Exosomes: Cell-Derived Nanoplatforms for the Delivery of Cancer Therapeutics
Abstract
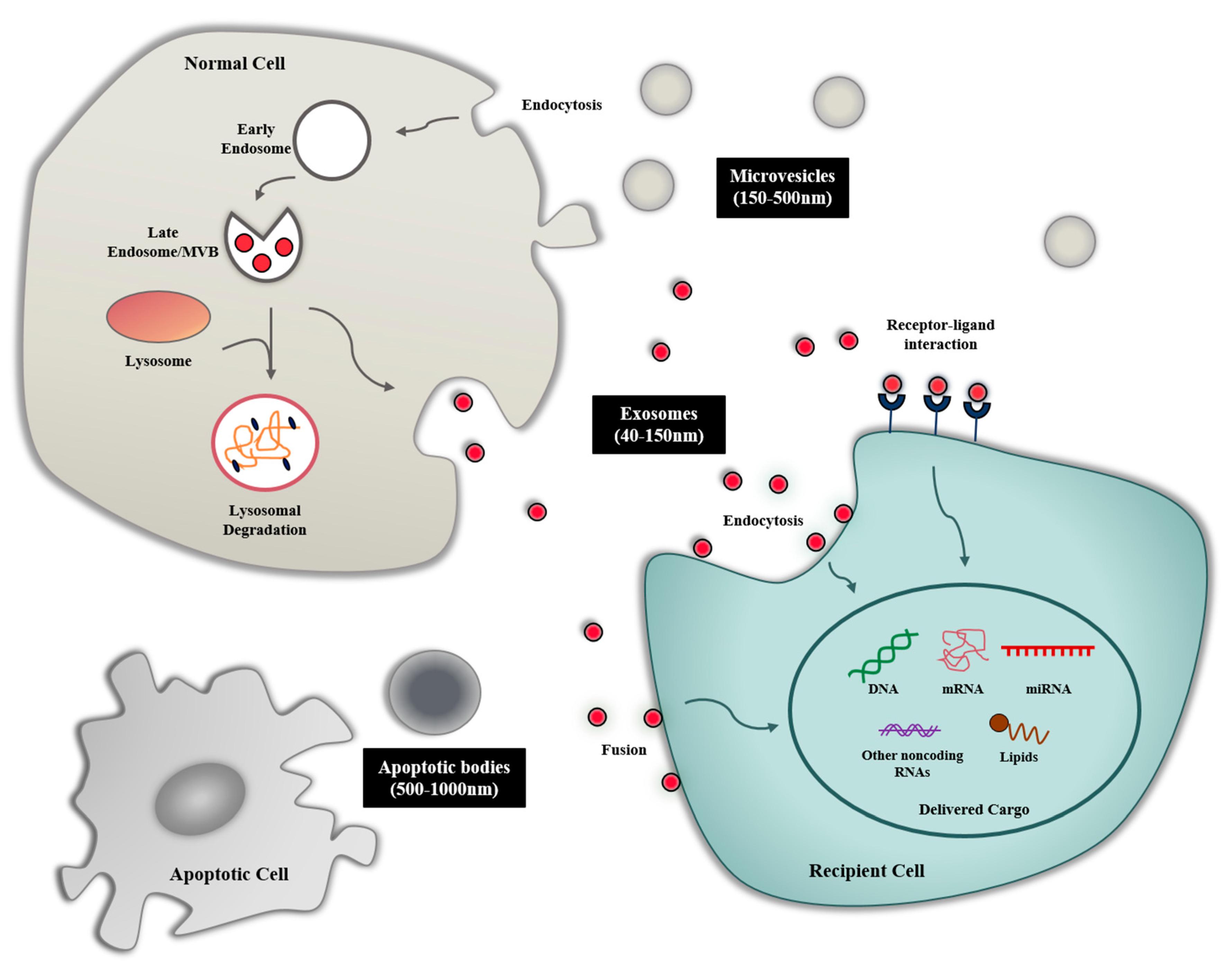
:1. Introduction
2. Biogenesis and Composition of Exosomes
2.1. Biogenesis of Exosome
2.2. Exosome Composition
3. Exosome-Mediated Intercellular Communication
3.1. Exosome Uptake
3.2. Exosome-Mediated Intercellular Communication in Tumor Microenvironment
4. Exosomes as Drug Delivery Vehicles
4.1. Encapsulation of Therapeutic Molecules
4.1.1. Co-Incubation
4.1.2. Membrane Permeability Enhancement
4.1.3. Cytoplasmic Abundance in Donor Cells
4.1.4. Selective Encapsulation in Exosome
4.2. Modification of Exosomal Membranes
5. Therapeutic Applications of Exosomes for Cancer Therapy
5.1. Tumor-Derived Exosomes
| Source of Exosome | Strategy | Outcome | Target Cancer Type | References |
|---|---|---|---|---|
| Tumor cell-derived exosome | ||||
| Human breast cancer (MDA-MB-231) | Loading with doxorubicin through electroporation | Inhibition of tumor progression and enhancing the cytotoxicity of doxorubicin | Breast | [63] |
| Human breast cancer (MDA-MB-231 and BT-474) | Carrying transforming growth factor-β (TGF-β) and interleukin-10 (IL-10) which exhibit the immune-regulatory functions, a potent immunosuppressive factor on T cell expression | Providing biomarkers for cancer diagnosis | Breast | [82] |
| Human glioblastoma (A172) | Carrying angiogenic proteins and RNA that induce new vessels at the early stage of cancer development | Providing biomarkers for cancer diagnosis | Glioblastoma | [38] |
| Non-small cell lung cancer (from NSCLC patients) | Carrying exosomal proteins such as alpha-2-HS-glycoprotein (AHSG) and extracellular matrix protein 1 (ECM1) | Providing biomarkers for cancer diagnosis | Non-small cell lung cancer (NSCLC) | [39] |
| Colorectal cancer | Increasing the transfer of small molecules including growth factors, chemokines, and RNAs | Providing biomarkers for cancer diagnosis | Colorectal cancer | [40] |
| Stem cell-derived exosome | ||||
| Mouse bone marrow MSCs (BM-MSCs) | Carrying MiR-16, which downregulates the expression of VEGF in the TME | Inhibition of angiogenesis | Breast cancer (4T1) | [84] |
| Human MSCs | Carrying MiR-100, which downregulates the expression of VEGF by modulating mTOR/HIF-1α signaling | Inhibition of angiogenesis | Breast cancer (MCF-7 and MDA-MB-231) | [85] |
| Human MSCs | Carrying MiR-124, which induces S-phase arrest through the downregulation of other CDKs | Inhibition of proliferation | Ovarian cancer | [86] |
| Immune cell-derived exosome | ||||
| Dendritic cell (DC) | Activating NK cells and T cells and inducing the secretion of interferon-γ (IFN-γ) | Inhibition of tumor progression | Melanoma (B16), Colon adenocarcinoma (MC38), Squamous cell carcinoma (KLN205) | [87,88,89] |
| Non-small cell lung cancer (NSCLC) | [90] | |||
| Loading factors that stimulate a wide range of immune cells to enhance antigen-specific T cell responses | Inhibition of tumor progression | Melanoma (B16/OVA) | [91] | |
| Enhancing antitumor immunity through TRL3 stimulation during the maturation of bone marrow derived DCs | Improve antitumor immunity and application in therapeutic cancer vaccines | Melanoma (B16F10) | [92] | |
| Expressing hepatocellular carcinoma antigen α-fetoprotein through lentivirus transfection | Inhibition of tumor progression | Hepatocellular carcinoma | [93] | |
| Natural-killer cell (NK) | Increasing the proliferation rate of NK cells involving FasL and perforin | Improve antitumor immunity | Melanoma | [94] |
| Treatment with dextran sulfate which block scavenger receptor A and preventing the ingestion of exosomes in the liver | Inhibition of tumor progression and improve antitumor immunity | Glioblastoma | [95] | |
| Activation of human NK cells with artificial antigen-presenting cells | Inhibition of proliferation | Acute lymphoblastic leukemia (SupB15, NALM-6), neuroblastoma (CHLA-136, CHLA-255), and breast cancer (MCF7) | [96] | |
| CD8 + T cells | Inducing the apoptosis of mesenchymal stem cells | Inhibition of tumor progression | CMS5a, CMS5m, CMS7, CT26, 4T1, B16 and B16F10 | [97] |
| Macrophages | Enhancing a pro-inflammatory cytokine, which induces the cytotoxic T cell immune response | Improve antitumor immunity and application in therapeutic cancer vaccines | Melanoma (B16F10) | [98] |
| Loading paclitaxel and doxorubicin through various methods to overcome multidrug resistance in MDCKMDR1 (Pgp+) cells | Enhancing the cytotoxicity of paclitaxel and doxorubicin | Murine Lewis lung carcinoma cell subline (3LL-M27) | [49] | |
| Other cells-derived exosome | ||||
| Human embryonic kidney 293T (HEK293T) | Carrying Imatinib or BCR-ABL siRNA which express IL-3 | Inhibition of tumor progression | Chronic myelogenous leukemia (LAMA84, K562) | [73] |
| Carrying super-repressor IκB (srIκB) to a therapeutic target | Inhibition of inflammatory responses | Monocytic THP-1 cells and human umbilical vein endothelial cells | [99] | |
| Carrying therapeutic GPI-anchored hyaluronidase to the overly accumulated ECM | Inhibition of tumor progression and activation of infiltration of T cells | Prostate cancer cell (PC3) | [100] | |
| Loading with doxorubicin through electroporation | Inhibition of tumor progression | Primary pulmonary artery smooth muscle cells | [101] | |
| Carrying MiR-497 which suppress cell proliferation, migration and angiogenesis of tumors | Inhibition of tumor progression | Non-small cell lung cancer (NSCLC) | [102] | |
| Adipocyte | Alleviating lung cancer metastasis by activating MMP9 | Promoting cancer cell invasion and metastasis | Lung cancer (3LL) | [103] |
| Inducing a metabolic reprogramming in tumor cell | Promoting cancer cell invasion and migration | Melanoma | [104] |
5.2. Stem Cell-Derived Exosomes
5.3. Immune Cell-Derived Exosomes
5.4. Other Cells-Derived Exosomes
6. Challenges and Perspectives
6.1. Large-Scale Production for the Therapeutic Use of Exosomes
6.2. The Heterogeneity of Exosomes
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EVs | Extracellular Vesicles |
| ILVs | Intraluminal Vesicles |
| ESCRT | Endosomal Sorting Complexes Required for Transport |
| MVBs | Multivesicular Bodies |
| MHC II | Myosin Heavy Chain Class II Proteins |
| miRNA | MicroRNA |
| mRNA | Messenger RNA |
| IFN-γ | Interferon γ |
| TEXs | Tumor-derived exosomes |
| NK | Natural-Killer |
| TME | Tumor Microenvironment |
| NSCLC | Non-Small Cell Lung Cancer |
| HGF | Hepatocyte Growth Factor |
| HEK293T | Human Embryonic Kidney 293T |
| OVA | Ovalbumin |
| IL3 | Interleukin 3 |
| SIRPα | Signal-Regulatory Protein Alpha |
| DCs | Dendritic Cells |
| MSCs | Mesenchymal Stem Cells |
| Dexs | Exosomes Derived from DCs |
References
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cossetti, C.; Iraci, N.; Mercer, T.R.; Leonardi, T.; Alpi, E.; Drago, D.; Alfaro-Cervello, C.; Saini, H.K.; Davis, M.P.; Schaeffer, J.; et al. Extracellular vesicles from neural stem cells transfer IFN-gamma via Ifngr1 to activate Stat1 signaling in target cells. Mol. Cell 2014, 56, 193–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pironti, G.; Strachan, R.T.; Abraham, D.; Mon-Wei Yu, S.; Chen, M.; Chen, W.; Hanada, K.; Mao, L.; Watson, L.J.; Rockman, H.A. Circulating Exosomes Induced by Cardiac Pressure Overload Contain Functional Angiotensin II Type 1 Receptors. Circulation 2015, 131, 2120–2130. [Google Scholar] [CrossRef] [PubMed]
- Lowenstein, P.R.; Mandel, R.J.; Xiong, W.D.; Kroeger, K.; Castro, M.G. Immune responses to adenovirus and adeno-associated vectors used for gene therapy of brain diseases: The role of immunological synapses in understanding the cell biology of neuroimmune interactions. Curr. Gene Ther. 2007, 7, 347–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.S.; Liu, F.; Huang, L. Implications of pharmacokinetic behavior of lipoplex for its inflammatory toxicity. Adv. Drug Deliv. Rev. 2005, 57, 689–698. [Google Scholar] [CrossRef]
- Ishida, T.; Ichihara, M.; Wang, X.; Yamamoto, K.; Kimura, J.; Majima, E.; Kiwada, H. Injection of PEGylated liposomes in rats elicits PEG-specific IgM, which is responsible for rapid elimination of a second dose of PEGylated liposomes. J. Control. Release 2006, 112, 15–25. [Google Scholar] [CrossRef]
- Sun, W.; Luo, J.D.; Jiang, H.; Duan, D.D. Tumor exosomes: A double-edged sword in cancer therapy. Acta Pharmacol. Sin. 2018, 39, 534–541. [Google Scholar] [CrossRef] [Green Version]
- Syn, N.L.; Wang, L.; Chow, E.K.; Lim, C.T.; Goh, B.C. Exosomes in Cancer Nanomedicine and Immunotherapy: Prospects and Challenges. Trends Biotechnol. 2017, 35, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Tan, E.; Sharghi-Namini, S.; Asada, H.H. Exosomes in Cancer Microenvironment and Beyond: Have we Overlooked these Extracellular Messengers? Cancer Microenviron. 2012, 5, 323–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booth, A.M.; Fang, Y.; Fallon, J.K.; Yang, J.M.; Hildreth, J.E.; Gould, S.J. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 2006, 172, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteomics 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Raposo, G. Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Thery, C. Biogenesis and secretion of exosomes. Curr Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Gusachenko, O.N.; Zenkova, M.A.; Vlassov, V.V. Nucleic acids in exosomes: Disease markers and intercellular communication molecules. Biochemistry 2013, 78, 1–7. [Google Scholar] [CrossRef]
- Urbanelli, L.; Magini, A.; Buratta, S.; Brozzi, A.; Sagini, K.; Polchi, A.; Tancini, B.; Emiliani, C. Signaling pathways in exosomes biogenesis, secretion and fate. Genes 2013, 4, 152–170. [Google Scholar] [CrossRef] [Green Version]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batagov, A.O.; Kurochkin, I.V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol. Direct 2013, 8, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 2010, 39, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Chen, L.; Huang, Y.; Li, K.; Jinye, A.; Fan, T.; Zhao, R.; Xia, X.; Shen, B.; Du, J.; et al. Exosome-delivered TRPP2 siRNA inhibits the epithelial-mesenchymal transition of FaDu cells. Oncol. Lett. 2019, 17, 1953–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannafon, B.N.; Gin, A.L.; Xu, Y.F.; Bruns, M.; Calloway, C.L.; Ding, W.Q. Metastasis-associated protein 1 (MTA1) is transferred by exosomes and contributes to the regulation of hypoxia and estrogen signaling in breast cancer cells. Cell Commun. Signal. 2019, 17, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lugini, L.; Cecchetti, S.; Huber, V.; Luciani, F.; Macchia, G.; Spadaro, F.; Paris, L.; Abalsamo, L.; Colone, M.; Molinari, A.; et al. Immune surveillance properties of human NK cell-derived exosomes. J. Immunol. 2012, 189, 2833–2842. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Li, X.; Wu, X.; Zhang, T.; Zhu, Q.; Wang, X.; Wang, H.; Wang, K.; Lin, Y.; Wang, X. Exosomes Released from Tumor-Associated Macrophages Transfer miRNAs That Induce a Treg/Th17 Cell Imbalance in Epithelial Ovarian Cancer. Cancer Immunol. Res. 2018, 6, 1578–1592. [Google Scholar] [CrossRef] [Green Version]
- Mahaweni, N.M.; Kaijen-Lambers, M.E.; Dekkers, J.; Aerts, J.G.; Hegmans, J.P. Tumour-derived exosomes as antigen delivery carriers in dendritic cell-based immunotherapy for malignant mesothelioma. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Clayton, A.; Mitchell, J.P.; Court, J.; Linnane, S.; Mason, M.D.; Tabi, Z. Human tumor-derived exosomes down-modulate NKG2D expression. J. Immunol. 2008, 180, 7249–7258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wieckowski, E.U.; Visus, C.; Szajnik, M.; Szczepanski, M.J.; Storkus, W.J.; Whiteside, T.L. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J. Immunol. 2009, 183, 3720–3730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; De Veirman, K.; Faict, S.; Frassanito, M.A.; Ribatti, D.; Vacca, A.; Menu, E. Multiple myeloma exosomes establish a favourable bone marrow microenvironment with enhanced angiogenesis and immunosuppression. J. Pathol. 2016, 239, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Dombroski, J.A.; King, M.R. Engineering of Exosomes to Target Cancer Metastasis. Cell Mol. Bioeng 2020, 13, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, T.M.; Alpaugh, M.L.; Barsky, S.H.; Rappa, G.; Lorico, A. Breast Cancer-Derived Extracellular Vesicles: Characterization and Contribution to the Metastatic Phenotype. Biomed. Res. Int. 2015, 2015, 634865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satake, T.; Suetsugu, A.; Nakamura, M.; Kunisada, T.; Saji, S.; Moriwaki, H.; Shimizu, M.; Hoffman, R.M. Color-coded Imaging of the Fate of Cancer-cell-derived Exosomes During Pancreatic Cancer Metastases in a Nude-mouse Model. Anticancer Res. 2019, 39, 4055–4060. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Li, B.; Wei, Y.; Zhao, Y.; Wang, L.; Zhang, P.; Yang, J.; He, W.; Chen, H.; Jiao, Z.; et al. Tumor-derived exosomes induce PD1(+) macrophage population in human gastric cancer that promotes disease progression. Oncogenesis 2018, 7, 41. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, M.; Rezaie, J. Tumor cells derived-exosomes as angiogenenic agents: Possible therapeutic implications. J. Transl. Med. 2020, 18, 249. [Google Scholar] [CrossRef]
- Niu, L.; Song, X.; Wang, N.; Xue, L.; Song, X.; Xie, L. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci. 2019, 110, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Mousavi, S.; Moallem, R.; Hassanian, S.M.; Sadeghzade, M.; Mardani, R.; Ferns, G.A.; Khazaei, M.; Avan, A. Tumor-derived exosomes: Potential biomarkers and therapeutic target in the treatment of colorectal cancer. J. Cell. Physiol. 2019, 234, 12422–12432. [Google Scholar] [CrossRef]
- Arranja, A.G.; Pathak, V.; Lammers, T.; Shi, Y. Tumor-targeted nanomedicines for cancer theranostics. Pharmacol. Res. 2017, 115, 87–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, X.; Xiang, X.; Grizzle, W.; Sun, D.; Zhang, S.; Axtell, R.C.; Ju, S.; Mu, J.; Zhang, L.; Steinman, L.; et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011, 19, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.C. Electroporation: A general phenomenon for manipulating cells and tissues. J. Cell. Biochem. 1993, 51, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Raiker, R.S.; Jay, S.M. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol. Pharm. 2015, 12, 3650–3657. [Google Scholar] [CrossRef] [Green Version]
- Jamur, M.C.; Oliver, C. Permeabilization of cell membranes. Methods Mol. Biol. 2010, 588, 63–66. [Google Scholar] [PubMed]
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Control. Release 2015, 205, 35–44. [Google Scholar] [CrossRef]
- Podolak, I.; Galanty, A.; Sobolewska, D. Saponins as cytotoxic agents: A review. Phytochem. Rev. 2010, 9, 425–474. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef] [Green Version]
- Shtam, T.A.; Kovalev, R.A.; Varfolomeeva, E.Y.; Makarov, E.M.; Kil, Y.V.; Filatov, M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. 2013, 11, 88. [Google Scholar] [CrossRef] [Green Version]
- Pascucci, L.; Cocce, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Vigano, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Bai, M.; Wang, J.; Zhu, K.; Liu, R.; Ge, S.; Li, J.; Ning, T.; Deng, T.; et al. Exosomes serve as nanoparticles to suppress tumor growth and angiogenesis in gastric cancer by delivering hepatocyte growth factor siRNA. Cancer Sci. 2018, 109, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, A.K.; Luciani, N.; Gazeau, F.; Aubertin, K.; Bonneau, S.; Chauvierre, C.; Letourneur, D.; Wilhelm, C. Combining magnetic nanoparticles with cell derived microvesicles for drug loading and targeting. Nanomedicine 2015, 11, 645–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Bonito, P.; Chiozzini, C.; Arenaccio, C.; Anticoli, S.; Manfredi, F.; Olivetta, E.; Ferrantelli, F.; Falcone, E.; Ruggieri, A.; Federico, M. Antitumor HPV E7-specific CTL activity elicited by in vivo engineered exosomes produced through DNA inoculation. Int. J. Nanomed. 2017, 12, 4579–4591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeelenberg, I.S.; Ostrowski, M.; Krumeich, S.; Bobrie, A.; Jancic, C.; Boissonnas, A.; Delcayre, A.; Le Pecq, J.B.; Combadiere, B.; Amigorena, S.; et al. Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Res. 2008, 68, 1228–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Gassart, A.; Trentin, B.; Martin, M.; Hocquellet, A.; Bette-Bobillo, P.; Mamoun, R.; Vidal, M. Exosomal sorting of the cytoplasmic domain of bovine leukemia virus TM Env protein. Cell Biol. Int. 2009, 33, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Yim, N.; Ryu, S.W.; Choi, K.; Lee, K.R.; Lee, S.; Choi, H.; Kim, J.; Shaker, M.R.; Sun, W.; Park, J.H.; et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun 2016, 7, 12277. [Google Scholar] [CrossRef]
- Batagov, A.O.; Kuznetsov, V.A.; Kurochkin, I.V. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics 2011, 12 (Suppl. 3), S18. [Google Scholar] [CrossRef] [Green Version]
- Teng, Y.; Ren, Y.; Hu, X.; Mu, J.; Samykutty, A.; Zhuang, X.; Deng, Z.; Kumar, A.; Zhang, L.; Merchant, M.L.; et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat. Commun. 2017, 8, 14448. [Google Scholar] [CrossRef]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Blomme, A.; Fahmy, K.; Peulen, O.; Costanza, B.; Fontaine, M.; Struman, I.; Baiwir, D.; de Pauw, E.; Thiry, M.; Bellahcene, A.; et al. Myoferlin is a novel exosomal protein and functional regulator of cancer-derived exosomes. Oncotarget 2016, 7, 83669–83683. [Google Scholar] [CrossRef] [PubMed]
- Hadla, M.; Palazzolo, S.; Corona, G.; Caligiuri, I.; Canzonieri, V.; Toffoli, G.; Rizzolio, F. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine 2016, 11, 2431–2441. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, N.; Lankar, D.; Faure, F.; Regnault, A.; Dumont, C.; Raposo, G.; Hivroz, C. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J. Immunol. 2002, 168, 3235–3241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamichhane, T.N.; Jeyaram, A.; Patel, D.B.; Parajuli, B.; Livingston, N.K.; Arumugasaamy, N.; Schardt, J.S.; Jay, S.M. Oncogene Knockdown via Active Loading of Small RNAs into Extracellular Vesicles by Sonication. Cell Mol. Bioeng. 2016, 9, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.T.; Umezaki, K.; Sawada, S.; Mukai, S.A.; Sasaki, Y.; Harada, N.; Shiku, H.; Akiyoshi, K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016, 6, 21933. [Google Scholar] [CrossRef] [Green Version]
- Yerneni, S.S.; Lathwal, S.; Shrestha, P.; Shirwan, H.; Matyjaszewski, K.; Weiss, L.; Yolcu, E.S.; Campbell, P.G.; Das, S.R. Rapid On-Demand Extracellular Vesicle Augmentation with Versatile Oligonucleotide Tethers. ACS Nano 2019, 13, 10555–10565. [Google Scholar] [CrossRef]
- Martinelli, C.; Gabriele, F.; Dini, E.; Carriero, F.; Bresciani, G.; Slivinschi, B.; Dei Giudici, M.; Zanoletti, L.; Manai, F.; Paolillo, M.; et al. Development of Artificial Plasma Membranes Derived Nanovesicles Suitable for Drugs Encapsulation. Cells 2020, 9, 1626. [Google Scholar] [CrossRef]
- Nasiri Kenari, A.; Cheng, L.; Hill, A.F. Methods for loading therapeutics into extracellular vesicles and generating extracellular vesicles mimetic-nanovesicles. Methods 2020, 177, 103–113. [Google Scholar] [CrossRef]
- Kalimuthu, S.; Gangadaran, P.; Rajendran, R.L.; Zhu, L.; Oh, J.M.; Lee, H.W.; Gopal, A.; Baek, S.H.; Jeong, S.Y.; Lee, S.W.; et al. A New Approach for Loading Anticancer Drugs into Mesenchymal Stem Cell-Derived Exosome Mimetics for Cancer Therapy. Front. Pharmacol. 2018, 9, 1116. [Google Scholar] [CrossRef]
- Nasiri Kenari, A.; Kastaniegaard, K.; Greening, D.W.; Shambrook, M.; Stensballe, A.; Cheng, L.; Hill, A.F. Proteomic and Post-Translational Modification Profiling of Exosome-Mimetic Nanovesicles Compared to Exosomes. Proteomics 2019, 19, e1800161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Hong, Y.; Cho, E.; Kim, G.B.; Kim, I.S. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell. Vesicles 2018, 7, 1440131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellavia, D.; Raimondo, S.; Calabrese, G.; Forte, S.; Cristaldi, M.; Patinella, A.; Memeo, L.; Manno, M.; Raccosta, S.; Diana, P.; et al. Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics 2017, 7, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Koh, E.; Lee, E.J.; Nam, G.H.; Hong, Y.; Cho, E.; Yang, Y.; Kim, I.S. Exosome-SIRPalpha, a CD47 blockade increases cancer cell phagocytosis. Biomaterials 2017, 121, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Choi, E.S.; Song, J.; Kang, Y.Y.; Mok, H. Mannose-Modified Serum Exosomes for the Elevated Uptake to Murine Dendritic Cells and Lymphatic Accumulation. Macromol. Biosci. 2019, 19, e1900042. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Yuan, D.; Deygen, I.; Klyachko, N.L.; Kabanov, A.V.; Batrakova, E.V. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: In vitro and in vivo evaluations. Nanomedicine 2018, 14, 195–204. [Google Scholar] [CrossRef]
- Lloret-Llinares, M.; Karadoulama, E.; Chen, Y.; Wojenski, L.A.; Villafano, G.J.; Bornholdt, J.; Andersson, R.; Core, L.; Sandelin, A.; Jensen, T.H. The RNA exosome contributes to gene expression regulation during stem cell differentiation. Nucleic Acids Res. 2018, 46, 11502–11513. [Google Scholar] [CrossRef]
- Gu, X.; Erb, U.; Buchler, M.W.; Zoller, M. Improved vaccine efficacy of tumor exosome compared to tumor lysate loaded dendritic cells in mice. Int. J. Cancer 2015, 136, E74–E84. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, J. Potential of cancer cell-derived exosomes in clinical application: A review of recent research advances. Clin. Ther. 2014, 36, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Rong, L.; Li, R.; Li, S.; Luo, R. Immunosuppression of breast cancer cells mediated by transforming growth factor-beta in exosomes from cancer cells. Oncol. Lett. 2016, 11, 500–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Park, S.R.; Jung, B.K.; Jeon, Y.K.; Lee, Y.S.; Kim, M.K.; Kim, Y.G.; Jang, J.Y.; Kim, C.W. Exosomes derived from mesenchymal stem cells suppress angiogenesis by down-regulating VEGF expression in breast cancer cells. PLoS ONE 2013, 8, e84256. [Google Scholar] [CrossRef] [Green Version]
- Pakravan, K.; Babashah, S.; Sadeghizadeh, M.; Mowla, S.J.; Mossahebi-Mohammadi, M.; Ataei, F.; Dana, N.; Javan, M. MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1alpha/VEGF signaling axis in breast cancer cells. Cell. Oncol. 2017, 40, 457–470. [Google Scholar] [CrossRef]
- Reza, A.; Choi, Y.J.; Yasuda, H.; Kim, J.H. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci. Rep. 2016, 6, 38498. [Google Scholar] [CrossRef]
- Viaud, S.; Terme, M.; Flament, C.; Taieb, J.; Andre, F.; Novault, S.; Escudier, B.; Robert, C.; Caillat-Zucman, S.; Tursz, T.; et al. Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: A role for NKG2D ligands and IL-15Ralpha. PLoS ONE 2009, 4, e4942. [Google Scholar] [CrossRef]
- Simhadri, V.R.; Reiners, K.S.; Hansen, H.P.; Topolar, D.; Simhadri, V.L.; Nohroudi, K.; Kufer, T.A.; Engert, A.; Pogge von Strandmann, E. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS ONE 2008, 3, e3377. [Google Scholar] [CrossRef] [Green Version]
- Munich, S.; Sobo-Vujanovic, A.; Buchser, W.J.; Beer-Stolz, D.; Vujanovic, N.L. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology 2012, 1, 1074–1083. [Google Scholar] [CrossRef] [Green Version]
- Besse, B.; Charrier, M.; Lapierre, V.; Dansin, E.; Lantz, O.; Planchard, D.; Le Chevalier, T.; Livartoski, A.; Barlesi, F.; Laplanche, A.; et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology 2016, 5, e1071008. [Google Scholar] [CrossRef] [Green Version]
- Gehrmann, U.; Hiltbrunner, S.; Georgoudaki, A.M.; Karlsson, M.C.; Naslund, T.I.; Gabrielsson, S. Synergistic induction of adaptive antitumor immunity by codelivery of antigen with alpha-galactosylceramide on exosomes. Cancer Res. 2013, 73, 3865–3876. [Google Scholar] [CrossRef] [Green Version]
- Damo, M.; Wilson, D.S.; Simeoni, E.; Hubbell, J.A. TLR-3 stimulation improves anti-tumor immunity elicited by dendritic cell exosome-based vaccines in a murine model of melanoma. Sci. Rep. 2015, 5, 17622. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Zuo, B.; Jing, R.; Gao, X.; Rao, Q.; Liu, Z.; Qi, H.; Guo, H.; Yin, H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 2017, 67, 739–748. [Google Scholar] [CrossRef]
- Zhu, L.; Kalimuthu, S.; Gangadaran, P.; Oh, J.M.; Lee, H.W.; Baek, S.H.; Jeong, S.Y.; Lee, S.W.; Lee, J.; Ahn, B.C. Exosomes Derived from Natural Killer Cells Exert Therapeutic Effect in Melanoma. Theranostics 2017, 7, 2732–2745. [Google Scholar] [CrossRef]
- Zhu, L.; Oh, J.M.; Gangadaran, P.; Kalimuthu, S.; Baek, S.H.; Jeong, S.Y.; Lee, S.W.; Lee, J.; Ahn, B.C. Targeting and Therapy of Glioblastoma in a Mouse Model Using Exosomes Derived from Natural Killer Cells. Front. Immunol. 2018, 9, 824. [Google Scholar] [CrossRef]
- Jong, A.Y.; Wu, C.H.; Li, J.; Sun, J.; Fabbri, M.; Wayne, A.S.; Seeger, R.C. Large-scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells. J. Extracell Vesicles 2017, 6, 1294368. [Google Scholar] [CrossRef] [Green Version]
- Seo, N.; Shirakura, Y.; Tahara, Y.; Momose, F.; Harada, N.; Ikeda, H.; Akiyoshi, K.; Shiku, H. Activated CD8(+) T cell extracellular vesicles prevent tumour progression by targeting of lesional mesenchymal cells. Nat. Commun. 2018, 9, 435. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Wang, Y.; Huang, L. Exosomes from M1-Polarized Macrophages Potentiate the Cancer Vaccine by Creating a Pro-inflammatory Microenvironment in the Lymph Node. Mol. Ther. 2017, 25, 1665–1675. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Kim, Y.; Mirzaaghasi, A.; Heo, J.; Kim, Y.N.; Shin, J.H.; Kim, S.; Kim, N.H.; Cho, E.S.; In Yook, J.; et al. Exosome-based delivery of super-repressor IkappaBalpha relieves sepsis-associated organ damage and mortality. Sci. Adv. 2020, 6, eaaz6980. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Nam, G.-H.; Koh, E.; Jeon, S.; Kim, G.B.; Jeong, C.; Kim, D.-H.; Yang, Y.; Kim, I.-S. Exosome as a Vehicle for Delivery of Membrane Protein Therapeutics, PH20, for Enhanced Tumor Penetration and Antitumor Efficacy. Adv. Funct. Mater. 2018, 28, 1703074. [Google Scholar] [CrossRef]
- Schindler, C.; Collinson, A.; Matthews, C.; Pointon, A.; Jenkinson, L.; Minter, R.R.; Vaughan, T.J.; Tigue, N.J. Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency. PLoS ONE 2019, 14, e0214545. [Google Scholar] [CrossRef] [Green Version]
- Jeong, K.; Yu, Y.J.; You, J.Y.; Rhee, W.J.; Kim, J.A. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab. Chip. 2020, 20, 548–557. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Guo, J.; Fei, X.; Yu, L.; Ma, S. Adipocyte-derived exosomes promote lung cancer metastasis by increasing MMP9 activity via transferring MMP3 to lung cancer cells. Oncotarget 2017, 8, 81880–81891. [Google Scholar] [CrossRef]
- Lazar, I.; Clement, E.; Dauvillier, S.; Milhas, D.; Ducoux-Petit, M.; LeGonidec, S.; Moro, C.; Soldan, V.; Dalle, S.; Balor, S.; et al. Adipocyte Exosomes Promote Melanoma Aggressiveness through Fatty Acid Oxidation: A Novel Mechanism Linking Obesity and Cancer. Cancer Res. 2016, 76, 4051–4057. [Google Scholar] [CrossRef] [Green Version]
- Crapnell, K.; Blaesius, R.; Hastings, A.; Lennon, D.P.; Caplan, A.I.; Bruder, S.P. Growth, differentiation capacity, and function of mesenchymal stem cells expanded in serum-free medium developed via combinatorial screening. Exp. Cell Res. 2013, 319, 1409–1418. [Google Scholar] [CrossRef]
- Shojaei, S.; Hashemi, S.M.; Ghanbarian, H.; Salehi, M.; Mohammadi-Yeganeh, S. Effect of mesenchymal stem cells-derived exosomes on tumor microenvironment: Tumor progression versus tumor suppression. J. Cell. Physiol. 2019, 234, 3394–3409. [Google Scholar] [CrossRef]
- Wen, C.; Seeger, R.C.; Fabbri, M.; Wang, L.; Wayne, A.S.; Jong, A.Y. Biological roles and potential applications of immune cell-derived extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1400370. [Google Scholar] [CrossRef] [Green Version]
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459. [Google Scholar] [CrossRef]
- Ewen, C.L.; Kane, K.P.; Bleackley, R.C. A quarter century of granzymes. Cell Death Differ. 2012, 19, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Kaspar, A.A.; Okada, S.; Kumar, J.; Poulain, F.R.; Drouvalakis, K.A.; Kelekar, A.; Hanson, D.A.; Kluck, R.M.; Hitoshi, Y.; Johnson, D.E.; et al. A distinct pathway of cell-mediated apoptosis initiated by granulysin. J. Immunol. 2001, 167, 350–356. [Google Scholar] [CrossRef]
- Keefe, D.; Shi, L.; Feske, S.; Massol, R.; Navarro, F.; Kirchhausen, T.; Lieberman, J. Perforin triggers a plasma membrane-repair response that facilitates CTL induction of apoptosis. Immunity 2005, 23, 249–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgents, J.E.; Moran, T.P.; West, M.L.; Davis, N.L.; Johnston, R.E.; Serody, J.S. The immunosuppressive tumor environment is the major impediment to successful therapeutic vaccination in Neu transgenic mice. J. Immunother. 2010, 33, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Gyorgy, B.; Hung, M.E.; Breakefield, X.O.; Leonard, J.N. Therapeutic applications of extracellular vesicles: Clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 439–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garzetti, L.; Menon, R.; Finardi, A.; Bergami, A.; Sica, A.; Martino, G.; Comi, G.; Verderio, C.; Farina, C.; Furlan, R. Activated macrophages release microvesicles containing polarized M1 or M2 mRNAs. J. Leukoc. Biol. 2014, 95, 817–825. [Google Scholar] [CrossRef]
- Charoenviriyakul, C.; Takahashi, Y.; Morishita, M.; Matsumoto, A.; Nishikawa, M.; Takakura, Y. Cell type-specific and common characteristics of exosomes derived from mouse cell lines: Yield, physicochemical properties, and pharmacokinetics. Eur. J. Pharm. Sci. 2017, 96, 316–322. [Google Scholar] [CrossRef]
- Willis, G.R.; Kourembanas, S.; Mitsialis, S.A. Toward Exosome-Based Therapeutics: Isolation, Heterogeneity, and Fit-for-Purpose Potency. Front. Cardiovasc. Med. 2017, 4, 63. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, T.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation. Eur. J. Pharm. Biopharm. 2016, 98, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Franzen, C.A.; Simms, P.E.; Van Huis, A.F.; Foreman, K.E.; Kuo, P.C.; Gupta, G.N. Characterization of uptake and internalization of exosomes by bladder cancer cells. Biomed. Res. Int. 2014, 2014, 619829. [Google Scholar] [CrossRef] [Green Version]
- Watson, D.C.; Bayik, D.; Srivatsan, A.; Bergamaschi, C.; Valentin, A.; Niu, G.; Bear, J.; Monninger, M.; Sun, M.; Morales-Kastresana, A.; et al. Efficient production and enhanced tumor delivery of engineered extracellular vesicles. Biomaterials 2016, 105, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Harmati, M.; Tarnai, Z.; Decsi, G.; Kormondi, S.; Szegletes, Z.; Janovak, L.; Dekany, I.; Saydam, O.; Gyukity-Sebestyen, E.; Dobra, G.; et al. Stressors alter intercellular communication and exosome profile of nasopharyngeal carcinoma cells. J. Oral. Pathol. Med. 2017, 46, 259–266. [Google Scholar] [CrossRef]
- Kanemoto, S.; Nitani, R.; Murakami, T.; Kaneko, M.; Asada, R.; Matsuhisa, K.; Saito, A.; Imaizumi, K. Multivesicular body formation enhancement and exosome release during endoplasmic reticulum stress. Biochem. Biophys. Res. Commun. 2016, 480, 166–172. [Google Scholar] [CrossRef] [PubMed]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, S.W.; Nguyen, J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J. Control. Release 2016, 228, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Baixauli, F.; Gutierrez-Vazquez, C.; Sanchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Willms, E.; Johansson, H.J.; Mager, I.; Lee, Y.; Blomberg, K.E.; Sadik, M.; Alaarg, A.; Smith, C.I.; Lehtio, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Chiang, C.Y.; Chen, C. Toward characterizing extracellular vesicles at a single-particle level. J. Biomed. Sci. 2019, 26, 9. [Google Scholar] [CrossRef]
- Corso, G.; Heusermann, W.; Trojer, D.; Gorgens, A.; Steib, E.; Voshol, J.; Graff, A.; Genoud, C.; Lee, Y.; Hean, J.; et al. Systematic characterization of extracellular vesicle sorting domains and quantification at the single molecule—single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J. Extracell. Vesicles 2019, 8, 1663043. [Google Scholar] [CrossRef]
- Lee, K.; Fraser, K.; Ghaddar, B.; Yang, K.; Kim, E.; Balaj, L.; Chiocca, E.A.; Breakefield, X.O.; Lee, H.; Weissleder, R. Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano 2018, 12, 494–503. [Google Scholar] [CrossRef]
- Escudier, B.; Dorval, T.; Chaput, N.; Andre, F.; Caby, M.P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [Green Version]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.A.; Delcayre, A.; et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, S.; Wei, D.; Wu, Z.; Zhou, X.; Wei, X.; Huang, H.; Li, G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol. Ther. 2008, 16, 782–790. [Google Scholar] [CrossRef] [PubMed]


| Method | Drug Loaded | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Passive Loading Methods | ||||
| Incubation with Drugs | Small hydrophobic molecules (curcumin and doxorubicin) | Simplest method Increasing solubility and stability of the hydrophobic drugs in blood circulation | Low drug-loading efficiency Not efficient for large molecules | [42,43] |
| Active Loading Methods | ||||
| Electroporation | Chemotherapeutic drug (doxorubicin and paclitaxel) | Loading with large molecules possible Applicable for nucleic acids | Low drug-loading efficiency (hydrophobic drugs) Cargo aggregation | [45,63,64] |
| Sonication | Chemotherapeutic drug (doxorubicin and paclitaxel), small RNAs | High drug-loading efficiency Applicable for nucleic acids | Deformation of membrane Low drug-loading efficiency (hydrophobic drugs) | [49,65] |
| Extrusion | Catalase | High drug-loading efficiency | Deformation of membrane Limitation of membrane | [50] |
| Freeze and Thaw Cycles | Proteins and peptides | Fusion of membranes possible | Low drug-loading efficiency Exosome aggregation | [50,66] |
| Click Chemistry | Drugs and nucleic acids | Quick and efficient reactions High specificity | Impairing the functionality of surface proteins | [67] |
| Exosome-mimic | ||||
| Mimetic Nanovesicles | Chemotherapeutic drug (doxorubicin and paclitaxel) | Easier to manufacture High the therapeutic delivery efficiency High yield production | Require to understand cargo loading (cellular uptake, cargo release, and fate of vesicles) | [68,69,70,71] |
| Cancer Type | Phase | Source of Exosomes | Results and Status | References |
|---|---|---|---|---|
| Melanoma | Phase I (n = 15) | Immature dendritic cells pulsed with MAGE 3 tumor peptides | Active, Not recruiting | [130] |
| Non-small cell lung cancer | Phase I (n = 4) | Immature dendritic cells pulsed with MAGE-A3, -A4, -A10, and MAGE-3DPO4 tumor peptide | Recruiting | [131] |
| Non-small cell lung cancer | Phase II (n = 22) | IFN-γ- matured dendritic cells pulsed with MAGE-A1, -A3, NY-ESO-1, Melan-A/MART1, MAGE-A3-DP04, EBV tumor peptides | Recruiting | [90] NCT01159288 |
| Colon cancer | Phase I (n = 40) | Autologous ascites combined with GM-CSF | Active, Not recruiting | [132] |
| Colon cancer | Phase I (n = 35) | Plant loaded with curcumin | Active, Not recruiting | NCT01294072 |
| Pancreatic cancer | Phase I (n = 28) | Mesenchymal stem cells | Not yet recruiting | NCT03608631 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Kim, E.H.; Kwak, G.; Chi, S.-G.; Kim, S.H.; Yang, Y. Exosomes: Cell-Derived Nanoplatforms for the Delivery of Cancer Therapeutics. Int. J. Mol. Sci. 2021, 22, 14. https://doi.org/10.3390/ijms22010014
Kim H, Kim EH, Kwak G, Chi S-G, Kim SH, Yang Y. Exosomes: Cell-Derived Nanoplatforms for the Delivery of Cancer Therapeutics. International Journal of Molecular Sciences. 2021; 22(1):14. https://doi.org/10.3390/ijms22010014
Chicago/Turabian StyleKim, Hyosuk, Eun Hye Kim, Gijung Kwak, Sung-Gil Chi, Sun Hwa Kim, and Yoosoo Yang. 2021. "Exosomes: Cell-Derived Nanoplatforms for the Delivery of Cancer Therapeutics" International Journal of Molecular Sciences 22, no. 1: 14. https://doi.org/10.3390/ijms22010014
APA StyleKim, H., Kim, E. H., Kwak, G., Chi, S.-G., Kim, S. H., & Yang, Y. (2021). Exosomes: Cell-Derived Nanoplatforms for the Delivery of Cancer Therapeutics. International Journal of Molecular Sciences, 22(1), 14. https://doi.org/10.3390/ijms22010014






