CKAP2L Knockdown Exerts Antitumor Effects by Increasing miR-4496 in Glioblastoma Cell Lines
Abstract
:1. Introduction
2. Results
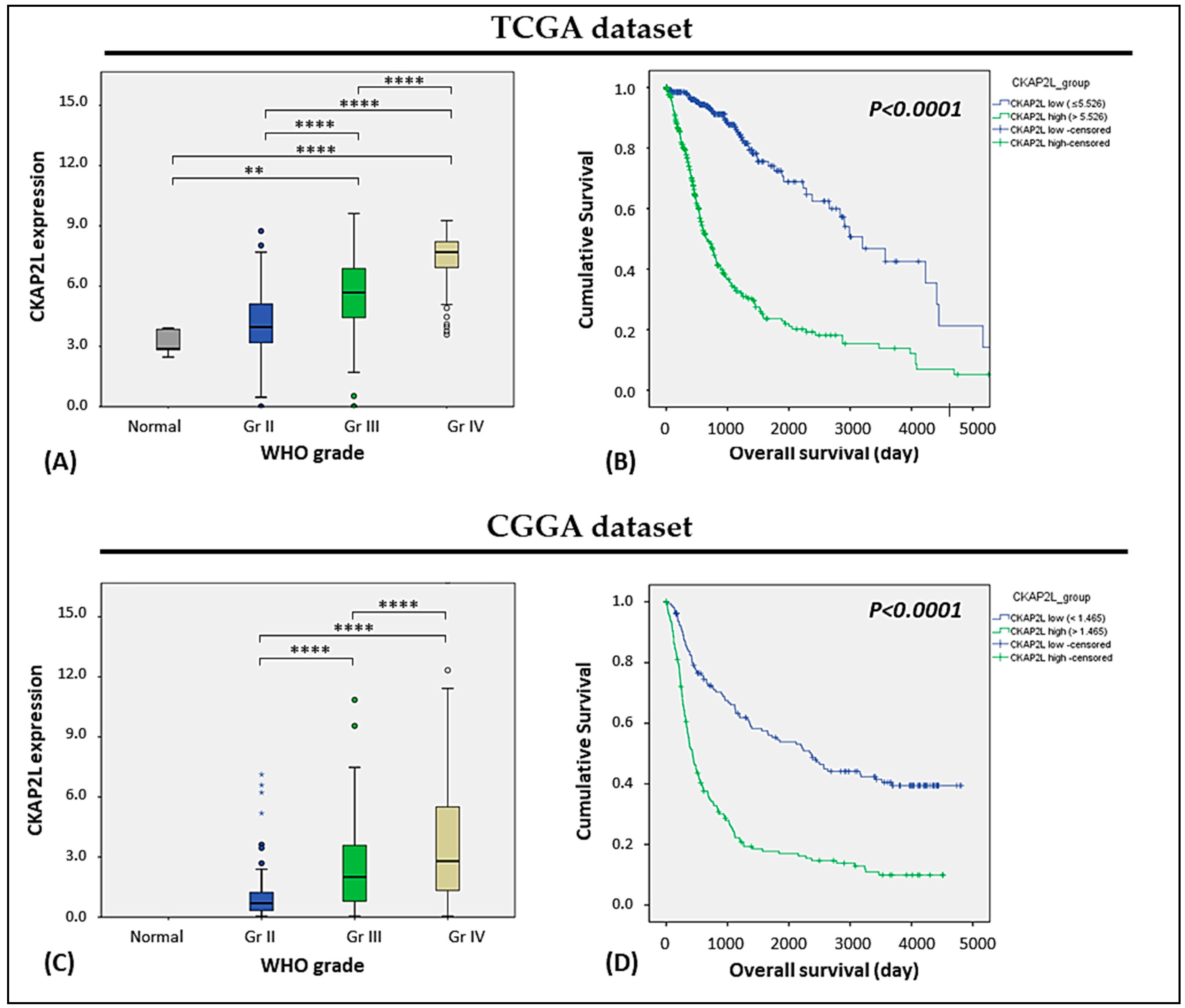
2.1. CKAP2L Expression Correlated with Tumor Grade and Overall Survival in Glioma
2.2. MITOTIC_SPINDLE Is the Key Enriched Phenotype in the High CKAP2L Expression Group
2.3. CKAP2L Staining Correlates with WHO Grade in Glioma Tissues
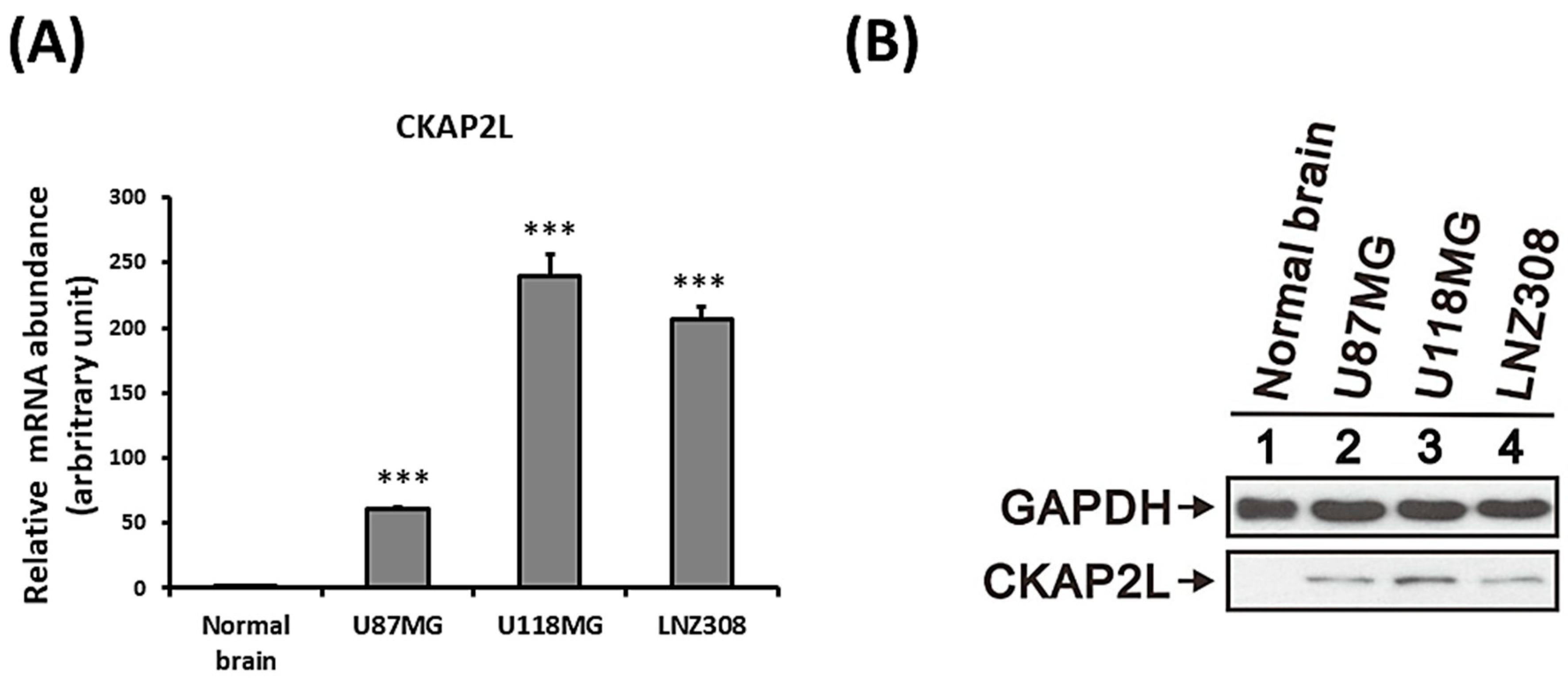
2.4. Increased CKAP2L Expression in Human Glioma Cells
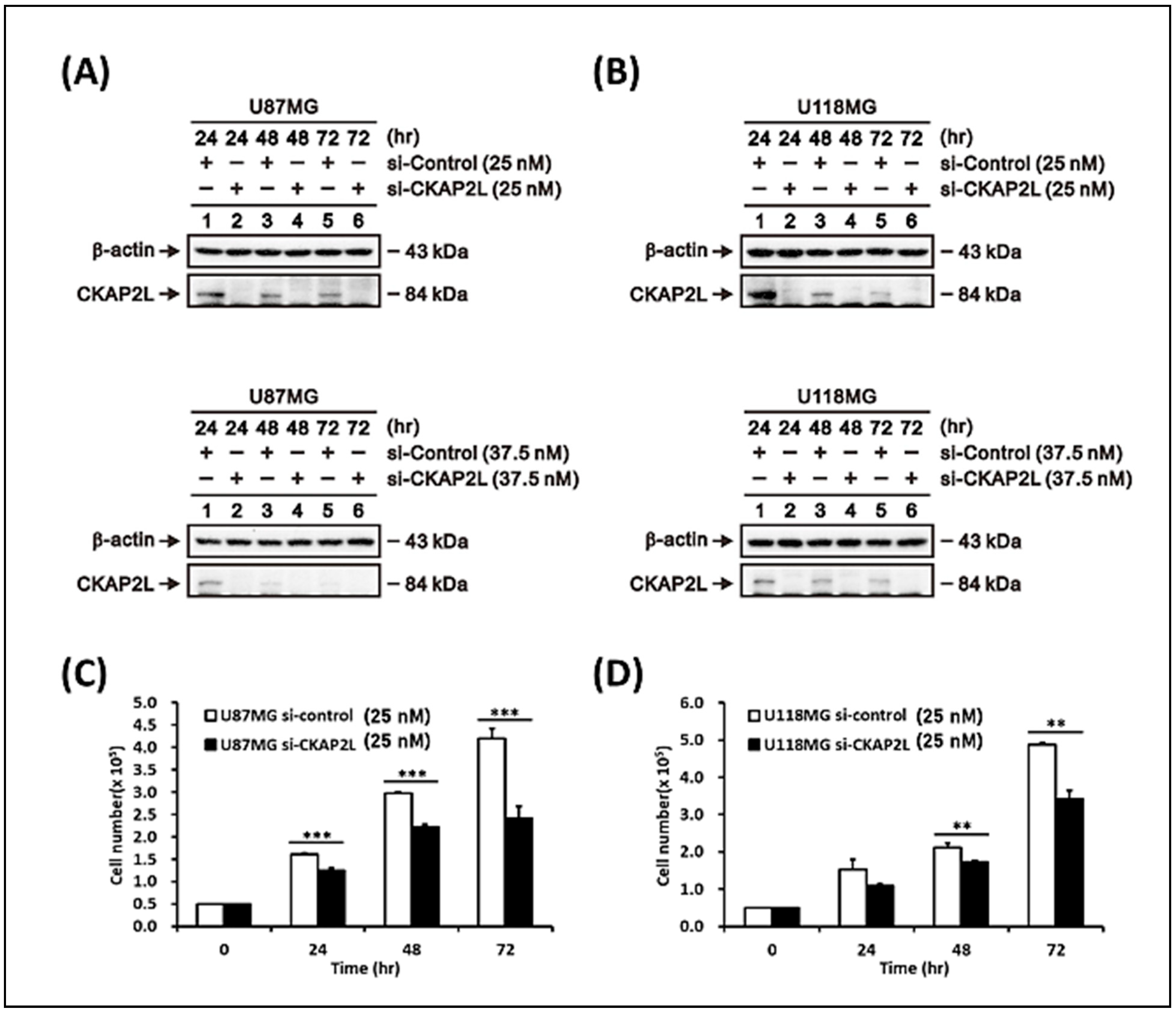
2.5. Suppression of CKAP2L Inhibits Glioma Cell Growth
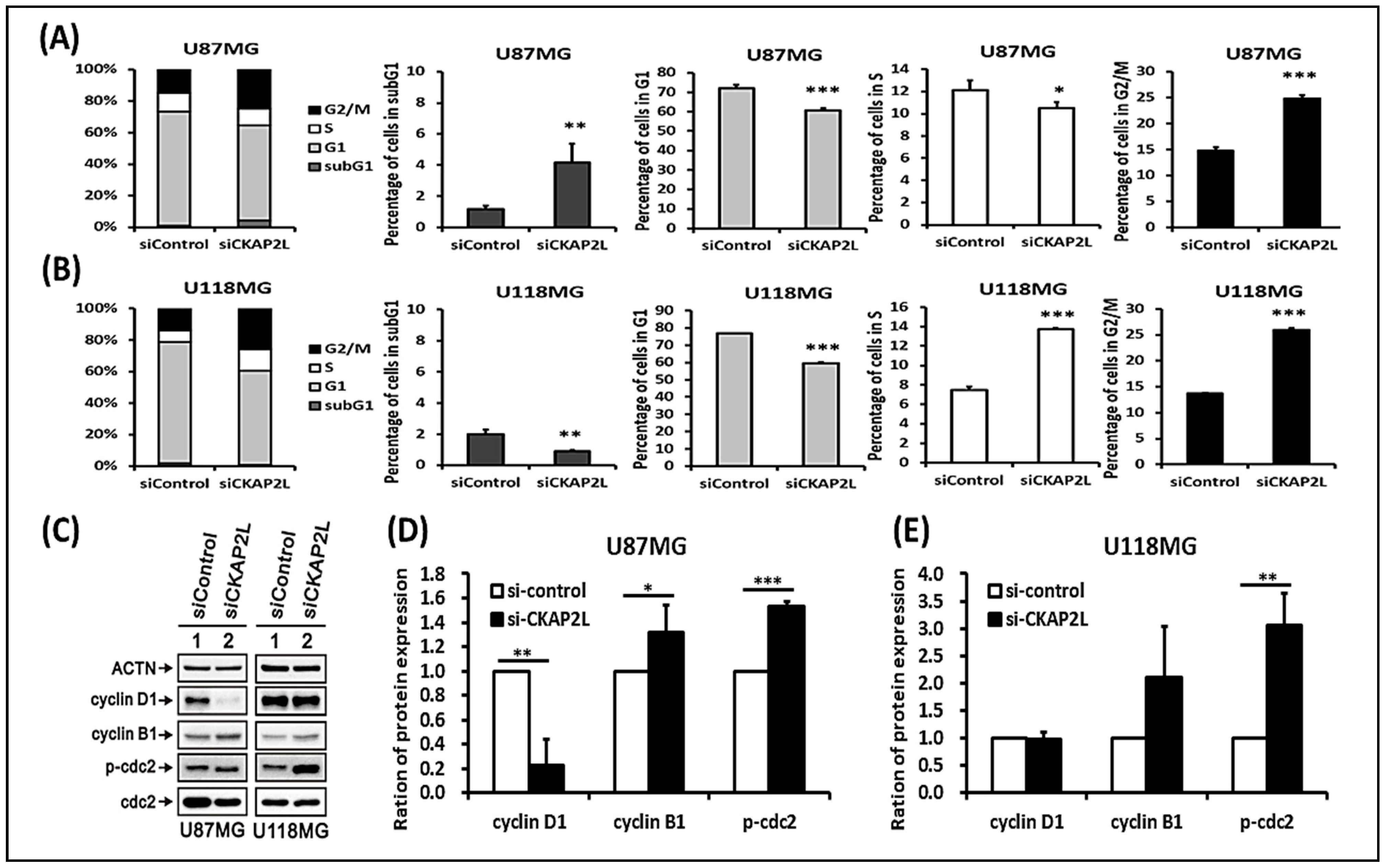
2.6. CKAP2L Knockdown Induces Cell Cycle G2/M Arrest
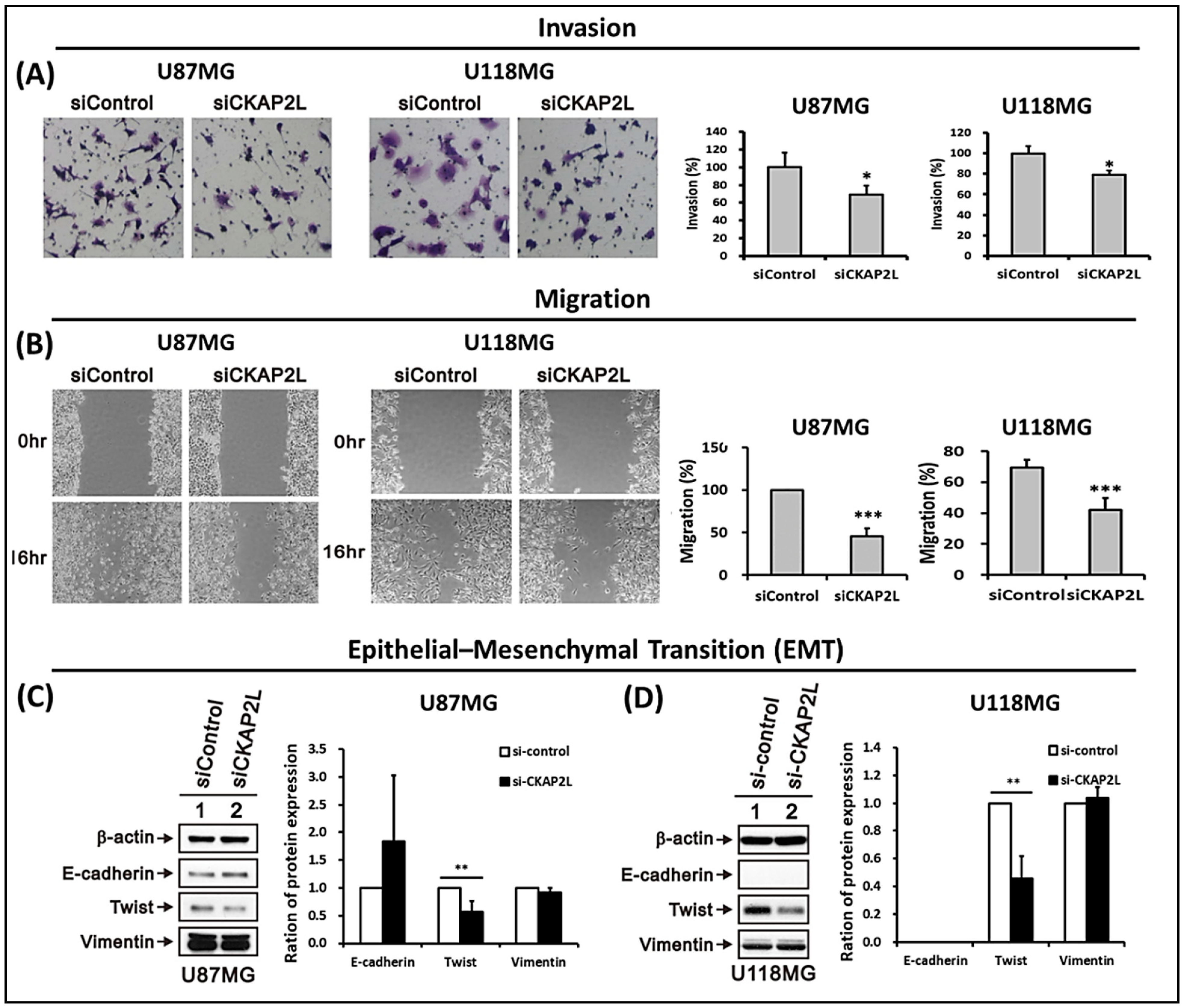
2.7. CKAP2L Knockdown Inhibits Invasiveness and Epithelial-Mesenchymal Transition (EMT) in Glioma
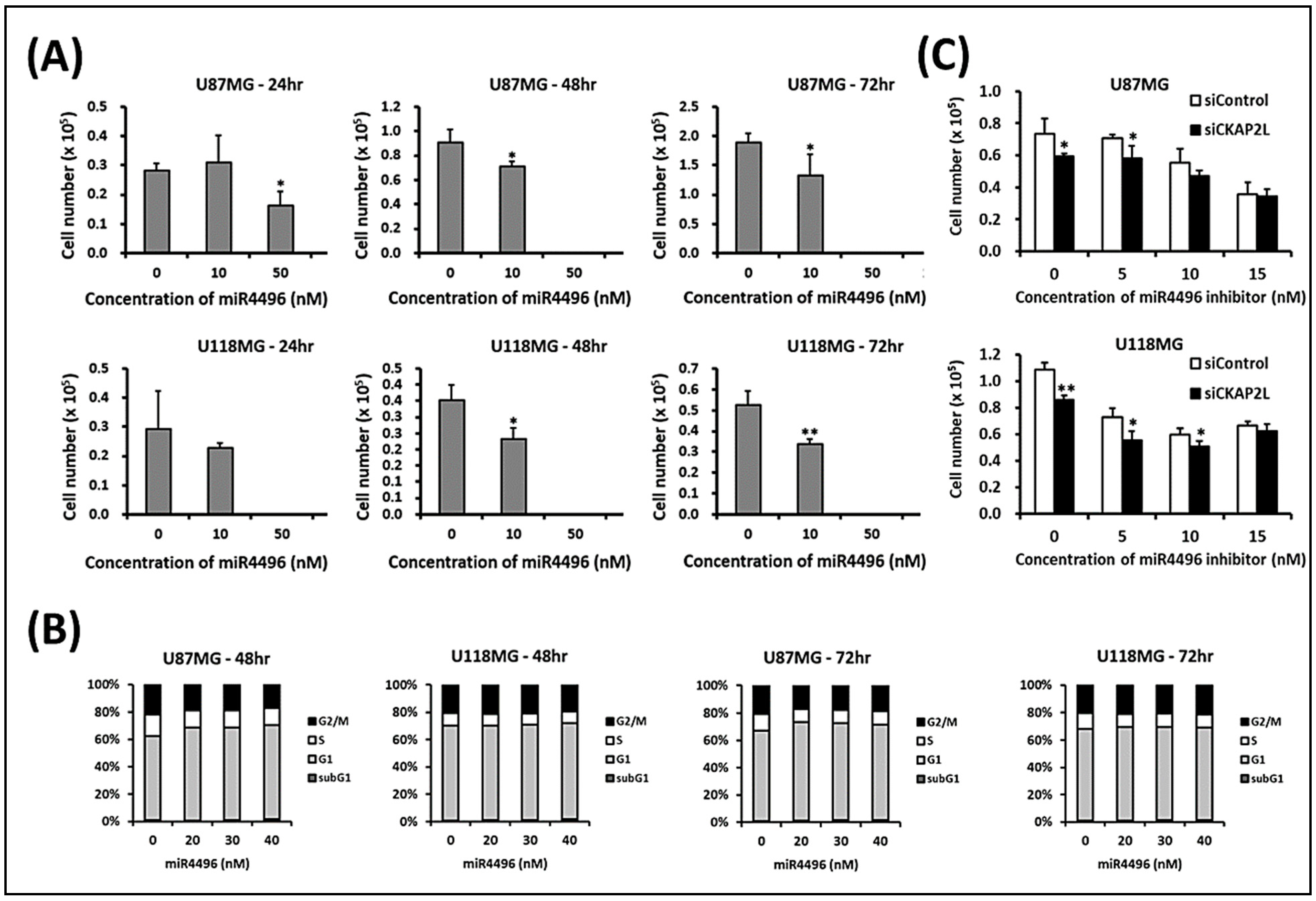
2.8. CKAP2L Knockdown Decreases Cell Numbers by Increasing miR-4496
3. Discussion
4. Materials and Methods
4.1. Analysis of Data from the TCGA and CGGA Databases
4.2. Gene Set Enrichment Analysis
4.3. Tissue Microarray Slide Preparation, Immunohistochemistry, and Scoring
4.4. Statistical Analysis of the Association between CKAP2L Expression and Other Factors
4.5. Human Glioma Cell Lines and Lysate Preparation
4.6. Transfection of siCKAP2L and siControl into Glioma Cell Lines and Cell Proliferation Assays
4.7. RNA Isolation and Real-Time Reverse Transcription-PCR
4.8. Western Blot Analysis
4.9. Cell Cycle Assessment
4.10. Wound Healing Assays
4.11. Cell Invasion Assays
4.12. miRNA Screening and Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CKAP2L | cytoskeleton-associated protein 2-like |
| EMT | epithelial-mesenchymal transition |
| FDR | False Discovery Rate |
| GBM | glioblastoma multiforme |
| IHC | immunohistochemistry |
| miR | microRNA |
| siRNA | small interfering RNA |
| WHO | World Health Organization |
References
- Patel, A.P.; Fisher, J.L.; Nichols, E.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; Abraha, H.N.; Agius, D.; Alahdab, F.; Alam, T.; et al. Global, regional, and national burden of brain and other CNS cancer, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 376–393. [Google Scholar] [CrossRef] [Green Version]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncology 2018, 20, iv1–iv86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Brat, D.J.; Aldape, K.; Colman, H.; Figrarella-Branger, D.; Fuller, G.N.; Giannini, C.; Holland, E.C.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.; Komori, T.; et al. cIMPACT-NOW update 5: Recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020, 139, 603–608. [Google Scholar] [CrossRef]
- Brat, D.J.; Aldape, K.; Colman, H.; Holland, E.C.; Louis, D.N.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.K.; Perry, A.; Reifenberger, G.; Stupp, R.; et al. cIMPACT-NOW update 3: Recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018, 136, 805–810. [Google Scholar] [CrossRef] [Green Version]
- Ellison, D.W.; Hawkins, C.; Jones, D.T.W.; Onar-Thomas, A.; Pfister, S.M.; Reifenberger, G.; Louis, D.N. cIMPACT-NOW update 4: Diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAF(V600E) mutation. Acta Neuropathol. 2019, 137, 683–687. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1andIDH2Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Pickles, J.C.; Fairchild, A.R.; Stone, T.J.; Brownlee, L.; Merve, A.; Yasin, S.A.; Avery, A.; Ahmed, S.W.; Ogunbiyi, O.; Zapata, J.G.; et al. DNA methylation-based profiling for paediatric CNS tumour diagnosis and treatment: A population-based study. Lancet Child Adolesc. Health 2020, 4, 121–130. [Google Scholar] [CrossRef]
- Nizamutdinov, D.; Stock, E.M.; Dandashi, J.A.; Vasquez, E.A.; Mao, Y.; Dayawansa, S.; Zhang, J.; Wu, E.; Fonkem, E.; Huang, J.H. Prognostication of Survival Outcomes in Patients Diagnosed with Glioblastoma. World Neurosurg. 2018, 109, e67–e74. [Google Scholar] [CrossRef] [PubMed]
- Delgado-López, P.D.; Corrales-García, E.M. Survival in glioblastoma: A review on the impact of treatment modalities. Clin. Transl. Oncol. 2016, 18, 1062–1071. [Google Scholar] [CrossRef]
- Balachandran, A.A.; Larcher, L.M.; Chen, S.; Veedu, R.N. Therapeutically Significant MicroRNAs in Primary and Metastatic Brain Malignancies. Cancers 2020, 12, 2534. [Google Scholar] [CrossRef]
- Buruiană, A.; FlorianȘ, I.; Florian, A.I.; Timiș, T.L.; Mihu, C.M.; Miclăuș, M.; Oșan, S.; Hrapșa, I.; Cataniciu, R.C.; Farcaș, M.; et al. The Roles of miRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations. Int. J. Mol. Sci. 2020, 21, 1950. [Google Scholar] [CrossRef] [Green Version]
- Pottoo, F.H.; Javed, M.N.; Rahman, J.U.; Abu-Izneid, T.; Khan, F.A. Targeted delivery of miRNA based therapeuticals in the clinical management of Glioblastoma Multiforme. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Yan, C.; Kong, X.; Gong, S.; Liu, F.; Zhao, Y. Recent advances of the regulation roles of MicroRNA in glioblastoma. Int. J. Clin. Oncol. 2020, 25, 1215–1222. [Google Scholar] [CrossRef]
- Lozada-Delgado, E.L.; Grafals-Ruiz, N.; Miranda-Román, M.A.; Santana-Rivera, Y.; Valiyeva, F.; Rivera-Díaz, M.; Marcos-Martínez, M.J.; Vivas-Mejía, P.E. Targeting MicroRNA-143 Leads to Inhibition of Glioblastoma Tumor Progression. Cancers 2018, 10, 382. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.K.; Lin, J.W.; Shih, J.W.; Chuang, H.Y.; Fong, I.H.; Yeh, C.T.; Lin, C.M. Targeting BC200/miR218-5p Signaling Axis for Overcoming Temozolomide Resistance and Suppressing Glioma Stemness. Cells 2020, 9, 1859. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, L.; Vanselow, K.; Skogs, M.; Toyoda, Y.; Lundberg, E.; Poser, I.; Falkenby, L.G.; Bennetzen, M.; Westendorf, J.; Nigg, E.A.; et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011, 30, 1520–1535. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.S.; Battaglia, A.; Szczepanski, S.; Kaygusuz, E.; Toliat, M.R.; Sakakibara, S.-I.; Altmüller, J.; Thiele, H.; Nürnberg, G.; Moosa, S.; et al. Mutations in CKAP2L, the Human Homolog of the Mouse Radmis Gene, Cause Filippi Syndrome. Am. J. Hum. Genet. 2014, 95, 622–632. [Google Scholar] [CrossRef] [Green Version]
- Xiong, G.; Li, L.; Chen, X.; Song, S.; Zhao, Y.; Cai, W.; Peng, J. Up-regulation of CKAP2L expression promotes lung adenocarcinoma invasion and is associated with poor prognosis. OncoTargets Ther. 2019, 12, 1171–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pouget, C.; Hergalant, S.; Lardenois, E.; Lacomme, S.; Houlgatte, R.; Carpentier, C.; Dehais, C.; Rech, F.; Taillandier, L.; Sanson, M.; et al. Ki-67 and MCM6 labeling indices are correlated with overall survival in anaplastic oligodendroglioma, IDH1-mutant and 1p/19q-codeleted: A multicenter study from the French POLA network. Brain Pathol. 2020, 30, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.Q.; Cheng, Z.J.; Zhang, H.P.; Wang, P.F.; Zhang, Y.; Hao, J.J.; Wang, M.R.; Wan, J.H. Overexpression of MCM6 predicts poor survival in patients with glioma. Hum. Pathol. 2018, 78, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Gauchotte, G.; Vigouroux, C.; Rech, F.; Battaglia-Hsu, S.-F.; Soudant, M.; Pinelli, C.; Civit, T.; Taillandier, L.; Vignaud, J.M.; Bressenot, A. Expression of Minichromosome Maintenance MCM6 Protein in Meningiomas is Strongly Correlated With Histologic Grade and Clinical Outcome. Am. J. Surg. Pathol. 2012, 36, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Case, C.M.; Sackett, D.L.; Wangsa, D.; Karpova, T.; McNally, J.G.; Ried, T.; Camps, J. CKAP2 Ensures Chromosomal Stability by Maintaining the Integrity of Microtubule Nucleation Sites. PLoS ONE 2013, 8, e64575. [Google Scholar] [CrossRef]
- Jin, Y.; Murakumo, Y.; Ueno, K.; Hashimoto, M.; Watanabe, T.; Shimoyama, Y.; Ichihara, M.; Takahashi, M. Identification of a mouse cytoskeleton-associated protein, CKAP2, with microtubule-stabilizing properties. Cancer Sci. 2004, 95, 815–821. [Google Scholar] [CrossRef]
- Sim, S.H.; Bae, C.D.; Kwon, Y.; Hwang, H.L.; Poojan, S.; Hong, H.I.; Kim, K.; Kang, S.H.; Kim, H.S.; Um, T.H.; et al. CKAP2 (cytoskeleton-associated protein2) is a new prognostic marker in HER2-negative luminal type breast cancer. PLoS ONE 2017, 12, e0182107. [Google Scholar] [CrossRef] [Green Version]
- Viticchiè, G.; Lena, A.M.; Latina, A.; Formosa, A.; Gregersen, L.H.; Lund, A.H.; Bernardini, S.; Mauriello, A.; Miano, R.; Spagnoli, L.G.; et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 2011, 10, 1121–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Lee, Y.C.; Cheng, C.J.; Wu, C.F.; Song, J.H.; Gallick, G.E.; Yu-Lee, L.Y.; Kuang, J.; Lin, S.H. RSK Promotes Prostate Cancer Progression in Bone through ING3, CKAP2, and PTK6-Mediated Cell Survival. Mol. Cancer Res. 2015, 13, 348–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, C.D.; Sung, Y.S.; Jeon, S.M.; Suh, Y.; Yang, H.K.; Kim, Y.I.; Park, K.H.; Choi, J.; Ahn, G.; Park, J. Up-regulation of cytoskeletal-associated protein 2 in primary human gastric adenocarcinomas. J. Cancer Res. Clin. Oncol. 2003, 129, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.W.; Eom, B.W.; Kook, M.C.; Kim, H.S.; Kim, M.K.; Hwang, H.L.; Chandra, V.; Poojan, S.; Song, Y.; Koh, J.S.; et al. Clinical implications of proliferation activity in T1 or T2 male gastric cancer patients. Exp. Mol. Med. 2015, 47, e193. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Zhao, L. CKAP2 Promotes Ovarian Cancer Proliferation and Tumorigenesis Through the FAK-ERK Pathway. DNA Cell Biol. 2017, 36, 983–990. [Google Scholar] [CrossRef]
- Wang, K.; Huang, R.; Li, G.; Zeng, F.; Zhao, Z.; Liu, Y.; Hu, H.; Jiang, T. CKAP2 expression is associated with glioma tumor growth and acts as a prognostic factor in high-grade glioma. Oncol. Rep. 2018, 40, 2036–2046. [Google Scholar] [CrossRef] [Green Version]
- Yumoto, T.; Nakadate, K.; Nakamura, Y.; Sugitani, Y.; Sugitani-Yoshida, R.; Ueda, S.; Sakakibara, S. Radmis, a Novel Mitotic Spindle Protein that Functions in Cell Division of Neural Progenitors. PLoS ONE 2013, 8, e79895. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Meng, F.; Wang, W.; Wang, Z.; Zhang, C.; Jiang, T. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci. Data 2017, 4, 170024. [Google Scholar] [CrossRef]
- Luo, W.; Chen, D.; Wang, H.; Hu, J. Stanniocalcin 1 is a prognostic biomarker in glioma. Oncol. Lett. 2020, 20, 2248–2256. [Google Scholar] [CrossRef]
- Zhu, W.; Giangrande, P.H.; Nevins, J.R. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 2004, 23, 4615–4626. [Google Scholar] [CrossRef]
- Morgan, D.O. Principles of CDK regulation. Nature 1995, 374, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Zhu, B.T. Critical role of cyclin B1/Cdc2 up-regulation in the induction of mitotic prometaphase arrest in human breast cancer cells treated with 2-methoxyestradiol. Biochim. Biophys. Acta 2012, 1823, 1306–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elledge, S.J.; Harper, J.W. Cdk inhibitors: On the threshold of checkpoints and development. Curr. Opin. Cell Biol. 1994, 6, 847–852. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, H.; Beach, D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993, 7, 1572–1583. [Google Scholar] [CrossRef] [Green Version]
- Louis, D.N.; Giannini, C.; Capper, D.; Paulus, W.; Figarella-Branger, D.; Lopes, M.B.; Batchelor, T.T.; Cairncross, J.G.; van den Bent, M.; Wick, W.; et al. cIMPACT-NOW update 2: Diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018, 135, 639–642. [Google Scholar] [CrossRef]
- Butler, M.; Pongor, L.; Su, Y.T.; Xi, L.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 2020, 6, 380–391. [Google Scholar] [CrossRef]
- Hatanpaa, K.J.; Burma, S.; Zhao, D.; Habib, A.A. Epidermal Growth Factor Receptor in Glioma: Signal Transduction, Neuropathology, Imaging, and Radioresistance. Neoplasia 2010, 12, 675–684. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, K.; Tachibana, O.; Sata, K.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Overexpression of the EGF Receptor and p53 Mutations are Mutually Exclusive in the Evolution of Primary and Secondary Glioblastomas. Brain Pathol. 1996, 6, 217–223. [Google Scholar] [CrossRef]
- Kang, D.W.; Choi, C.Y.; Cho, Y.-H.; Tian, H.; Di Paolo, G.; Choi, K.-Y.; Min, D.S. Targeting phospholipase D1 attenuates intestinal tumorigenesis by controlling β-catenin signaling in cancer-initiating cells. J. Exp. Med. 2015, 212, 1219–1237. [Google Scholar] [CrossRef]
- Kang, D.W.; Yang, E.S.; Noh, Y.N.; Hwang, W.C.; Jo, S.Y.; Suh, Y.A.; Park, W.S.; Choi, K.Y.; Min, D.S. MicroRNA-320a and microRNA-4496 attenuateHelicobacter pylori cytotoxin-associated gene A (CagA)-induced cancer-initiating potential and chemoresistance by targeting β-catenin and ATP-binding cassette, subfamily G, member 2. J. Pathol. 2017, 241, 614–625. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.F.; Wang, Z.F.; Cheng, Q.; Wang, G.R.; Bai, Z.M. Androgen receptor suppresses prostate cancer cell invasion via altering the miR-4496/β-catenin signals. Biochem. Biophys. Res. Commun. 2018, 504, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Hueng, D.Y.; Tsai, W.C. Proteolipid Protein 2 Overexpression Indicates Aggressive Tumor Behavior and Adverse Prognosis in Human Gliomas. Int. J. Mol. Sci. 2018, 19, 3353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, W.C.; Hueng, D.Y.; Lin, C.R.; Yang, T.C.; Gao, H.W. Nrf2 Expressions Correlate with WHO Grades in Gliomas and Meningiomas. Int. J. Mol. Sci. 2016, 17, 722. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Average Intensity | Average % Tumor | Average Score | Correlation * | |
|---|---|---|---|---|
| Non-neoplastic brain tissue | 0.67 | 10 | 10 | |
| Classification of gliomas | ||||
| Pilocytic astrocytoma | 1 | 5 | 5 | No correlation (p = 0.057) |
| Diffuse astrocytoma, IDH1-mutant | 1 | 20 | 20 | |
| Diffuse astrocytoma, IDH1-wildtype | 1 | 26.82 | 26.82 | |
| Anaplastic astrocytoma, IDH1-mutant | 1 | 23.33 | 23.33 | |
| Anaplastic astrocytoma, IDH1-wildtype | 1 | 40 | 40 | |
| Glioblastoma, IDH1-mutant | 1 | 42.5 | 42.5 | |
| Glioblastoma, IDH1-wildtype | 1 | 36.72 | 39.06 | |
| Oligodendroglioma, NOS | 0.75 | 13.75 | 13.75 | No correlation (p = 0.960) |
| Anaplastic oligodendroglioma, NOS | 0.67 | 13.33 | 13.33 | |
| Diffuse midline glioma, H3 K27M-mutant | 1.08 | 39.58 | 50.42 | |
| WHO grades of gliomas | ||||
| WHO grade I | 1 | 5 | 5 | Positive correlation (p = 0.005 *) |
| WHO grade II | 0.94 | 23.13 | 23.13 | |
| WHO grade III | 0.86 | 25 | 25 | |
| WHO grade IV | 1.02 | 38.10 | 42.20 |
| Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|
| Variable | Total | OR (95% CI) | p Value | OR (95% CI) | p Value |
| Age | |||||
| <50 | 32 | 1 | |||
| ≥50 | 39 | 1.72 (1.48–2.28) | 0.010 * | 1.72 (1.48–2.26) | 0.042 * |
| MGMT | |||||
| Unmethylated | 31 | 1 | |||
| Methylated | 40 | 1.07 (1.03–1.16) | 0.714 | 1.08 (1.04–1.18) | 0.008 * |
| EGFRvIII | |||||
| Negative | 54 | 1 | |||
| Positive | 17 | 1.42 (1.29–1.50) | 0.089 | 1.49 (1.39–1.55) | 0.012 * |
| P53 | |||||
| Negative | 35 | 1 | |||
| overexpression | 36 | 1.39 (1.31–1.55) | 0.081 | 1.44 (1.32–1.68) | 0.007 * |
| NF1 | |||||
| Negative | 37 | 1 | |||
| Positive | 34 | 1.39 (1.32–1.53) | 0.081 | 1.40 (1.32–1.58) | 0.013 * |
| AxL | |||||
| Negative | 25 | 1 | |||
| Positive | 46 | 1.23 (1.12–1.48) | 0.297 | 1.27 (1.12–1.62) | < 0.001 * |
| p-AxL | |||||
| Negative | 18 | 1 | |||
| Positive | 53 | 1.16 (0.98–1.63) | 0.512 | 1.12 (0.96–1.55) | 0.017 * |
| H3Lys27 | |||||
| Negative | 9 | 1 | |||
| Positive | 62 | 2.39 (0.38–1.36) | 0.020 * | 2.47 (1.23–1.31) | 0.010 * |
| Multivariate Analysis | |||
|---|---|---|---|
| Variable | Hazard Ratio | 95% Confidence Interval | p Value |
| Sex (male/female) | 1.69 | 0.70–4.06 | 0.240 |
| Age (<50/≥50) | 6.05 | 2.36–15.52 | <0.001 * |
| CKAP2L (≤20/>20) | 0.43 | 0.17–1.05 | 0.063 |
| IDH1 R132H (Negative/Positive) | 2.87 | 1.05–2.87 | 0.040 * |
| ATRX (Preserve/Loss) | 0.36 | 0.17–0.79 | 0.010 * |
| H3K27M (Negative/Positive) | 0.60 | 0.19–1.96 | 0.398 |
| MGMT (Unmethylated/ Methylated) | 0.46 | 0.19–1.08 | 0.073 |
| EGFR (Negative/Positive) | 5.99 | 0.96–37.56 | 0.056 |
| EGFRvIII (Negative/Positive) | 1.10 | 0.44–2.77 | 0.845 |
| P53 (Negative/Overexpression) | 1.89 | 0.85–4.21 | 0.119 |
| Neurofilament (Negative/Positive) | 0.28 | 0.09–0.81 | 0.019 |
| NF1 (Negative/Positive) | 1.69 | 0.73–3.89 | 0.222 |
| AxL (Negative/Positive) | 2.75 | 1.17–6.51 | 0.021 * |
| p-AxL (Negative/Positive) | 4.49 | 1.53–13.16 | 0.006 * |
| NUR77 (Negative/Positive) | 0.35 | 0.15–0.83 | 0.018 * |
| H3Lys27 (Negative/Positive) | 0.33 | 0.11–0.99 | 0.049 * |
| PDGFRA (Negative/Positive) | 9.73 | 1.74–54.48 | <0.001 * |
| U87 | Normalized Intensity | Coefficient of Variation | log2FC * | p-Value | ||
|---|---|---|---|---|---|---|
| Name of miR | U87 with siControl | U87 with siCKAP2L | U87 with siControl | U87 with siCKAP2L | (siCKAP2L/siControl) | (siCKAP2L/siControl) |
| hsa-miR-642a-3p | 467.39 | 3253.615 | 0.08381377 | 0.23918642 | 2.79934478 | 0.036951922 |
| hsa-miR-4496 | 90.5 | 405 | 0.16408003 | 0.21300501 | 2.16193221 | 0.036620139 |
| hsa-miR-642b-3p | 144.875 | 391.245 | 0.18913124 | 0.1822687 | 1.43326365 | 0.04486455 |
| hsa-miR-132-3p | 56 | 101.75 | 0.22728432 | 0.02432308 | 0.86153006 | 0.03789425 |
| hsa-miR-4740-3p | 33.5 | 55.5 | 0.02110767 | 0.01274066 | 0.72832668 | 0.00103146 |
| hsa-miR-4712-3p | 65 | 106 | 0.13054279 | 0.05336655 | 0.70555264 | 0.02956881 |
| hsa-miR-10a-3p | 48 | 76.5 | 0.02946278 | 0.06470258 | 0.67242534 | 0.015924106 |
| hsa-miR-491-3p | 52.5 | 81 | 0.0404061 | 0.03491885 | 0.62560449 | 0.007606987 |
| hsa-miR-4723-5p | 4147.415 | 6316.36 | 0.08147367 | 0.01306662 | 0.60688099 | 0.01261677 |
| U118 | Normalized Intensity | Coefficient of Variation | log2FC * | p-Value | ||
| Name of miR | U118 with siControl | U118 with siCKAP2L | U118 with siControl | U118 with siCKAP2L | (siCKAP2L/siControl) | (siCKAP2L/siControl) |
| hsa-miR-668-3p | 475.865 | 766.455 | 0.06281068 | 0.01006522 | 0.68764874 | 0.005594934 |
| hsa-miR-4740-3p | 33.5 | 63 | 0.02110767 | 0.0673435 | 0.91119073 | 0.010462609 |
| hsa-miR-642b-3p | 116.165 | 390.805 | 0.087228 | 0.02241797 | 1.75027347 | 0.001187312 |
| hsa-miR-4496 | 89.5 | 469.16 | 0.05530444 | 0.04473299 | 2.39012043 | 0.001608935 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-F.; Tsai, W.-C.; Chou, C.-H.; Huang, L.-C.; Huang, S.-M.; Hueng, D.-Y.; Tsai, C.-K. CKAP2L Knockdown Exerts Antitumor Effects by Increasing miR-4496 in Glioblastoma Cell Lines. Int. J. Mol. Sci. 2021, 22, 197. https://doi.org/10.3390/ijms22010197
Li Y-F, Tsai W-C, Chou C-H, Huang L-C, Huang S-M, Hueng D-Y, Tsai C-K. CKAP2L Knockdown Exerts Antitumor Effects by Increasing miR-4496 in Glioblastoma Cell Lines. International Journal of Molecular Sciences. 2021; 22(1):197. https://doi.org/10.3390/ijms22010197
Chicago/Turabian StyleLi, Yao-Feng, Wen-Chiuan Tsai, Chung-Hsing Chou, Li-Chun Huang, Shih-Ming Huang, Dueng-Yuan Hueng, and Chia-Kuang Tsai. 2021. "CKAP2L Knockdown Exerts Antitumor Effects by Increasing miR-4496 in Glioblastoma Cell Lines" International Journal of Molecular Sciences 22, no. 1: 197. https://doi.org/10.3390/ijms22010197






