rashid, A.; Lee, J.-H.; Kim, S.G.; Hamilton, S.R.; Wu, T.-T. Frequent CpG Island Methylation in Serrated Adenomas of the Colorectum. Am. J. Pathol. 2003, 162, 815–822. [Google Scholar] [CrossRef]
- Karen Curtin, M.L.S.; Wade, S. Samowitz CpG Island Methylation in Colorectal Cancer: Past, Present and Future. Pathol. Res. Inter. 2011, 2011, 8. [Google Scholar]
- Nazemalhosseini Mojarad, E.; Kuppen, P.J.; Aghdaei, H.A.; Zali, M.R. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol. Hepatol. Bed. Bench. 2013, 6, 120–128. [Google Scholar]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [CrossRef]
- Siddiqui, H.; Al-Ghafari, A.; Choudhry, H.; Al Doghaither, H. Roles of long non-coding RNAs in colorectal cancer tumorigenesis: A Review. Mol. Clin. Oncol. 2019, 11, 167–172. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Z.; Wang, X.; Huang, Z.; He, Z.; Chen, Y. Long non-coding RNA: A new player in cancer. J. Hematol. Oncol. 2013, 6, 37. [Google Scholar] [CrossRef]
- Galamb, O.; Barták, B.K.; Kalmár, A.; Nagy, Z.B.; Szigeti, K.A.; Tulassay, Z.; Igaz, P.; Molnár, B. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J. Gastroenterol. 2019, 25, 5026–5048. [Google Scholar] [CrossRef]
- Svoboda, M.; Slyskova, J.; Schneiderova, M.; Makovicky, P.; Bielik, L.; Levy, M.; Lipska, L.; Hemmelova, B.; Kala, Z.; Protivankova, M.; et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 2014, 35, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Alaiyan, B.; Ilyayev, N.; Stojadinovic, A.; Izadjoo, M.; Roistacher, M.; Pavlov, V.; Tzivin, V.; Halle, D.; Pan, H.; Trink, B.; et al. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer 2013, 13, 196. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Tan, X.; Wang, X.; Jin, H.; Liu, L.; Ma, L.; Yu, H.; Fan, Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumor Biology 2014, 35, 12181–12188. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yu, Y.; Xu, B.; Zhang, M.; Li, Q.; Miao, L. Pivotal prognostic and diagnostic role of the long noncoding RNA colon cancerassociated transcript 1 expression in human cancer (Review). Mol. Med. Rep. 2019, 19, 771–782. [Google Scholar] [PubMed]
- Tsang, W.P.; Ng, E.K.O.; Ng, S.S.M.; Jin, H.; Yu, J.; Sung, J.J.Y.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2009, 31, 350–358. [Google Scholar] [CrossRef]
- Ohtsuka, M.; Ling, H.; Ivan, C.; Pichler, M.; Matsushita, D.; Goblirsch, M.; Stiegelbauer, V.; Shigeyasu, K.; Zhang, X.; Chen, M.; et al. H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-beta-Catenin Signaling in Colorectal Cancer. Ebio. Med. 2016, 13, 113–124. [Google Scholar]
- Yoruker, E.E.; Keskin, M.; Kulle, C.B.; Holdenrieder, S.; Gezer, U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed. Rep. 2018, 9, 181–186. [Google Scholar]
- Yoshimizu, T.; Miroglio, A.; Ripoche, M.A.; Gabory, A.; Vernucci, M.; Riccio, A.; Colnot, S.; Godard, C.; Terris, B.; Jammes, H.; et al. The H19 locus acts in vivo as a tumor suppressor. Proc. Natl. Acad. Sci. USA 2008, 105, 12417–12422. [Google Scholar] [CrossRef]
- Graham, L.D.; Pedersen, S.K.; Brown, G.S.; Ho, T.; Kassir, Z.; Moynihan, A.T.; Vizgoft, E.K.; Dunne, R.; Pimlott, L.; Young, G.P.; et al. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Gen. Cancer 2011, 2, 829–840. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, X.; Gao, S.; Jing, F.; Yang, Y.; Du, L.; Zheng, G.; Li, P.; Li, C.; Wang, C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 2016, 7, 85551–85563. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, Y.; Ai, M.; Wang, H.; Duan, Z.; Wang, H.; Zhao, L.; Yu, J.; Ding, Y.; Wang, S. Long Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death Dis. 2017, 8, e2862. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Ye, X.; Du, Q.; Zhu, B.; Zhai, Q.; Li, X.X. The Long Non-Coding RNA CRNDE Promotes Colorectal Carcinoma Progression by Competitively Binding miR-217 with TCF7L2 and Enhancing the Wnt/beta-Catenin Signaling Pathway. Cell. Physiol. Biochem. 2017, 41, 2489–2502. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Li, B. Role of miRNA in transformation from normal tissue to colorectal adenoma and cancer. J. Cancer Res. Ther. 2019, 15, 278–285. [Google Scholar] [PubMed]
- Vymetalkova, V.; Vodicka, P.; Vodenkova, S.; Alonso, S.; Schneider-Stock, R. DNA methylation and chromatin modifiers in colorectal cancer. Mol. Aspects. Med. 2019, 69, 73–92. [Google Scholar] [CrossRef] [PubMed]
- Tsikitis, V.L.; Potter, A.; Mori, M.; Buckmeier, J.A.; Preece, C.R.; Harrington, C.A.; Bartley, A.N.; Bhattacharyya, A.K.; Hamilton, S.R.; Lance, M.P.; et al. MicroRNA Signatures of Colonic Polyps on Screening and Histology. Cancer Prev. Res. 2016, 9, 942–949. [Google Scholar] [CrossRef]
- Kanth, P.; Hazel, M.W.; Boucher, K.M.; Yang, Z.; Wang, L.; Bronner, M.P.; Boylan, K.E.; Burt, R.W.; Westover, M.; Neklason, D.W.; et al. Small RNA sequencing of sessile serrated polyps identifies microRNA profile associated with colon cancer. Genes Chromosomes Cancer 2019, 58, 23–33. [Google Scholar] [CrossRef]
- Aslam, M.I.; Hussein, S.; West, K.; Singh, B.; Jameson, J.S.; Pringle, J.H. MicroRNAs associated with initiation and progression of colonic polyp: A feasibility study. Int. J. Surg. 2015, 13, 272–279. [Google Scholar] [CrossRef]
- Oberg, A.L.; French, A.J.; Sarver, A.L.; Subramanian, S.; Morlan, B.W.; Riska, S.M.; Borralho, P.M.; Cunningham, J.M.; Boardman, L.A.; Wang, L.; et al. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS ONE 2011, 6, e20465. [Google Scholar] [CrossRef]
- Ito, M.; Mitsuhashi, K.; Igarashi, H.; Nosho, K.; Naito, T.; Yoshii, S.; Takahashi, H.; Fujita, M.; Sukawa, Y.; Yamamoto, E.; et al. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int. J. Cancer 2014, 135, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Ren, L.L.; Zheng, P.; Zheng, H.M.; Yu, Y.N.; Wang, J.L.; Lin, Y.W.; Chen, Y.X.; Ge, Z.Z.; Chen, X.Y.; et al. miR-194 as a predictor for adenoma recurrence in patients with advanced colorectal adenoma after polypectomy. Cancer. Prev. Res. (Phila) 2014, 7, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Ardila, H.J.; Sanabria-Salas, M.C.; Meneses, X.; Rios, R.; Huertas-Salgado, A.; Serrano, M.L. Circulating miR-141-3p, miR-143-3p and miR-200c-3p are differentially expressed in colorectal cancer and advanced adenomas. Mol. Clin. Oncol. 2019, 11, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Z.B.; Wichmann, B.; Kalmar, A.; Galamb, O.; Bartak, B.K.; Spisak, S.; Tulassay, Z.; Molnar, B. Colorectal adenoma and carcinoma specific miRNA profiles in biopsy and their expression in plasma specimens. Clin. Epigenetics 2017, 9, 22. [Google Scholar] [CrossRef]
- Uratani, R.; Toiyama, Y.; Kitajima, T.; Kawamura, M.; Hiro, J.; Kobayashi, M.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Mori, T.; et al. Diagnostic Potential of Cell-Free and Exosomal MicroRNAs in the Identification of Patients with High-Risk Colorectal Adenomas. PLoS ONE 2016, 11, e0160722. [Google Scholar] [CrossRef]
- Yau, T.O.; Tang, C.M.; Harriss, E.K.; Dickins, B.; Polytarchou, C. Faecal microRNAs as a non-invasive tool in the diagnosis of colonic adenomas and colorectal cancer: A meta-analysis. Sci. Rep. 2019, 9, 9491. [Google Scholar] [CrossRef]
- Wu, C.W.; Ng, S.C.; Dong, Y.; Tian, L.; Ng, S.S.; Leung, W.W.; Law, W.T.; Yau, T.O.; Chan, F.K.; Sung, J.J.; et al. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin. Cancer Res. 2014, 20, 2994–3002. [Google Scholar] [CrossRef]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama 2008, 299, 425–436. [Google Scholar] [CrossRef]
- Diosdado, B.; van de Wiel, M.A.; Terhaar Sive Droste, J.S.; Mongera, S.; Postma, C.; Meijerink, W.J.; Carvalho, B.; Meijer, G.A. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br. J. Cancer 2009, 101, 707–714. [Google Scholar] [CrossRef]
- Kanaan, Z.; Roberts, H.; Eichenberger, M.R.; Billeter, A.; Ocheretner, G.; Pan, J.; Rai, S.N.; Jorden, J.; Williford, A.; Galandiuk, S. A plasma microRNA panel for detection of colorectal adenomas: A step toward more precise screening for colorectal cancer. Ann. Surg. 2013, 258, 400–408. [Google Scholar] [CrossRef]
- Tsikitis, V.L.; White, I.; Mori, M.; Potter, A.; Bhattcharyya, A.; Hamilton, S.R.; Buckmeier, J.; Lance, P.; Thompson, P. Differential expression of microRNA-320a, -145, and -192 along the continuum of normal mucosa to high-grade dysplastic adenomas of the colorectum. Am. J. Surg. 2014, 207, 717–722, discussion 722. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Du, L.; Yang, X.; Zhang, X.; Wang, L.; Yang, Y.; Li, J.; Wang, C. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br. J. Cancer 2014, 111, 1985–1992. [Google Scholar] [CrossRef] [PubMed]
- de Groen, F.L.; Timmer, L.M.; Menezes, R.X.; Diosdado, B.; Hooijberg, E.; Meijer, G.A.; Steenbergen, R.D.; Carvalho, B. Oncogenic Role of miR-15a-3p in 13q Amplicon-Driven Colorectal Adenoma-to-Carcinoma Progression. PLoS ONE 2015, 10, e0132495. [Google Scholar] [CrossRef] [PubMed]
- Gattolliat, C.H.; Uguen, A.; Pesson, M.; Trillet, K.; Simon, B.; Doucet, L.; Robaszkiewicz, M.; Corcos, L. MicroRNA and targeted mRNA expression profiling analysis in human colorectal adenomas and adenocarcinomas. Eur. J. Cancer 2015, 51, 409–420. [Google Scholar] [CrossRef]
- Hibino, Y.; Sakamoto, N.; Naito, Y.; Goto, K.; Oo, H.Z.; Sentani, K.; Hinoi, T.; Ohdan, H.; Oue, N.; Yasui, W. Significance of miR-148a in Colorectal Neoplasia: Downregulation of miR-148a Contributes to the Carcinogenesis and Cell Invasion of Colorectal Cancer. Pathobiology 2015, 82, 233–241. [Google Scholar] [CrossRef]
- Ho, G.Y.F.; Jung, H.J.; Schoen, R.E.; Wang, T.; Lin, J.; Williams, Z.; Weissfeld, J.L.; Park, J.Y.; Loudig, O.; Suh, Y. Differential expression of circulating microRNAs according to severity of colorectal neoplasia. Transl. Res. 2015, 166, 225–232. [Google Scholar] [CrossRef]
- Tadano, T.; Kakuta, Y.; Hamada, S.; Shimodaira, Y.; Kuroha, M.; Kawakami, Y.; Kimura, T.; Shiga, H.; Endo, K.; Masamune, A.; et al. MicroRNA-320 family is downregulated in colorectal adenoma and affects tumor proliferation by targeting CDK6. World J. Gastrointest. Oncol. 2016, 8, 532–542. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Ding, Y.; Fan, Z.; Zhang, J.; Zhang, H.; Jiang, B.; Zhu, Y. Serum MicroRNA profile in patients with colon adenomas or cancer. BMC Med. Genomics 2017, 10, 23. [Google Scholar] [CrossRef]
- Zhang, J.; Raju, G.S.; Chang, D.W.; Lin, S.H.; Chen, Z.; Wu, X. Global and targeted circulating microRNA profiling of colorectal adenoma and colorectal cancer. Cancer 2018, 124, 785–796. [Google Scholar] [CrossRef]
- Aherne, S.T.; Madden, S.F.; Hughes, D.J.; Pardini, B.; Naccarati, A.; Levy, M.; Vodicka, P.; Neary, P.; Dowling, P.; Clynes, M. Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression. BMC Cancer 2015, 15, 329. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Jiraskova, K.; Hughes, D.J.; Brezina, S.; Gumpenberger, T.; Veskrnova, V.; Buchler, T.; Schneiderova, M.; Levy, M.; Liska, V.; Vodenkova, S.; et al. Functional Polymorphisms in DNA Repair Genes Are Associated with Sporadic Colorectal Cancer Susceptibility and Clinical Outcome. Int J. Mol. Sci. 2018, 20, 97. [Google Scholar] [CrossRef] [PubMed]
- Cervena, K.; Vodicka, P.; Vymetalkova, V. Diagnostic and prognostic impact of cell-free DNA in human cancers: Systematic review. Mutat. Res. 2019, 781, 100–129. [Google Scholar] [CrossRef] [PubMed]
- Myint, N.N.M.; Verma, A.M.; Fernandez-Garcia, D.; Sarmah, P.; Tarpey, P.S.; Al-Aqbi, S.S.; Cai, H.; Trigg, R.; West, K.; Howells, L.M.; et al. Circulating tumor DNA in patients with colorectal adenomas: Assessment of detectability and genetic heterogeneity. Cell Death Dis. 2018, 9, 894. [Google Scholar] [CrossRef]
- Aravanis, A.M.; Lee, M.; Klausner, R.D. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017, 168, 571–574. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Mead, R.; Duku, M.; Bhandari, P.; Cree, I.A. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br. J. Cancer. 2011, 105, 239–245. [Google Scholar] [CrossRef]
- Galanopoulos, M.; Papanikolaou, I.; Zografos, E.; Viazis, N.; Papatheodoridis, G.; Karamanolis, D.; Marinos, E.; Mantzaris, G.; Gazouli, M. Comparative Study of Mutations in Single Nucleotide Polymorphism Loci of KRAS and BRAF Genes in Patients Who Underwent Screening Colonoscopy, With and Without Premalignant Intestinal Polyps. Anticancer Res. 2017, 37, 651–658. [Google Scholar] [CrossRef]
- Kopreski, M.S.; Benko, F.A.; Borys, D.J.; Khan, A.; McGarrity, T.J.; Gocke, C.D. Somatic Mutation Screening: Identification of Individuals Harboring K-ras Mutations With the Use of Plasma DNA. JNCI: J. Natl. Cancer Inst. 2000, 92, 918–923. [Google Scholar] [CrossRef]
- Perrone, F.; Lampis, A.; Bertan, C.; Verderio, P.; Ciniselli, C.M.; Pizzamiglio, S.; Frattini, M.; Nucifora, M.; Molinari, F.; Gallino, G.; et al. Circulating Free DNA in a Screening Program for Early Colorectal Cancer Detection. Tumori 2014, 100, 115–121. [Google Scholar] [CrossRef]
- Gausachs, M.; Borras, E.; Chang, K.; Gonzalez, S.; Azuara, D.; Delgado Amador, A.; Lopez-Doriga, A.; San Lucas, F.A.; Sanjuan, X.; Paules, M.J.; et al. Mutational Heterogeneity in APC and KRAS Arises at the Crypt Level and Leads to Polyclonality in Early Colorectal Tumorigenesis. Clin. Cancer Res. 2017, 23, 5936–5947. [Google Scholar] [CrossRef] [PubMed]
- Thirlwell, C.; Will, O.C.; Domingo, E.; Graham, T.A.; McDonald, S.A.; Oukrif, D.; Jeffrey, R.; Gorman, M.; Rodriguez-Justo, M.; Chin-Aleong, J.; et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology 2010, 138, 1441–1454.e7. [Google Scholar] [CrossRef] [PubMed]
- Fleshner, P.; Braunstein, G.D.; Ovsepyan, G.; Tonozzi, T.R.; Kammesheidt, A. Tumor-associated DNA mutation detection in individuals undergoing colonoscopy. Cancer Med. 2018, 7, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta. 2007, 1775, 181–232. [Google Scholar] [CrossRef]
- Martincorena, I.; Roshan, A.; Gerstung, M.; Ellis, P.; Van Loo, P.; McLaren, S.; Wedge, D.C.; Fullam, A.; Alexandrov, L.B.; Tubio, J.M.; et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015, 348, 880–886. [Google Scholar] [CrossRef]
- Zhu, D.; Keohavong, P.; Finkelstein, S.D.; Swalsky, P.; Bakker, A.; Weissfeld, J.; Srivastava, S.; Whiteside, T.L. K-ras gene mutations in normal colorectal tissues from K-ras mutation-positive colorectal cancer patients. Cancer Res. 1997, 57, 2485–2492. [Google Scholar]
- Takayama, T.; Katsuki, S.; Takahashi, Y.; Ohi, M.; Nojiri, S.; Sakamaki, S.; Kato, J.; Kogawa, K.; Miyake, H.; Niitsu, Y. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N. Engl. J. Med. 1998, 339, 1277–1284. [Google Scholar] [CrossRef]
- Tobi, M.; Luo, F.C.; Ronai, Z. Detection of K-ras mutation in colonic effluent samples from patients without evidence of colorectal carcinoma. J. Natl. Cancer Inst. 1994, 86, 1007–1010. [Google Scholar]
- Chen, A.; Braunstein, G.; Anselmo, M.; Jaboni, J.; Viloria, F.; Neidich, J.; Li, X.; Kammesheidt, A. Mutation detection with a liquid biopsy 96 mutation assay in cancer patients and healthy donors. Cancer Trans. Med. 2017, 3, 39. [Google Scholar]
- Gocke, C.D.; Benko, F.A.; Kopreski, M.S.; McGarrity, T.J. p53 and APC mutations are detectable in the plasma and serum of patients with colorectal cancer (CRC) or adenomas. Ann. N. Y. Acad. Sci. 2000, 906, 44–50. [Google Scholar] [CrossRef]
- Berger, B.M.; Ahlquist, D.A. Stool DNA screening for colorectal neoplasia: Biological and technical basis for high detection rates. Pathology 2012, 44, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Lidgard, G.P.; Domanico, M.J.; Bruinsma, J.J.; Light, J.; Gagrat, Z.D.; Oldham–Haltom, R.L.; Fourrier, K.D.; Allawi, H.; Yab, T.C.; Taylor, W.R.; et al. Clinical Performance of an Automated Stool DNA Assay for Detection of Colorectal Neoplasia. Clin. Gastroenterol. Hepatol. 2013, 11, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef] [PubMed]
- Hardingham, J.E.; Hewett, P.J.; Sage, R.E.; Finch, J.L.; Nuttall, J.D.; Kotasek, D.; Dobrovic, A. Molecular detection of blood-borne epithelial cells in colorectal cancer patients and in patients with benign bowel disease. Int. J. Cancer 2000, 89, 8–13. [Google Scholar] [CrossRef]
- Racila, E.; Euhus, D.; Weiss, A.J.; Rao, C.; McConnell, J.; Terstappen, L.W.; Uhr, J.W. Detection and characterization of carcinoma cells in the blood. Proc. Natl. Acad. Sci. USA 1998, 95, 4589–4594. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef]
- Lim, S.H.S.; Becker, T.M.; Chua, W.; Ng, W.L.; de Souza, P.; Spring, K.J. Circulating tumour cells and the epithelial mesenchymal transition in colorectal cancer. J. Clin. Pathol. 2014, 67, 848–853. [Google Scholar] [CrossRef]
- Sotelo, M.J.; Sastre, J.; Maestro, M.L.; Veganzones, S.; Vieitez, J.M.; Alonso, V.; Gravalos, C.; Escudero, P.; Vera, R.; Aranda, E.; et al. Role of circulating tumor cells as prognostic marker in resected stage III colorectal cancer. Ann. Oncol. 2015, 26, 535–541. [Google Scholar] [CrossRef]
- Tan, Y.; Wu, H. The significant prognostic value of circulating tumor cells in colorectal cancer: A systematic review and meta-analysis. Cur. Prob. Cancer. 2018, 42, 95–106. [Google Scholar] [CrossRef]
- Huang, X.; Gao, P.; Song, Y.; Sun, J.; Chen, X.; Zhao, J.; Liu, J.; Xu, H.; Wang, Z. Relationship between circulating tumor cells and tumor response in colorectal cancer patients treated with chemotherapy: A meta-analysis. BMC Cancer 2014, 14, 976. [Google Scholar] [CrossRef]
- Guadagni, S.; Fiorentini, G.; De Simone, M.; Masedu, F.; Zoras, O.; Mackay, A.R.; Sarti, D.; Papasotiriou, I.; Apostolou, P.; Catarci, M.; et al. Precision oncotherapy based on liquid biopsies in multidisciplinary treatment of unresectable recurrent rectal cancer: A retrospective cohort study. J. Cancer Res. Clin. Oncol. 2020, 146, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Guadagni, S.; Clementi, M.; Mackay, A.R.; Ricevuto, E.; Fiorentini, G.; Sarti, D.; Palumbo, P.; Apostolou, P.; Papasotiriou, I.; Masedu, F.; et al. Real-life multidisciplinary treatment for unresectable colorectal cancer liver metastases including hepatic artery infusion with chemo-filtration and liquid biopsy precision oncotherapy: Observational cohort study. J. Cancer Res. Clin. Oncol. 2020, 146, 1273–1290. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-S.; You, J.-F.; Hung, H.-Y.; Hsieh, P.-S.; Hsieh, B.; Lenz, H.-J.; Idos, G.; Friedland, S.; Yi-Jiun Pan, J.; Shao, H.-J.; et al. Novel Circulating Tumor Cell Assay for Detection of Colorectal Adenomas and Cancer. Clin. Trans. Gastroenterol. 2019, 10, e00088. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-Y.; Berndt, S.I.; Shiels, M.S.; Katki, H.A.; Chaturvedi, A.K.; Wentzensen, N.; Trabert, B.; Kemp, T.J.; Pinto, L.A.; Hildesheim, A.; et al. Circulating inflammation markers and colorectal adenoma risk. Carcinogenesis 2019, 40, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Godos, J.; Biondi, A.; Galvano, F.; Basile, F.; Sciacca, S.; Giovannucci, E.L.; Grosso, G. Markers of systemic inflammation and colorectal adenoma risk: Meta-analysis of observational studies. World J. Gastroenterol. 2017, 23, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Basavaraju, U.; Shebl, F.M.; Palmer, A.J.; Berry, S.; Hold, G.L.; El-Omar, E.M.; Rabkin, C.S. Cytokine gene polymorphisms, cytokine levels and the risk of colorectal neoplasia in a screened population of Northeast Scotland. Eur. J. Cancer. Prev. 2015, 24, 296–304. [Google Scholar] [CrossRef]
- Henry, C.J.; Sedjo, R.L.; Rozhok, A.; Salstrom, J.; Ahnen, D.; Levin, T.R.; D’Agostino, R.; Haffner, S.; DeGregori, J.; Byers, T. Lack of significant association between serum inflammatory cytokine profiles and the presence of colorectal adenoma. BMC Cancer 2015, 15, 123. [Google Scholar] [CrossRef]
- Murphy, N.; Cross, A.J.; Huang, W.-Y.; Rajabzadeh-Heshejin, V.; Stanczyk, F.; Hayes, R.; Gunter, M.J. A prospective evaluation of C-peptide levels and colorectal adenoma incidence. Cancer Epidemiology 2015, 39, 160–165. [Google Scholar] [CrossRef]
- Comstock, S.S.; Xu, D.; Hortos, K.; Kovan, B.; McCaskey, S.; Pathak, D.R.; Fenton, J.I. Association of serum cytokines with colorectal polyp number and type in adult males. Eur. J. Cancer. Prev. 2016, 25, 173–181. [Google Scholar] [CrossRef]
- Peacock, S.D.; Massey, T.E.; Vanner, S.J.; King, W.D. Telomere length in the colon is related to colorectal adenoma prevalence. PLoS ONE 2018, 13, e0205697. [Google Scholar] [CrossRef]
- Kroupa, M.; Rachakonda, S.K.; Liska, V.; Srinivas, N.; Urbanova, M.; Jiraskova, K.; Schneiderova, M.; Vycital, O.; Vymetalkova, V.; Vodickova, L.; et al. Relationship of telomere length in colorectal cancer patients with cancer phenotype and patient prognosis. Br. J. Cancer 2019, 121, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Tomasova, K.; Kroupa, M.; Forsti, A.; Vodicka, P.; Vodickova, L. Telomere maintenance in interplay with DNA repair in pathogenesis and treatment of colorectal cancer. Mutagenesis 2020, geaa005. [Google Scholar] [CrossRef]
- Zöchmeister, C.; Brezina, S.; Hofer, P.; Baierl, A.; Bergmann, M.M.; Bachleitner-Hofmann, T.; Karner-Hanusch, J.; Stift, A.; Gerger, A.; Leeb, G.; et al. Leukocyte telomere length throughout the continuum of colorectal carcinogenesis. Oncotarget 2018, 9, 13582–13592. [Google Scholar] [CrossRef] [PubMed]
- Valls Bautista, C.; Pinol Felis, C.; Rene Espinet, J.M.; Buenestado Garcia, J.; Vinas Salas, J. Telomerase activity and telomere length in the colorectal polyp-carcinoma sequence. Rev. Esp. Enferm. Dig. 2009, 101, 179–186. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roger, L.; Jones, R.E.; Heppel, N.H.; Williams, G.T.; Sampson, J.R.; Baird, D.M. Extensive Telomere Erosion in the Initiation of Colorectal Adenomas and Its Association With Chromosomal Instability. J. Natl. Cancer Inst. 2013, 105, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Kim, Y.J.; Kim, H.J.; Kim, S.K.; Lee, J.H. Telomere length changes in colorectal cancers and polyps. J. Korean Med. Sci. 2002, 17, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Suraweera, N.; Mouradov, D.; Li, S.; Jorissen, R.N.; Hampson, D.; Ghosh, A.; Sengupta, N.; Thaha, M.; Ahmed, S.; Kirwan, M.; et al. Relative telomere lengths in tumor and normal mucosa are related to disease progression and chromosome instability profiles in colorectal cancer. Oncotarget 2016, 7, 36474–36488. [Google Scholar] [CrossRef][Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).