Construction of an Electrochemical Receptor Sensor Based on Graphene/Thionine for the Sensitive Determination of β-Lactam Antibiotics Content in Milk
Abstract
:1. Introduction
2. Results and Discussion
2.1. Purification of Protein
2.2. Identification of Enzyme Markers
2.3. Characterization of GO/TH Composite Materials
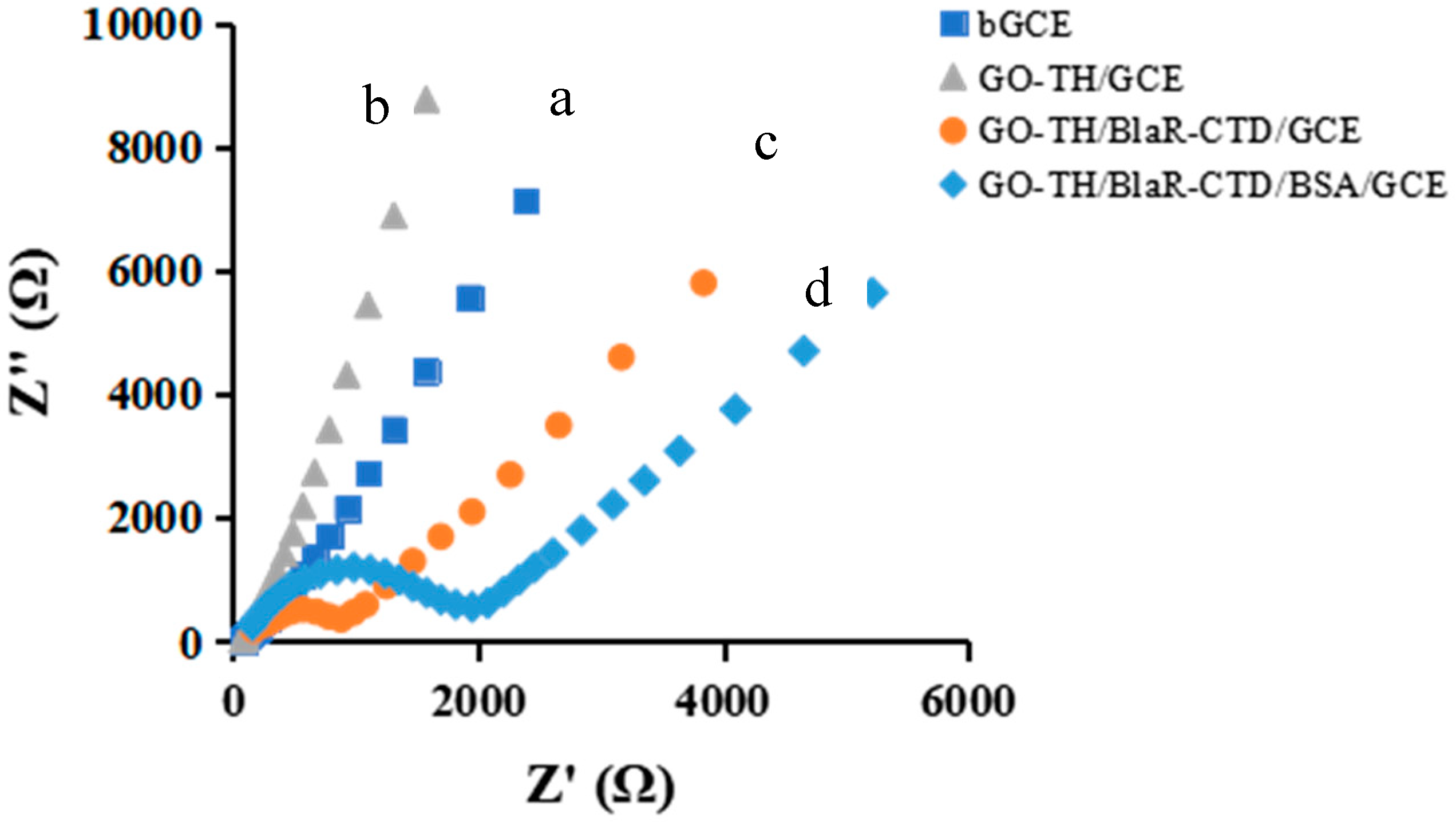
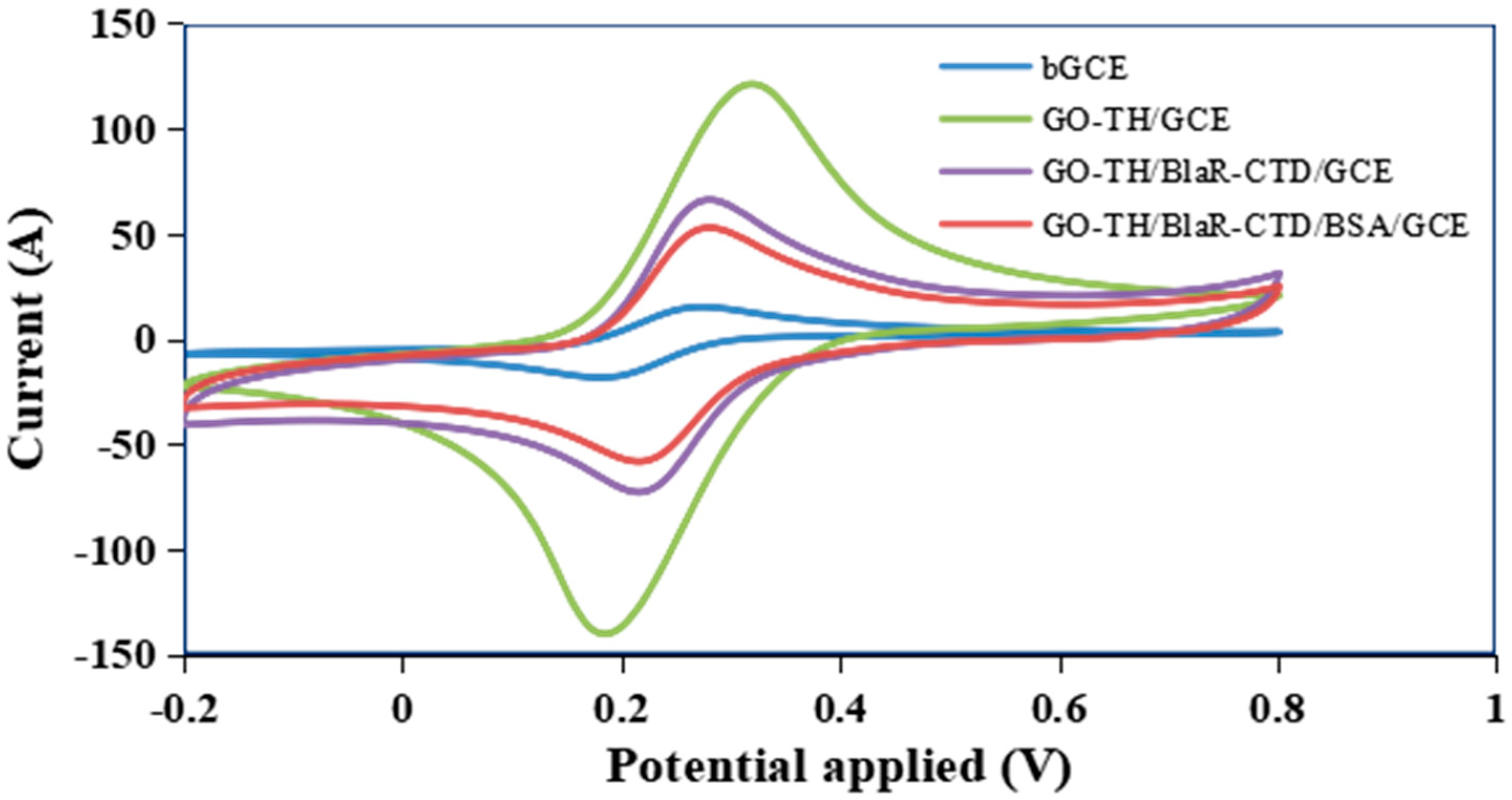
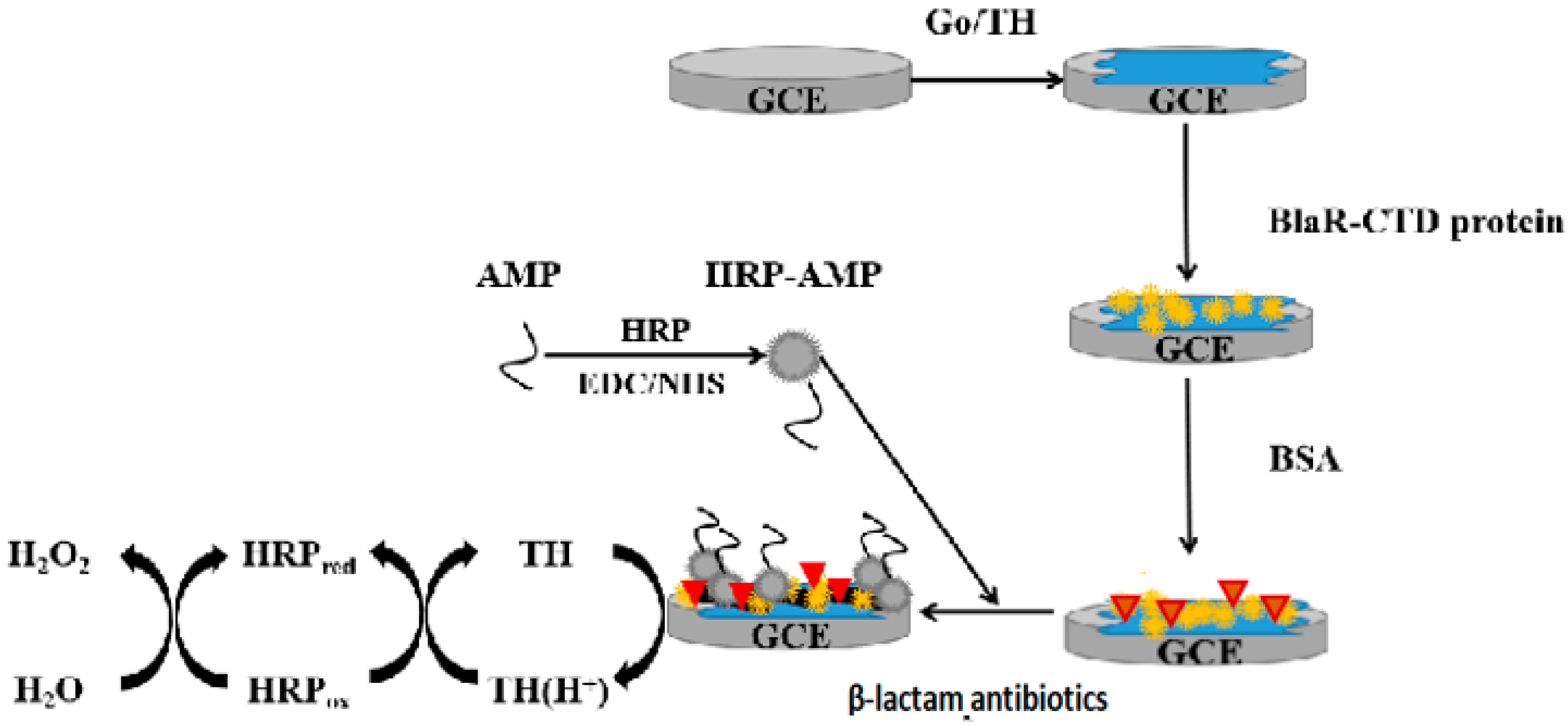
2.4. Characterization of the GO/TH/GCE
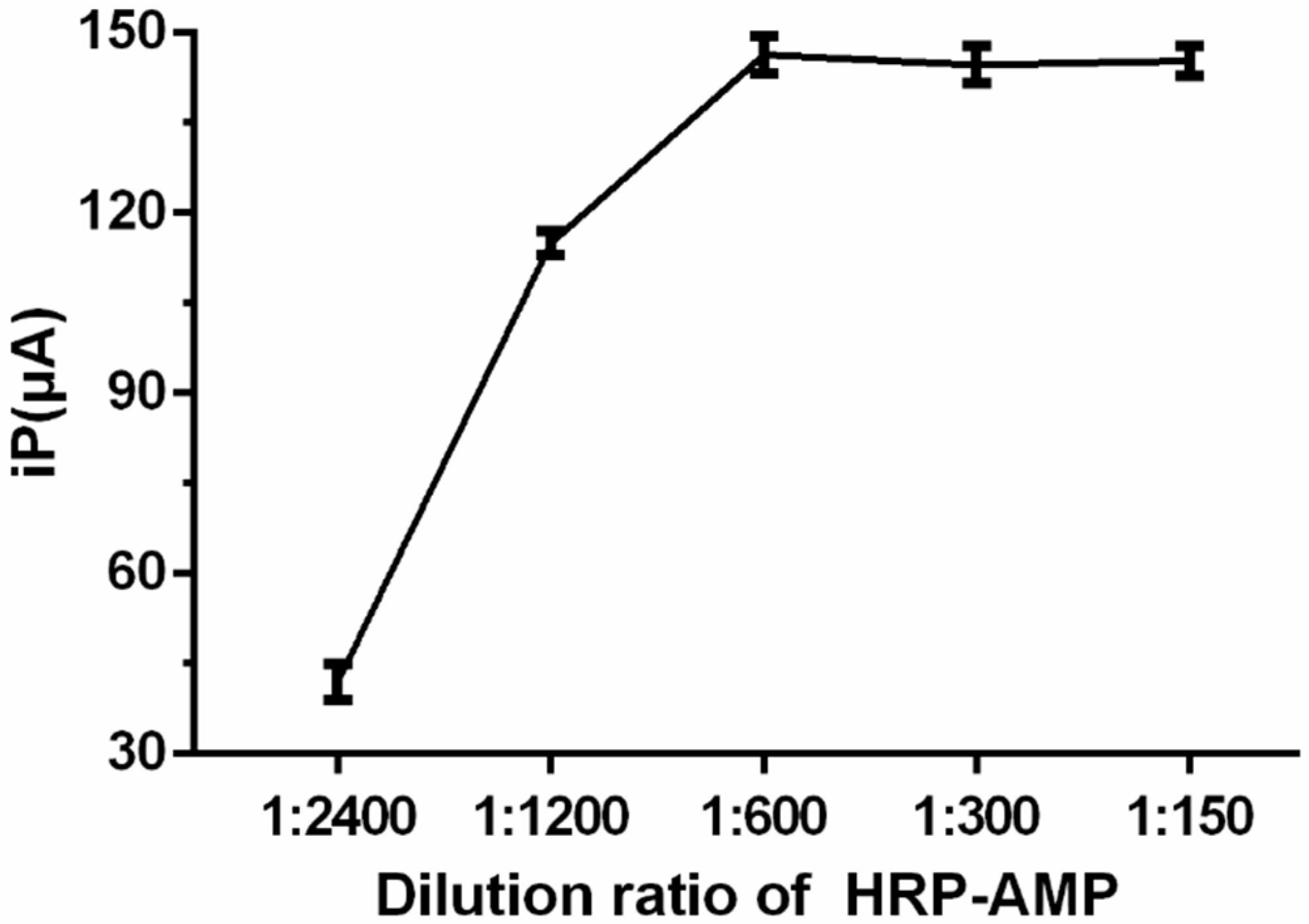
2.5. Optimization of the Detection Conditions
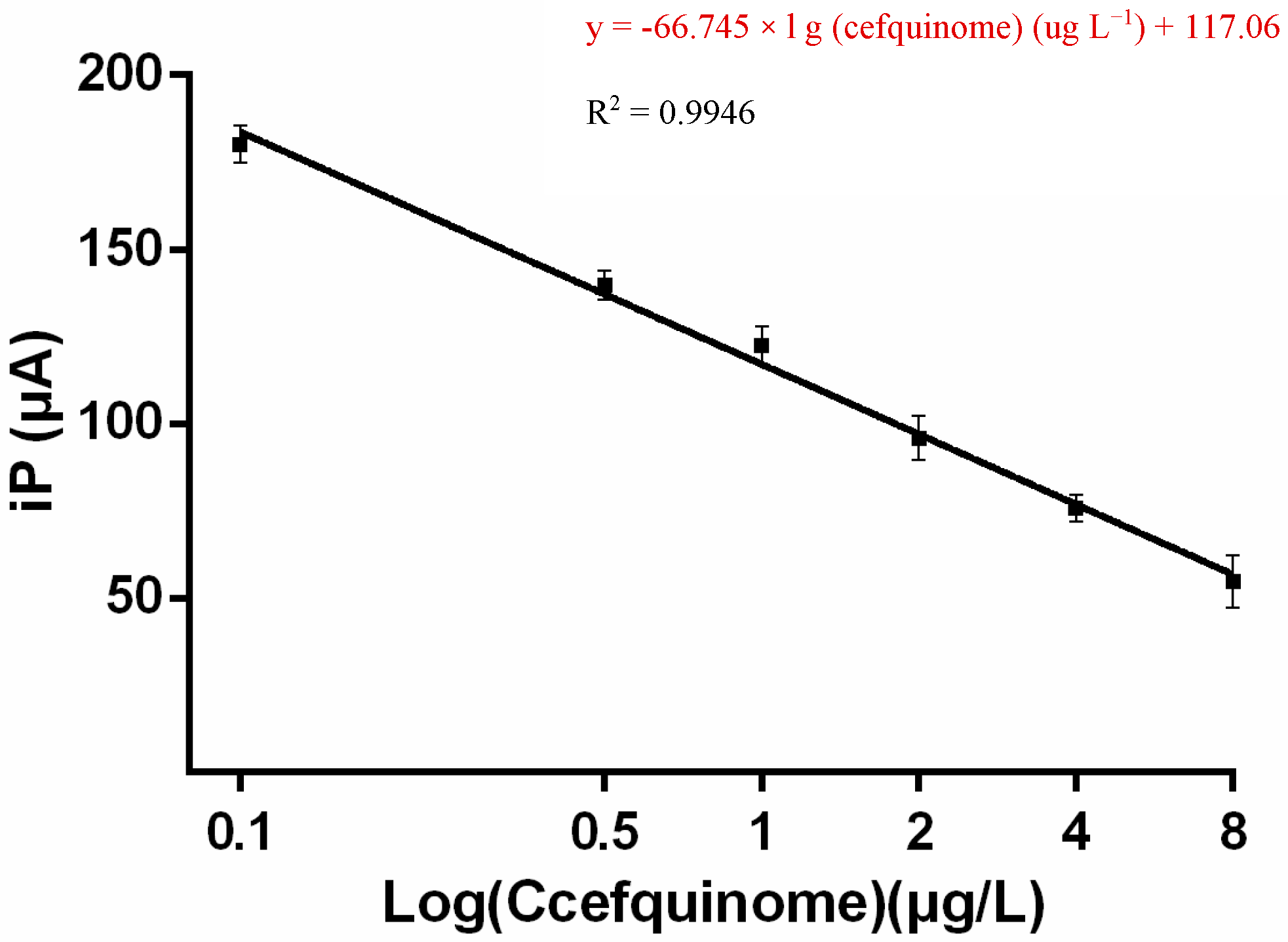
2.6. Linear Range and LOD
2.7. Reproducibility, Repeatability and Stability of the Sensor
2.8. Real Sample Analysis
3. Materials and Methods
3.1. Materials
3.2. Apparatus
3.3. Preparation and Purification of Protein Samples
3.4. Preparation of Enzyme Markers
3.5. Preparation and Characterization of GO/TH
3.6. Preparation of Electrochemical Receptor Sensor
3.7. Electrochemical Measurements
3.8. Sample Preparation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cherian, P.T.; Cheramie, M.N.; Marreddy Ravi, K.R.; Fernando, D.M.; Hurdle, J.G.; Lee, R.E. New β-lactam-Tetramic acid hybrids show promising antibacterial activities. Bioorganic Med. Chem. Lett. 2018, 28, 3015–3112. [Google Scholar] [CrossRef] [PubMed]
- Ronquillo, M.G.; Hernandez, J.C.A. Antibiotic and synthetic growth promoters in animal diets: Review of impact and analytical methods. Food Control 2016, 72, 255–267. [Google Scholar] [CrossRef]
- Baynes, R.E.; Dedonder, K.; Kissell, L.; Mzyk, D.T.; Marmulak, T.; Smith, G.; Tell, L.; Gehring, R.; Davis, J.; Riviere, J.E. Health concerns and management of select veterinary drug residues. Food Chem. Toxicol. 2016, 88, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhammad, A.; Yusof, N.A.; Hajian, R.; Abdullah, J. Construction of an Electrochemical Sensor Based on Carbon Nanotubes/Gold Nanoparticles for Trace Determination of Amoxicillin in Bovine Milk. Sensors 2016, 16, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barceló, D.; Farré, M. Analytical methodologies for the detection of β-lactam antibiotics in milk and feed samples. TRAC Trends Anal. Chem. 2009, 28, 729–744. [Google Scholar]
- Ohmori, T.; Suzuki, A.; Niwa, T.; Ushikoshi, H.K.; Shirai, K.; Yoshida, S.; Ogura, S.; Itoh, Y. Simultaneous determination of eight β-lactam antibiotics in human serum by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2011, 879, 1038–1042. [Google Scholar] [CrossRef]
- Briscoe, S.E.; Mcwhinney, B.C.; Lipman, J.; Roberts, J.A.; Ungerer, J.P. A method for determining the free (unbound) concentration of ten beta-lactam antibiotics in human plasma using high performance liquid chromatography with ultraviolet detection. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 907, 178–184. [Google Scholar] [CrossRef]
- Samanidou, V.F.; Evaggelopoulou, E.N.; Papadoyannis, I.N. Development of a validated HPLC method for the determination of four penicillin antibiotics in pharmaceuticals and human biological fluids. J. Sep. Sci. 2015, 29, 1550–1560. [Google Scholar] [CrossRef]
- Seok, K.Y.; Ahmad Raston, N.H.; Bock Gu, B.M. Aptamer-based nanobiosensors. Biosens. Bioelectron 2016, 76, 2–19. [Google Scholar] [CrossRef]
- Li, Z.; Liu, C.; Sarpong, V.; Gu, Z. Multisegment nanowire/nanoparticle hybrid arrays as electrochemical biosensors for simultaneous detection of antibiotics. Biosens. Bioelectron. 2019, 126, 632–639. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, T.; Cheng, F.; Zhang, J.; Zhu, J. Toward the Early Evaluation of Therapeutic Effects: An Electrochemical Platform for Ultrasensitive Detection of Apoptotic Cells. Anal. Chem. 2011, 83, 7902–7909. [Google Scholar] [CrossRef] [PubMed]
- Merola, G.; Martini, E.; Tomassetti, M.; Campanella, L. Simple and suitable immunosensor for β-lactam antibiotics analysis in real matrixes: Milk, serum, urine. J. Pharm. Biomed. Anal. 2015, 106, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Majdinasab, M.; Yaqub, M.; Rahim, A.; Catanante, G.; Hayat, A.; Marty, J.L. An Overview on Recent Progress in Electrochemical Biosensors for Antimicrobial Drug Residues in Animal-Derived Food. Sensors 2017, 17, 1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacciatore, G.; Petz, M.; Rachid, S.; Hakenbeck, R.; Bergwerff, A.A. Development of an optical biosensor assay for detection of beta-lactam antibiotics in milk using the penicillin-binding protein 2x. Anal. Chim. Acta 2004, 520, 105–115. [Google Scholar] [CrossRef]
- Ahmed, S.; Ning, J.; Cheng, G.; Ahmad, I.; Li, J.; Mingyue, L.; Qu, W.; Iqbal, M.; Shabbir, M.A.B.; Yuan, Z. Receptor-based screening assays for the detection of antibiotics residues–A review. Talanta 2017, 166, 176–186. [Google Scholar] [CrossRef]
- Kerff, F.; Charlier, P.; Colombo, M.L.; Sauvage, E.; Brans, A.; Frere, J.M.; Joris, B.; Fonze, E. Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis. Biochemistry 2003, 42, 12835–12843. [Google Scholar] [CrossRef]
- Duval, V.; Swinnen, M.; Lepage, S.; Brans, A.; Granier, B.; Franssen, C.; Frere, J.M.; Joris, B. The kinetic properties of the carboxy terminal domain of the Bacillus licheniformis 749/I BlaR penicillin-receptor shed a new light on the derepression of beta-lactamase synthesis. Mol. Microbiol. 2003, 48, 1553–1564. [Google Scholar] [CrossRef] [Green Version]
- Golemi-Kotra, D.; Cha, J.Y.; Meroueh, S.O.; Vakulenko, S.B.; Mobashery, S. Resistance to beta-lactam antibiotics and its mediation by the sensor domain of the transmembrane BlaR signaling pathway in Staphylococcus aureus. J. Biol. Chem. 2003, 278, 18419–18425. [Google Scholar] [CrossRef] [Green Version]
- Zapun, A.; Contrerasmartel, C.; Vernet, T. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 2008, 32, 361–385. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xu, X.X.; Liu, L.; Kuang, H.; Xu, L.; Xu, C. Rapid Detection of 21 β-Lactams using Immunochromatographic Assay Based on Mutant BlaR-CTD Protein from Bacillus Licheniformis. Analyst 2020, 145, 3257–3265. [Google Scholar] [CrossRef]
- Peng, J.; Cheng, G.; Huang, L.; Wang, Y.; Hao, H.; Peng, D.; Liu, Z.; Yuan, Z. Development of a direct ELISA based on carboxy-terminal of penicillin-binding protein BlaR for the detection of beta-lactam antibiotics in foods. Anal. Bioanal. Chem. 2013, 405, 8925–8933. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.M.; Anbarasu, A.; Ramaiah, S. Molecular docking and molecular dynamics studies on β-lactamases and penicillin binding proteins. Mol. Biosyst. 2014, 10, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Z.; Wen, K.; Liang, X.; Shen, J. Penicillin-binding protein3 of Streptococcus pneumoniae and its application in screening of β-lactams in milk. Anal. Biochem. 2013, 442, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Ahmed, S.; Cheng, G.; Chen, T.; Wang, Y.; Peng, D.; Yuan, Z. Analysis of the stability and affinity of BlaR-CTD protein to β-lactam antibiotics based on docking and mutagenesis studies. J. Biol. Eng. 2019, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, H.; Zhao, Y.; Zhang, F.; Yang, F.; Tang, J.; He, P. A label-free electrochemiluminescence aptasensor for thrombin detection based on host-guest recognition between tris(bipyridine)ruthenium(II)-β-cyclodextrin and aptamer. Biosens. Bioelectron. 2014, 54, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, C.R.; Wang, W.C.; Xue, J.; Huang, Y.L.; Yang, X.X.; Tan, B.; Zhou, X.P.; Shao, C.; Ding, S.J.; et al. A novel electrochemical immunosensor for highly sensitive detection of Aflatoxin B1 in corn using single-walled carbon nanotubes/chitosan. Food Chem. 2016, 192, 197–202. [Google Scholar] [CrossRef]
- He, B.; Yan, S. Voltammetric kanamycin aptasensor based on the use of thionine incorporated into Au@Pt core-shell nanoparticles. Microchim. Acta 2019, 186, 77. [Google Scholar] [CrossRef]
- Lu, L.; Seenivasan, R.; Wang, Y.C.; Yu, J.H.; Gunasekaran, S. An Electrochemical Immunosensor for Rapid and Sensitive Detection of Mycotoxins Fumonisin B1 and Deoxynivalenol. Electrochim Acta 2016, 213, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Yuge, R.; Nihey, F.; Toyama, K.; Yudasaka, M. Preparation and Characterization of Newly Discovered Fibrous Aggregates of Single-Walled Carbon Nanohorns. Adv. Mater. 2016, 28, 7174–7177. [Google Scholar] [CrossRef]
- Yuge, R.; Manako, T.; Nakahara, K.; Yasui, M.; Iwasa, S.; Yoshitake, T. The production of an electrochemical capacitor electrode using holey single-wall carbon nanohorns with high specifific surface area. Carbon 2012, 50, 5569–5573. [Google Scholar] [CrossRef]
- Yu, T.X.; Cheng, W.; Li, Q.; Luo, C.H.; Yan, L.; Zhang, D.C.; Yin, Y.B.; Ding, S.J.; Ju, H.X. Electrochemical immunosensor for competitive detection of neuron specifific enolase using functional carbon nanotubes and gold nanoprobe. Talanta 2012, 93, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Hashkavayi, A.B.; Raoof, J.B.; Azimi, R.; Ojani, R. Label-free and sensitive aptasensor based on dendritic gold nanostructures on functionalized SBA-15 for determination of chloramphenicol. Anal. Bioanal. Chem. 2016, 408, 2557–2565. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Gupta, V.B. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Drug Discov. Ther. 2011, 2, 21–25. [Google Scholar] [CrossRef]
- Dhand, C.; Sumana, G.; Datta, M.; Malhotra, B.D. Electrophoretically deposited nano-structured polyaniline film for glucose sensing. Thin Solid Film. 2010, 519, 1145–1150. [Google Scholar] [CrossRef]



| Method | LOD (μg/kg) | Sample | Recovery (%) | Ref |
|---|---|---|---|---|
| Receptor sensor | 0.06–17.16 | milk | 95.7–106.9 | In this study |
| Receptor assay | 0.05–21.46 | milk | 64.47–108.35 | [24] |
| Medicine | Average Determination Value of 20 Blank Samples (μg/L) | Standard Deviation SD | LOD (μg/L) | LOQ (μg/L) | MRL (μg/L) |
|---|---|---|---|---|---|
| Cefalexin | 4.79 | 3.03 | 14.88 | 36.09 | 100 |
| Cefaquinoxime | 1.2 | 0.42 | 2.46 | 5.4 | 20 |
| Ceftiofur | 6.75 | 3.47 | 17.16 | 41.45 | 100 |
| Penicillin G | 0.03 | 0.01 | 0.06 | 0.13 | 4 |
| Ampicillin | 0.12 | 0.03 | 0.21 | 0.42 | 4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhang, L.; Wang, Y.; Ou, Y.; Wang, X.; Pan, Y.; Wang, Y.; Huang, L.; Cheng, G.; Xie, S.; et al. Construction of an Electrochemical Receptor Sensor Based on Graphene/Thionine for the Sensitive Determination of β-Lactam Antibiotics Content in Milk. Int. J. Mol. Sci. 2020, 21, 3306. https://doi.org/10.3390/ijms21093306
Wang L, Zhang L, Wang Y, Ou Y, Wang X, Pan Y, Wang Y, Huang L, Cheng G, Xie S, et al. Construction of an Electrochemical Receptor Sensor Based on Graphene/Thionine for the Sensitive Determination of β-Lactam Antibiotics Content in Milk. International Journal of Molecular Sciences. 2020; 21(9):3306. https://doi.org/10.3390/ijms21093306
Chicago/Turabian StyleWang, Lei, Liyun Zhang, Yuke Wang, Yahong Ou, Xu Wang, Yuanhu Pan, Yulian Wang, Lingli Huang, Guyue Cheng, Shuyu Xie, and et al. 2020. "Construction of an Electrochemical Receptor Sensor Based on Graphene/Thionine for the Sensitive Determination of β-Lactam Antibiotics Content in Milk" International Journal of Molecular Sciences 21, no. 9: 3306. https://doi.org/10.3390/ijms21093306






