In Vivo Simultaneous Analysis of Gene Expression by Dual-Color Luciferases in Caenorhabditis elegans
Abstract
1. Introduction
2. Results
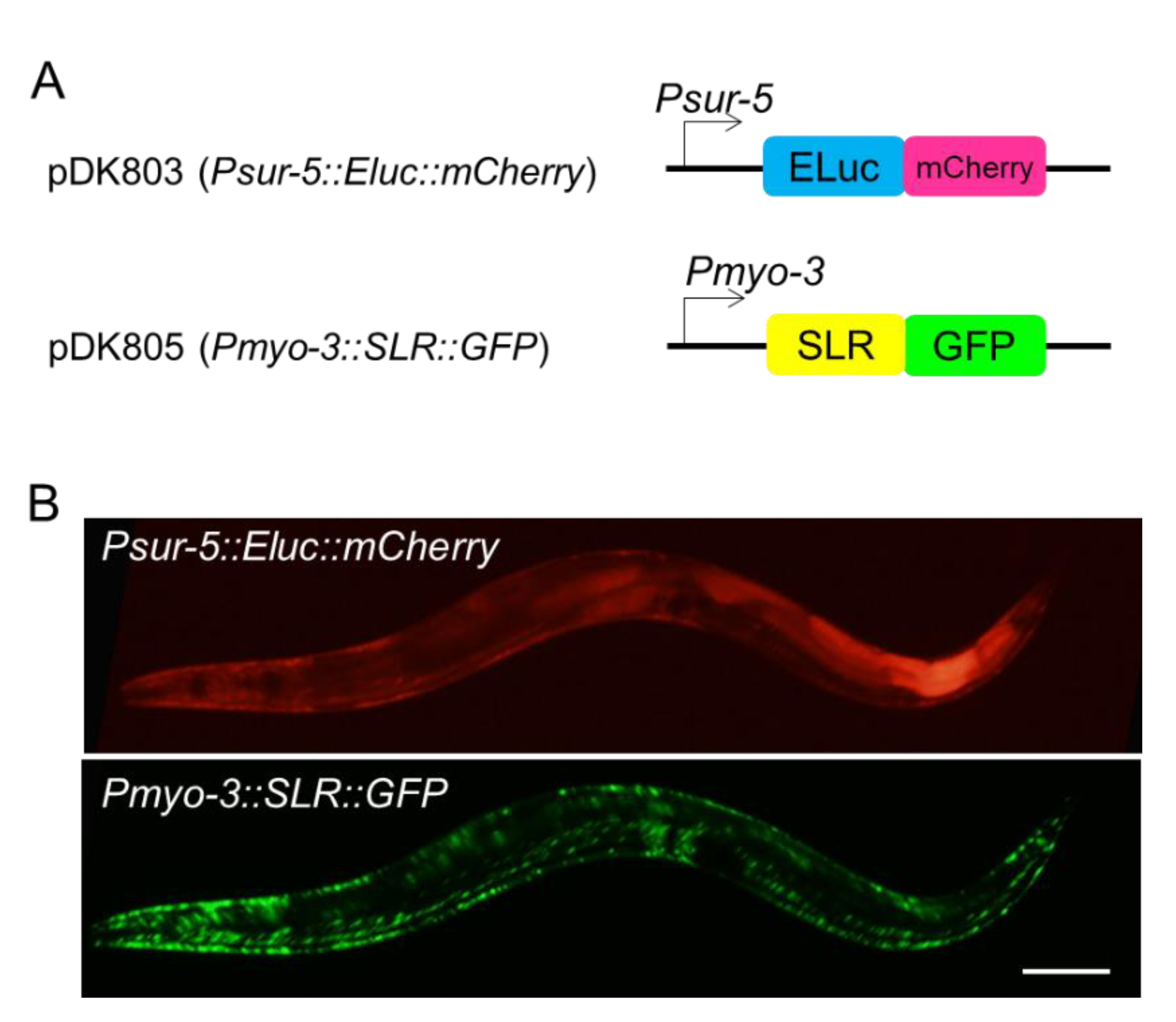
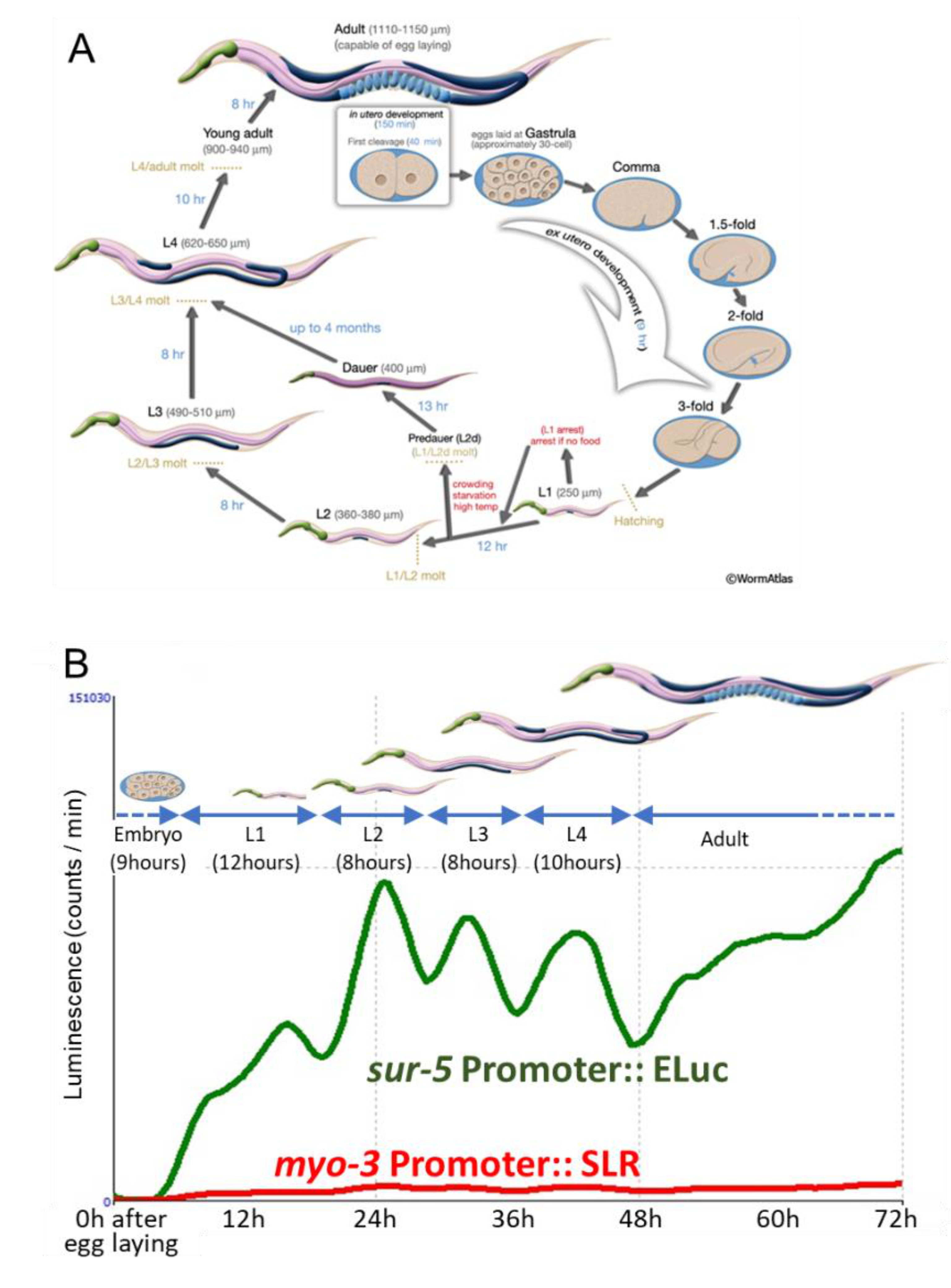
2.1. Real-Time Monitoring Two Promoter Activities in C. elegans
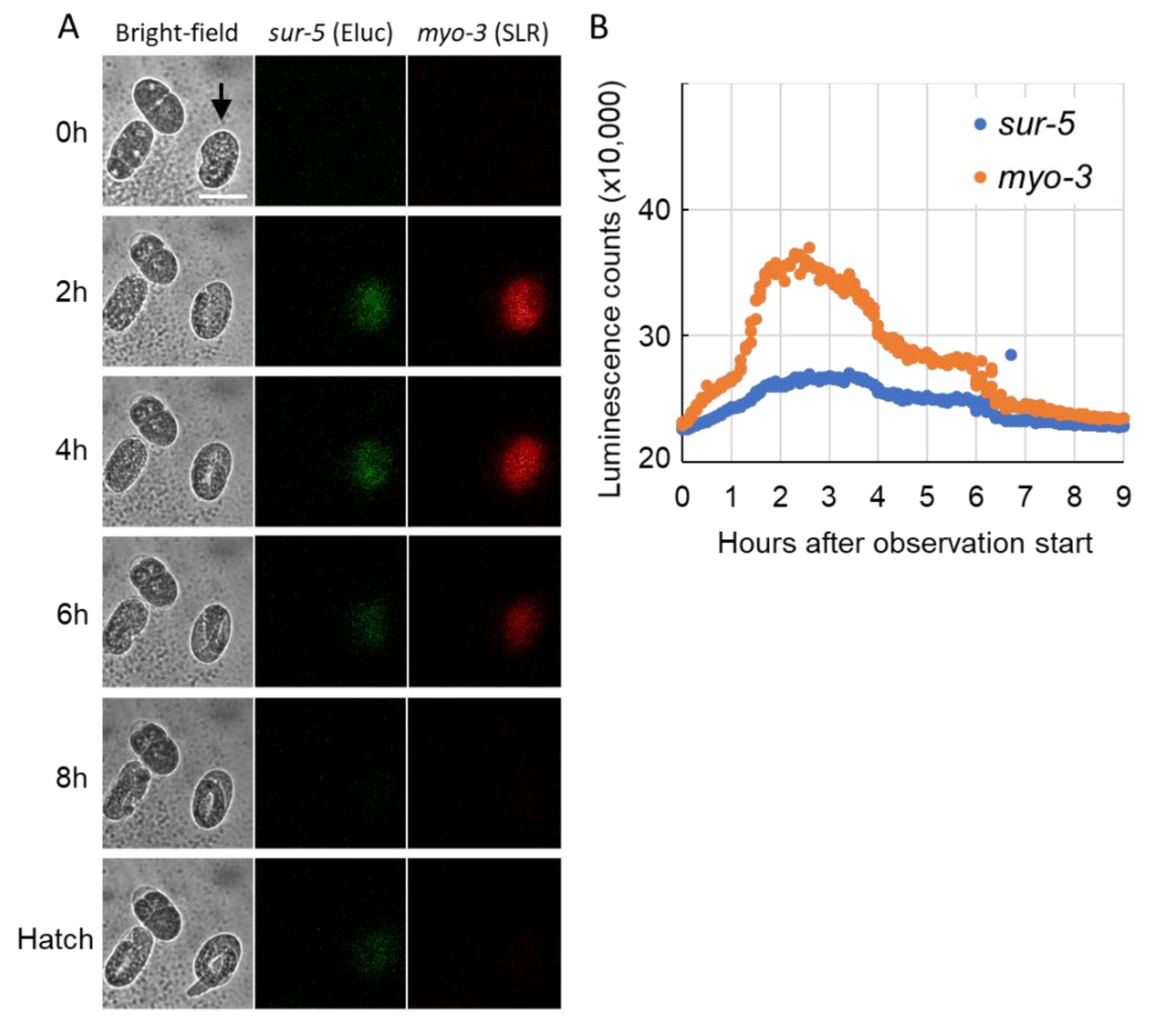
2.2. Bioluminescent Imaging in the Developing Embryo of C. elegans
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. Molecular Biology and Transgenic Animals
4.3. Real-Time Monitoring of Gene Expression by Bioluminescent Imaging
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Badr, C.E.; Tannous, B.A. Bioluminescence imaging: Progress and applications. Trends Biotechnol. 2011, 29, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Mezzanotte, L.; van’t Root, M.; Karatas, H.; Goun, E.A.; Lowik, C. In Vivo Molecular Bioluminescence Imaging: New Tools and Applications. Trends Biotechnol. 2017, 35, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Rathbun, C.M.; Prescher, J.A. Bioluminescent Probes for Imaging Biology beyond the Culture Dish. Biochemistry 2017, 56, 5178–5184. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Kimura, T.; Sugata, K.; Enomoto, T.; Asakawa, A.; Kubota, H.; Ikeda, M.; Ohmiya, Y. Multicolor luciferase assay system: One-step monitoring of multiple gene expressions with a single substrate. Biotechniques 2005, 38, 891–894. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.; Ikeda, M.; Ohmiya, Y.; Nakajima, Y. Simultaneous monitoring of independent gene expression patterns in two types of cocultured fibroblasts with different color-emitting luciferases. BMC Biotechnol. 2008, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Enomoto, T.; Shimogawara, M.; Yasuda, K.; Nakajima, Y.; Ohmiya, Y. Bioluminescence imaging of dual gene expression at the single-cell level. Biotechniques 2010, 48, 460–462. [Google Scholar] [CrossRef] [PubMed]
- Yasunaga, M.; Nakajima, Y.; Ohmiya, Y. Dual-color bioluminescence imaging assay using green- and red-emitting beetle luciferases at subcellular resolution. Anal. Bioanal Chem. 2014, 406, 5735–5742. [Google Scholar] [CrossRef]
- Noguchi, T.; Michihata, T.; Nakamura, W.; Takumi, T.; Shimizu, R.; Yamamoto, M.; Ikeda, M.; Ohmiya, Y.; Nakajima, Y. Dual-color luciferase mouse directly demonstrates coupled expression of two clock genes. Biochemistry 2010, 49, 8053–8061. [Google Scholar] [CrossRef]
- Kapitan, M.; Eichhof, I.; Lagadec, Q.; Ernst, J.F. Click beetle luciferases as dual reporters of gene expression in Candida albicans. Microbiology 2016, 162, 1310–1320. [Google Scholar] [CrossRef]
- Ogura, R.; Matsuo, N.; Wako, N.; Tanaka, T.; Ono, S.; Hiratsuka, K. Multi-color luciferases as reporters for monitoring transient gene expression in higher plants. Plant Biotechnol. 2005, 22, 151–155. [Google Scholar] [CrossRef][Green Version]
- Olmedo, M.; Geibel, M.; Artal-Sanz, M.; Merrow, M. A High-Throughput Method for the Analysis of Larval Developmental Phenotypes in Caenorhabditis elegans. Genetics 2015, 201, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Goya, M.E.; Romanowski, A.; Caldart, C.S.; Benard, C.Y.; Golombek, D.A. Circadian rhythms identified in Caenorhabditis elegans by in vivo long-term monitoring of a bioluminescent reporter. Proc. Natl. Acad. Sci. USA 2016, 113, E7837–E7845. [Google Scholar] [CrossRef] [PubMed]
- Lagido, C.; McLaggan, D.; Flett, A.; Pettitt, J.; Glover, L.A. Rapid sublethal toxicity assessment using bioluminescent Caenorhabditis elegans, a novel whole-animal metabolic biosensor. Toxicol. Sci. 2009, 109, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Orita, S.; Han, M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell Biol. 1998, 18, 4556–4564. [Google Scholar] [CrossRef]
- Dibb, N.J.; Maruyama, I.N.; Krause, M.; Karn, J. Sequence analysis of the complete Caenorhabditis elegans myosin heavy chain gene family. J. Mol. Biol. 1989, 205, 603–613. [Google Scholar] [CrossRef]
- Okkema, P.G.; Harrison, S.W.; Plunger, V.; Aryana, A.; Fire, A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 1993, 135, 385–404. [Google Scholar]
- Yasunaga, M.; Murotomi, K.; Abe, H.; Yamazaki, T.; Nishii, S.; Ohbayashi, T.; Oshimura, M.; Noguchi, T.; Niwa, K.; Ohmiya, Y.; et al. Highly sensitive luciferase reporter assay using a potent destabilization sequence of calpain 3. J. Biotechnol. 2015, 194, 115–123. [Google Scholar] [CrossRef]
- Mazo-Vargas, A.; Park, H.; Aydin, M.; Buchler, N.E. Measuring fast gene dynamics in single cells with time-lapse luminescence microscopy. Mol. Biol. Cell 2014, 25, 3699–3708. [Google Scholar] [CrossRef]
- Fox, R.M.; Watson, J.D.; Von Stetina, S.E.; McDermott, J.; Brodigan, T.M.; Fukushige, T.; Krause, M.; Miller, D.M., 3rd. The embryonic muscle transcriptome of Caenorhabditis elegans. Genome Biol. 2007, 8, R188. [Google Scholar] [CrossRef]
- Levin, M.; Hashimshony, T.; Wagner, F.; Yanai, I. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev. Cell 2012, 22, 1101–1108. [Google Scholar] [CrossRef]
- Mello, C.C.; Kramer, J.M.; Stinchcomb, D.; Ambros, V. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991, 10, 3959–3970. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doi, M.; Sato, M.; Ohmiya, Y. In Vivo Simultaneous Analysis of Gene Expression by Dual-Color Luciferases in Caenorhabditis elegans. Int. J. Mol. Sci. 2021, 22, 119. https://doi.org/10.3390/ijms22010119
Doi M, Sato M, Ohmiya Y. In Vivo Simultaneous Analysis of Gene Expression by Dual-Color Luciferases in Caenorhabditis elegans. International Journal of Molecular Sciences. 2021; 22(1):119. https://doi.org/10.3390/ijms22010119
Chicago/Turabian StyleDoi, Motomichi, Megumi Sato, and Yoshihiro Ohmiya. 2021. "In Vivo Simultaneous Analysis of Gene Expression by Dual-Color Luciferases in Caenorhabditis elegans" International Journal of Molecular Sciences 22, no. 1: 119. https://doi.org/10.3390/ijms22010119
APA StyleDoi, M., Sato, M., & Ohmiya, Y. (2021). In Vivo Simultaneous Analysis of Gene Expression by Dual-Color Luciferases in Caenorhabditis elegans. International Journal of Molecular Sciences, 22(1), 119. https://doi.org/10.3390/ijms22010119




