Characterization of New Proteomic Biomarker Candidates in Mucopolysaccharidosis Type IVA
Abstract
1. Introduction
2. Results
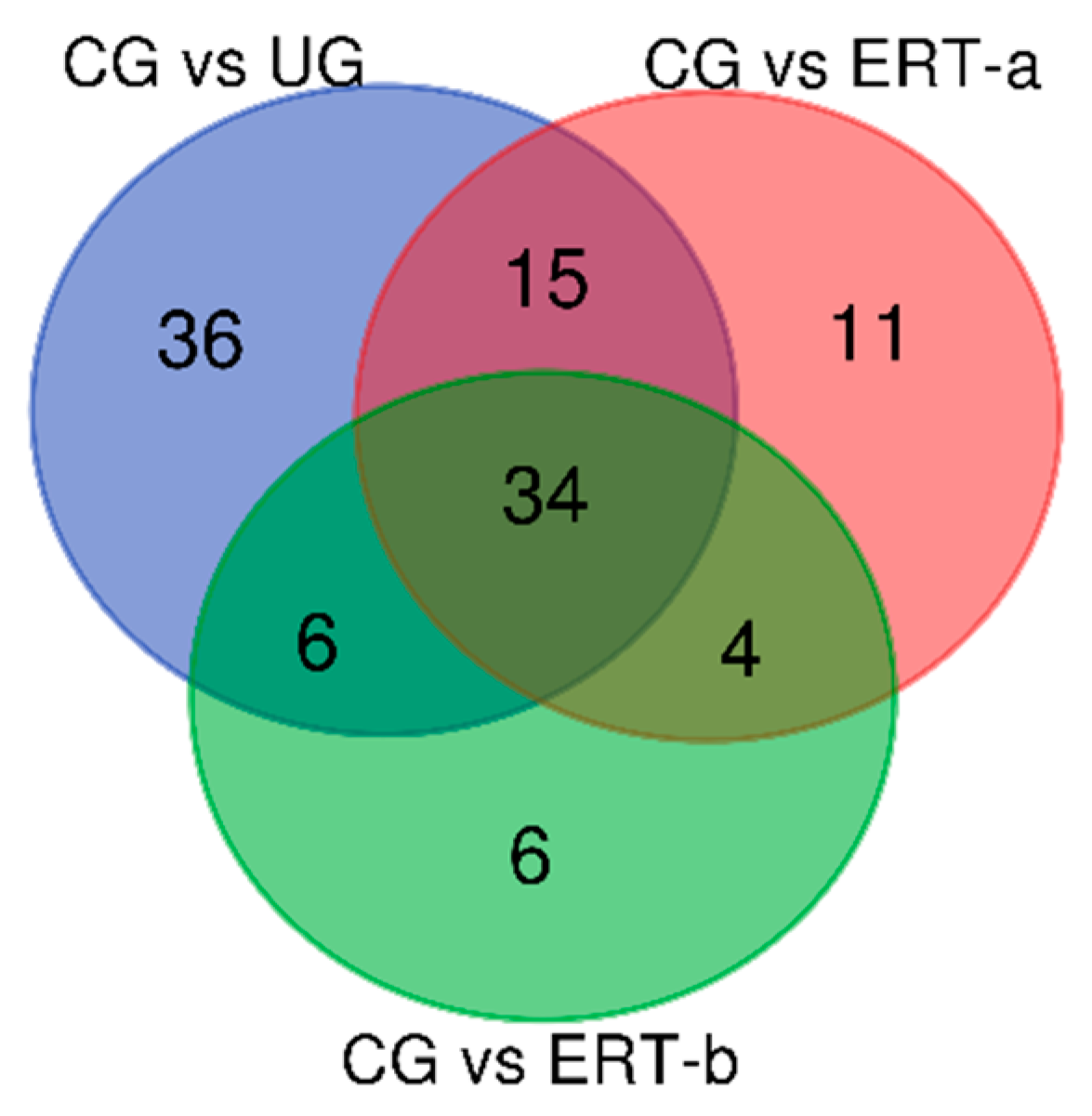
2.1. Qualitative Analysis of Proteins
2.2. Quantitative Analysis of Proteins by SWATH
2.2.1. Proteins Down Regulated in MPS IVA Patients Relative to Healthy Controls
2.2.2. Proteins Upregulated in Untreated MPS IVA Patients Relative to Untreated Patients and Controls
2.2.3. Proteins Not Detected in Healthy Controls but Are Detected in MPSIV Patients
3. Discussion
3.1. Limitations of the Study
3.2. Conclusions
4. Materials and Methods
4.1. Study Work Flow
4.2. Samples
4.3. Protein Extraction
4.4. Enzyme Activity Test
4.5. Proteomic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hendriksz, C.J.; Berger, K.I.; Giugliani, R.; Harmatz, P.; Kampmann, C.; Mackenzie, W.G.; Raiman, J.; Villarreal, M.S.; Savarirayan, R. International Guidelines for the Management and Treatment of Morquioa Syndrome. Am. J. Med. Genet. A 2015, 167A, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Alméciga-Díaz, C.J.; Sawamoto, K.; Mackenzie, W.G.; Theroux, M.C.; Pizarro, C.; Mason, R.W.; Orii, T.; Tomatsu, S. Mucopolysaccharidosis IVA and glycosaminoglycans. Mol. Genet. Metab. 2017, 120, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Peracha, H.; Sawamoto, K.; Averill, L.; Kecskemethy, H.; Theroux, M.; Thacker, M.; Nagao, K.; Pizarro, C.; Mackenzie, W.; Kobayashi, H.; et al. Diagnosis and prognosis of Mucopolysaccharidosis IVA. Mol. Genet. Metab. 2018, 125, 18–37. [Google Scholar] [CrossRef] [PubMed]
- Melbouci, M.; Mason, R.W.; Suzuki, Y.; Fukao, T.; Orii, T.; Tomatsu, S. Growth impairment in mucopolysaccharidoses. Mol. Genet. Metab. 2018, 124, 1–10. [Google Scholar] [CrossRef]
- Montaño, A.M.; Tomatsu, S.; Gottesman, G.S.; Smith, M.; Orii, T. International Morquio A Registry: Clinical manifestation and natural course of Morquio A disease. J. Inherit. Metab. Dis. 2007, 30, 165–174. [Google Scholar] [CrossRef]
- Sawamoto, K.; Álvarez González, J.V.; Piechnik, M.; Otero, F.J.; Couce, M.L.; Suzuki, Y.; Tomatsu, S. Mucopolysaccharidosis IVA: Diagnosis, Treatment, and Management. Int. J. Mol. Sci. 2020, 21, 1517. [Google Scholar] [CrossRef]
- Tomatsu, S.; Montaño, A.M.; Oikawa, H.; Smith, M.; Barrera, L.; Chinen, Y.; Thacker, M.M.; Mackenzie, W.G.; Suzuki, Y.; Orii, T. Mucopolysaccharidosis type IVA (Morquio A disease): Clinical review and current treatment. Curr. Pharm. Biotechnol. 2011, 12, 931–945. [Google Scholar] [CrossRef]
- Lavery, C.; Hendriksz, C. Mortality in patients with morquio syndrome a. JIMD Rep. 2015, 15, 59. [Google Scholar]
- Hendriksz, C.; Santra, S.; Jones, S.A.; Geberhiwot, T.; Jesaitis, L.; Long, B.; Qi, Y.; Hawley, S.M.; Decker, C. Safety, immunogenicity, and clinical outcomes in patients with Morquio A syndrome participating in 2 sequential open-label studies of elosulfasealfa enzyme replacement therapy (MOR-002/MOR-100), representing 5 years of treatment. Mol. Genet. Metab. 2018, 123, 479–487. [Google Scholar] [CrossRef]
- Hendriksz, C.J. Elosulfasealfa (BMN 110) for the treatment of mucopolysaccharidosis IVA (Morquio A Syndrome). Expert Rev. Clin. Pharmacol. 2016, 9, 1521–1532. [Google Scholar] [CrossRef]
- Qi, Y.; Musson, D.G.; Schweighardt, B.; Tompkins, T.; Jesaitis, L.; Shaywitz, A.J.; Yang, K.; O’Neill, C.A. Pharmacokinetic and Pharmacodynamic Evaluation of Elosulfase Alfa, an Enzyme Replacement Therapy in Patients with Morquio a Syndrome. Clin. Pharmacokinet. 2014, 53, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Tomatsu, S.; Sawamoto, K.; Shimada, T.; Bober, M.B.; Kubaski, F.; Yasuda, E.; Mason, R.W.; Khan, S.; Alméciga-Díaz, C.J.; Barrera, L.A.; et al. Enzyme replacement therapy for treating mucopolysaccharidosis type IVA (Morquio A syndrome): Effect and limitations. Expert Opin. Orphan Drugs 2015, 3, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Tomatsu, S.; Montaño, A.M.; Dung, V.C.; Ohashi, A.; Oikawa, H.; Oguma, T.; Orii, T.; Barrera, L.; Sly, W.S. Enhancement of drug delivery: Enzyme-replacement therapy for murine Morquio A syndrome. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Tomatsu, S.; Sawamoto, K.; Alméciga-Díaz, C.J.; Shimada, T.; Bober, M.B.; Chinen, Y.; Yabe, H.; Montaño, A.M.; Giugliani, R.; Kubaski, F.; et al. Impact of enzyme replacement therapy and hematopoietic stem cell transplantation in patients with Morquio A syndrome. Drug Des. Devel. Ther. 2015, 9, 1937. [Google Scholar] [CrossRef] [PubMed]
- Sawamoto, K.; Stapleton, M.; Alméciga-Díaz, C.J.; Espejo-Mojica, A.J.; Losada, J.C.; Suarez, D.A.; Tomatsu, S. Therapeutic Options for Mucopolysaccharidoses: Current and Emerging Treatments. Drugs 2019, 79, 1103–1134. [Google Scholar] [CrossRef]
- Long, B.; Tompkins, T.; Decker, C.; Jesaitis, L.; Khan, S.; Slasor, P.; Harmatz, P.; O’Neill, C.A.; Schweighardt, B. Long-term Immunogenicity of Elosulfase Alfa in the Treatment of Morquio A Syndrome: Results From MOR-005, a Phase III Extension Study. Clin. Ther. 2017, 39, 118–129. [Google Scholar] [CrossRef]
- Doherty, C.; Stapleton, M.; Piechnik, M.; Mason, R.W.; Mackenzie, W.G.; Yamaguchi, S.; Kobayashi, H.; Suzuki, Y.; Tomatsu, S. Effect of enzyme replacement therapy on the growth of patients with Morquio A. J. Hum. Genet. 2019, 64, 625–635. [Google Scholar] [CrossRef]
- Tomatsu, S.; Averill, L.W.; Sawamoto, K.; Mackenzie, W.G.; Bober, M.B.; Pizarro, C.; Gogg, C.J.; Xie, L.; Orii, T.; Theroux, M. Obstructive airway in Morquio A syndrome, the past, the present and the future. Mol. Genet. Metab. 2016, 117, 150–156. [Google Scholar] [CrossRef]
- Regier, S.D.; Tanpaiboon, P. Role of elosulfase alfa in mucopolysaccharidosis IV A. Appl. Clin. Genet. 2016, 9, 67–74. [Google Scholar] [CrossRef]
- Fujitsuka, H.; Sawamoto, K.; Peracha, H.; Mason, R.W.; Mackenzie, W.; Kobayashi, H.; Yamaguchi, S.; Suzuki, Y.; Orii, K.; Orii, T.; et al. Biomarkers in patients with mucopolysaccharidosis type II and IV. Mol. Genet. Metab. Rep. 2019, 19, 100455. [Google Scholar] [CrossRef]
- Tomatsu, S.; Yasuda, E.; Patel, P.; Ruhnke, K.; Shimada, T.; Mackenzie, W.G.; Mason, R.; Thacker, M.M.; Theroux, M.; Montaño, A.M.; et al. Morquio A syndrome: Diagnosis and current and future therapies. Pediatr. Endocrinol. Rev. 2014, 12, S141–S151. [Google Scholar]
- Rabilloud, T.; Chevallet, M.; Luche, S.; Lelong, C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J. Proteom. 2010, 73, 2064–2077. [Google Scholar] [CrossRef] [PubMed]
- Kapphahn, R.J.; Richards, M.J.; Ferrington, D.A.; Fliesler, S.J. Lipid-derived and other oxidative modifications of retinal proteins in a rat model of Smith-Lemli-Opitz syndrome. Exp. Eye Res. 2019, 178, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Santana, A.M.; Thomas, F.C.; Silva, D.G.; McCulloch, E.; Vidal, A.M.C.; Burchmore, R.J.S.; Fagliari, J.J.; Eckersall, P.D. Reference 1D and 2D electrophoresis maps for potential disease related proteins in milk whey from lactating buffaloes and blood serum from buffalo calves (Water buffalo, Bubalusbubalis). Res. Vet. Sci. 2018, 118, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Vidova, V.; Spacil, Z. A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition. Anal. Chim. Acta 2017, 964, 7–23. [Google Scholar] [CrossRef]
- Couselo-Seijas, M.; López-Canoa, J.N.; Agra-Bermejo, R.M.; Díaz-Rodriguez, E.; Fernandez, A.L.; Martinez-Cereijo, J.M.; Bravo, S.B.; Velo, A.; González-Melchor, L.; Fernández-López, X.A.; et al. Cholinergic activity regulates the secretome of epicardial adipose tissue: Association with atrial fibrillation. J. Cell Physiol. 2019, 234, 10512–10522. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.P.; López, A.C.; Bravo, S.B.; Vázquez-Estévez, S.; Acea-Nebril, B.; Núñez, C. Proteomic analysis of the bio-corona formed on the surface of (Au, Ag, Pt)-nanoparticles in human serum. Colloids Surf. B Biointerf. 2019, 177, 141–148. [Google Scholar] [CrossRef]
- Shilov, I.V.; Seymour, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Hunter, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The Paragon Algorithm, a Next Generation Search Engine That Uses Sequence Temperature Values and Feature Probabilities to Identify Peptides from Tandem Mass Spectra. Mol. Cell Proteom. 2007, 6, 1638–1655. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.-S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim., A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.-S.; Askenase, P.; Batagov, A.O.; Benito-Martin, A.; Camussi, G.; Clayton, A.; et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Barallobre-Barreiro, J.; Chung, Y.L.; Mayr, M. Proteomics and Metabolomics for Mechanistic Insights and Biomarker Discovery in Cardiovascular Disease. Rev. Española Cardiol. Rev. Esp. de Cardiol. 2013, 66, 657–661. [Google Scholar] [CrossRef]
- Gregorich, Z.R.; Ge, Y. Top-down proteomics in health and disease: Challenges and opportunities. Proteomics 2014, 14, 1195–1210. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.T.; Chung, M.C.M. Label-Free Quantitative Phosphoproteomics Reveals Regulation of Vasodilator-Stimulated Phosphoprotein upon Stathmin-1 Silencing in a Pair of Isogenic Colorectal Cancer Cell Lines. Proteomics 2018, 18, e1700242. [Google Scholar] [CrossRef]
- Braccia, C.; Espinal, M.P.; Pini, M.; De PietriTonelli, D.; Armirotti, A. A new SWATH ion library for mouse adult hippocampal neural stem cells. Data Br. 2018, 18, 1–8. [Google Scholar] [CrossRef]
- Jayabalan, N.; Lai, A.; Nair, S.; Guanzon, D.; Scholz-Romero, K.; Palma, C.; McIntyre, H.D.; Lappas, M.; Salomon, C. Quantitative proteomics by SWATH-MS suggest an association between circulating exosomes and maternal metabolic changes in gestational diabetes mellitus. Proteomics 2019, 19, e1800164. [Google Scholar] [CrossRef] [PubMed]
- Jamwal, R.; Barlock, B.J.; Adusumalli, S.; Ogasawara, K.; Simons, B.L..; Akhlaghi, F. Multiplex and Label-Free Relative Quantification Approach for Studying Protein Abundance of Drug Metabolizing Enzymes in Human Liver Microsomes Using SWATH-MS. J. Proteom. Res. 2017, 16, 4134–4143. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, J.; Lu, C.; Zhang, Q.; Xie, W.; Sun, P.; Dong, X.; Yue, L.; Sun, Y.; Yi, X.; et al. Identification of Protein Abundance Changes in Hepatocellular Carcinoma Tissues Using PCT–SWATH. Proteom. Clin. Appl. 2019, 13, e1700179. [Google Scholar] [CrossRef]
- Li, H.; Mao, Y.; Xiong, Y.; Zhao, H.; Shen, F.; Gao, X.; Yang, P.; Xiaohui, L.; Fu, D. A Comprehensive Proteome Analysis of Peripheral Blood Mononuclear Cells (PBMCs) to Identify Candidate Biomarkers of Pancreatic Cancer. Cancer Genom. Proteom. 2019, 16, 81–89. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Etheridge, N.; Nouwens, A.S.; Dodd, P.R. SWATH analysis of the synaptic proteome in Alzheimer’s disease. Neurochem. Int. 2015, 87, 1–12. [Google Scholar] [CrossRef]
- Ebhardt, H.A.; Degen, S.; Tadini, V.; Schilb, A.; Johns, N.; Greig, C.A.; Fearon, K.; Aebersold, R.; Jacobi, C. Comprehensive proteome analysis of human skeletal muscle in cachexia and sarcopenia: A pilot study. J. Cachexia Sarcopenia Muscle 2017, 8, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Walsh, T.; Atherton, J.J.; Kostner, K.; Schulz, B.; Punyadeera, C. Identification and validation of a salivary protein panel to detect heart failure early. Theranostics 2017, 7, 4350–4358. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, A.; Gieselmann, V. Lysosomal disorders: From storage to cellular damage. Biochim. Biophys. Acta 2009, 1793, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Pierzynowska, K.; Gaffke, L.; Podlacha, M.; Brokowska, J.; Węgrzyn, G. Mucopolysaccharidosis and Autophagy: Controversies on the Contribution of the Process to the Pathogenesis and Possible Therapeutic Applications. Neuromol. Med. 2020, 22, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Morrone, A.; Tylee, K.L.; Al-Sayed, M.; Brusius-Facchin, A.C.; Caciotti, A.; Church, H.J.; Coll, M.J.; Davidson, K.; Fietz, M.J.; Gort, L.; et al. Molecular Testing of 163 Patients with Morquio A (Mucopolysaccharidosis IVA) Identifies 39 Novel GALNS Mutations. Mol. Genet. Metab. 2014, 112, 160–170. [Google Scholar] [CrossRef]
- Shimada, T.; Tomatsu, S.; Yasuda, E.; Mason, R.W.; Mackenzie, W.G.; Shibata, Y.; Kubaski, F.; Giugliani, R.; Yamaguchi, S.; Suzuki, Y.; et al. Chondroitin 6-sulfate as a novel biomarker for mucopolysaccharidosis IVA and VII. JIMD Rep. 2014, 16, 15–24. [Google Scholar]
- Shimada, T.; Tomatsu, S.; Mason, R.W.; Yasuda, E.; Mackenzie, W.G.; Hossain, J.; Shibata, Y.; Montaño, A.M.; Kubaski, F.; Giuliani, R.; et al. Di-sulfated Keratan Sulfate as a Novel Biomarker for Mucopolysaccharidosis II, IVA, and IVB. JIMD Rep. 2015, 21, 1–13. [Google Scholar]
- Khan, S.A.; Mason, R.W.; Giugliani, R.; Orii, K.; Fukao, T.; Suzuki, Y.; Yamaguchi, S.; Kobayashi, H.; Orii, T.; Tomatsu, S. Glycosaminoglycans analysis in blood and urine of patients with mucopolysaccharidosis. Mol. Genet. Metab. 2018, 125, 44–52. [Google Scholar] [CrossRef]
- Marques, A.R.A.; Saftig, P. Lysosomal storage disorders—Challenges, concepts and avenues for therapy: Beyond rare diseases. J. Cell Sci. 2019, 132, jcs221739. [Google Scholar] [CrossRef]
- Anding, A.L.; Baehrecke, E.H. Cleaning House: Selective Autophagy of Organelles. Dev. Cell. 2017, 41, 10–22. [Google Scholar] [CrossRef]
- Bartel, K.; Pein, H.; Popper, B.; Schmitt, S.; Janaki-Raman, S.; Schulze, A.; Lengauer, F.; Koeberle, A.; Werz, O.; Zischka, H.; et al. Connecting lysosomes and mitochondria—A novel role for lipid metabolism in cancer cell death. Cell Commun. Signal 2019, 17, 87. [Google Scholar] [CrossRef] [PubMed]
- Styers, M.L.; Salazar, G.; Love, R.; Peden, A.A.; Kowalczyk, A.P.; Faundez, V. The Endo-Lysosomal Sorting Machinery Interacts with the Intermediate Filament Cytoskeleton. Mol. Biol. Cell. 2004, 15, 5369–5382. [Google Scholar] [CrossRef]
- Kurz, T.; Terman, A.; Gustafsson, B.; Brunk, U.T. Lysosomes in iron metabolism, ageing and apoptosis. Histochem. Cell Biol. 2008, 129, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Osellame, L.D.; Duchen, M.R. Quality control gone wrong: Mitochondria, lysosomal storage disorders and neurodegeneration. Br. J. Pharmacol. 2014, 171, 1958–1972. [Google Scholar] [CrossRef] [PubMed]
- Fraldi, A.; Annunziata, F.; Lombardi, A.; Kaiser, H.J.; Medina, D.L.; Spampanato, C.; OlindFedele, A.; Polishchuk, R.; Sorrentino, N.C.; Simons, K.; et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010, 29, 3607–3620. [Google Scholar] [CrossRef]
- Cooper, G.M. Lysosomes. In The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Fiorenza, M.T.; Moro, E.; Erickson, R.P. The pathogenesis of lysosomal storage disorders: Beyond the engorgement of lysosomes to abnormal development and neuroinflammation. Hum. Mol. Genet. 2018, 27, R119–R129. [Google Scholar] [CrossRef]
- Carroll, B.; Dunlop, E.A. The lysosome: A crucial hub for AMPK and mTORC1 signalling. Biochem. J. 2017, 474, 1453–1466. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Sabatini, D.M. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014, 24, 400–406. [Google Scholar] [CrossRef]
- Langeveld, M.; Ghauharali, K.M.; Sauerwein, H.P.; Ackermans, M.T.; Groener, C.; Aerts, J.M.; Serlie, M.J. Type I Gaucher Disease, a Glycosphingolipid Storage Disorder, Is Associated with Insulin Resistance. J. Clin. Endocrinol. Metab. 2008, 93, 845–851. [Google Scholar] [CrossRef]
- Álvarez, J.V.; Bravo, S.B.; García-Vence, M.; De Castro, M.J.; Luzardo, A.; Colón, C.; Tomatsu, S.; Otero-Espinar, F.J.; Couce, M.L. Proteomic Analysis in Morquio A Cells Treated with Immobilized Enzymatic Replacement Therapy on Nanostructured Lipid Systems. Int. J. Mol. Sci. 2019, 20, 4610. [Google Scholar] [CrossRef]
- Donida, B.; Marchetti, D.P.; Jacques, C.E.D.; Ribas, G.; Deon, M.; Manini, P.; da Rosa, H.T.; Moura, D.J.; Saffi, J.; Giugliani, R.; et al. Oxidative profile exhibited by Mucopolysaccharidosis type IVA patients at diagnosis: Increased keratan urinary levels. Mol. Genet. Metab. Rep. 2017, 11, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Caccavo, D.; Pellegrino, N.M.; Altamura, M.; Rigon, A.; Amati, L.; Amoroso, A.; Jirillo, E. Antimicrobial and immunoregulatory functions of lactoferrin and its potential therapeutic application. J. Endotoxin Res. 2002, 8, 403–417. [Google Scholar] [PubMed]
- Adlerova, L.; Bartoskova, A.; Faldyna, M. Lactoferrin: A review. Vet. Med. 2008, 53, 457–468. [Google Scholar] [CrossRef]
- Cornish, J.; Callon, K.E.; Naot, D.; Palmano, K.P.; Banovic, T.; Bava, U.; Watson, M.; Lin, J.M.; Tong, P.C.; Chen, Q.; et al. Lactoferrinis a Potent Regulator of Bone Cell Activity and Increases Bone Formation in Vivo. Endocrinology 2004, 145, 4366–4374. [Google Scholar] [CrossRef]
- Kruzel, M.L.; Zimecki, M.; Actor, J.K. Lactoferrin in a Context of Inflammation-Induced Pathology. Front. Immunol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Pick, R.; Begandt, D.; Stocker, T.J.; Salvermoser, M.; Thome, S.; Böttcher, R.T.; Montanez, E.; Harrison, U.; Forné, I.; Khandoga, A.G.; et al. Coronin 1A, a novel player in integrin biology, controls neutrophil trafficking in innate immunity. Blood 2017, 130, 847–858. [Google Scholar] [CrossRef]
- Ohmae, S.; Noma, N.; Toyomoto, M.; Shinohar, M.; Takeiri, M.; Fuji, H.; Takemoto, K.; Iwaisako, K.; Fujita, T.; Takeda, N.; et al. Actin-binding protein coronin 1A controls osteoclastic bone resorption by regulating lysosomal secretion of cathepsin K. Sci. Rep. 2017, 7, 41710. [Google Scholar] [CrossRef]
- Arnett, T.R. Osteoclast Biology. In Osteoporosis, 4th ed.; Marcus, R., Dempster, D., Cauley, J., Feldman, D., Eds.; Elsevier Enhanced Reader: London, UK, 2013; Chapter 8; pp. 149–160. [Google Scholar]
- Wilson, S.; Brömme, D. Potential role of cathepsin K in the pathophysiology of mucopolysaccharidoses. J. Pediatr. Rehabil. Med. 2010, 3, 139–146. [Google Scholar] [CrossRef]
- Pelletier, M.F.; Marcil, A.; Sevigny, G.; Jakob, C.A.; Tessier, D.C.; Chevet, E.; Menard, R.; Bergeron, J.J.; Thomas, D.Y. The heterodimeric structure of glucosidase II is required for its activity, solubility, and localization in vivo. Glycobiology 2000, 10, 815–827. [Google Scholar] [CrossRef]
- Schvartz, I.; Seger, D.; Shaltiel, S. Vitronectin. Int. J. Biochem. Cell Biol. 1999, 31, 539–544. [Google Scholar] [CrossRef]
- Cattaneo, M. Inherited Disorders of Platelet Function. In Platelets, 4th ed.; Michelson, A.D., Ed.; Academic Press: Cambridge, MA, USA, 2019; Chapter 57; pp. 877–904. [Google Scholar]
- Boskey, A.L.; GehronRobey, P. The Regulatory Role of Matrix Proteins in Mineralization of Bone. In Osteoporosis, 4th ed.; Marcus, R., Dempster, D., Cauley, J., Feldman, D., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 35–255. [Google Scholar]
- Sperb-Ludwig, F.; Heineck, B.L.; Michelin-Tirelli, K.; Alegra, T.; Schwartz, I.V.D. Chitotriosidase on treatment-naïve patients with Gaucher disease: A genotype vs. phenotype study. Clin. Chim. Acta 2019, 492, 1–6. [Google Scholar] [CrossRef] [PubMed]
- De Franceschi, L.; Roseti, L.; Desando, G.; Facchini, A.; Grigolo, B. A molecular and histological characterization of cartilage from patients with Morquio syndrome. Osteoarthr. Cartil. 2007, 15, 1311–1317. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bank, R.A.; Groener, J.E.M.; van Gemund, J.J.; Maaswinkel, P.D.; Hoeben, K.A.; Schut, H.A.; Everts, V. Deficiency in N-acetylgalactosamine-6-sulfate sulfatase results in collagen perturbations in cartilage of Morquio syndrome A patients. Mol. Genet. Metab. 2009, 97, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Camelier, M.V.; Burin, M.G.; De Mari, J.; Vieira, T.A.; Marasca, G.; Giugliani, R. Practical and reliable enzyme test for the detection of mucopolysaccharidosis IVA (Morquio Syndrome type A) in dried blood samples. Clin. Chim. Acta 2011, 412, 1805–1808. [Google Scholar] [CrossRef]
- Varela-Rodríguez, B.M.; Juiz-Valiña, P.; Varela, L.; Outeiriño-Blanco, E.; Bravo, S.B.; García-Brao, M.J.; Mena, E.; Noguera, J.F.; Valero-Gasalla, J.; Cordido, F.; et al. Beneficial Effects of Bariatric Surgery-Induced by Weight Loss on the Proteome of Abdominal Subcutaneous Adipose Tissue. J. Clin. Med. 2020, 9, 213. [Google Scholar] [CrossRef]
- Camino, T.; Lago-Baameiro, N.; Bravo, S.B.; Sueiro, A.; Couto, I.; Santos, F.; Baltar, J.; Casanueva, F.; Pardo, M. Vesicles Shed by Pathological Murine Adipocytes Spread Pathology: Characterization and Functional Role of Insulin Resistant/Hypertrophied Adiposomes. Int. J. Mol. Sci. 2020, 21, 2252. [Google Scholar] [CrossRef]
- Hermida-Nogueira, L.; Barrachina, M.N.; Izquierdo, I.; García-Vence, M.; Lacerenza, S.; Bravo, S.; Castrillo, A.; García, A. Proteomic analysis of extracellular vesicles derived from platelet concentrates treated with Mirasol® identifies biomarkers of platelet storage lesion. J. Proteom. 2020, 210, 103529. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.P.; López, A.C.; Vence, M.G.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.G.; Jardiel, T.; Bravo, S.B.; Núñez, C. Proteomic investigation on bio-corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212, 103581. [Google Scholar] [CrossRef]








| Patient ID | Sex | Age at Diagnosis (y) | ERT | Age at Start of Treatment (y) | Current Characteristics | ||||
|---|---|---|---|---|---|---|---|---|---|
| Age (y) | Height (cm) | 6 Minute Walk Test (m) | FVC (mL) | FEV1 (mL) | |||||
| 1 | F | 1 | No | - | 31 | 98 | 250 | 600 | 500 |
| 2 | M | 2 | No | - | 31 | 113 | 305 | 870 | 700 |
| 3 | M | 2 | No | - | 21 | 95 | ND * | 380 | 260 |
| 4 | M | 2 | No | 40 | 99 | ND * | 480 | 360 | |
| 5 | F | 4 | No | - | 15 | 103 | 341 | 920 | 820 |
| 6 | F | 3 | No | - | 29 | 99 | ND * | ND ** | ND ** |
| 7 | F | 1 | No | - | 18 | 119 | 272 | 110 | 900 |
| 8 | M | unknown | No | - | 21 | 103 | ND * | 920 | 700 |
| 9 | M | 1 | Yes | 12 | 16 | 100 | 105 | 690 | 450 |
| 10 | M | 2 | Yes | 2 | 6 | 104 | 450 | 770 | 720 |
| 11 | M | 3 | Yes | 13 | 18 | 113.5 | 472 | 1390 | 1330 |
| 12 | M | 3 | Yes | 11 | 19 | 113 | 234 | 1350 | 1160 |
| 13 | M | 5 | Yes | 18 | 22 | 110 | 344 | 870 | 730 |
| Patients and Control Groups- | Sample ID | Proteins Identified Per Sample (n) | Proteins Identified in All or All but One Samples (n) |
|---|---|---|---|
| Untreated Group | UG 1 | 460 | 235 |
| UG 2 | 330 | ||
| UG 3 | 367 | ||
| UG 4 | 612 | ||
| UG 5 | NA | ||
| UG 6 | NA | ||
| UG 7 | 341 | ||
| UG 8 | 304 | ||
| ERT-a Group | ERT-a 1 | 480 | 301 |
| ERT-a 2 | 338 | ||
| ERT-a 3 | 369 | ||
| ERT-a 4 | 406 | ||
| ERT-a 5 | 400 | ||
| ERT-b Group | ERT-b 1 | 492 | 222 |
| ERT-b 2 | 190 | ||
| ERT-b 3 | 437 | ||
| ERT-b 4 | 252 | ||
| ERT-b 5 | 557 | ||
| Healthy Control Group | CG 1 | 144 | 164 |
| CG 2 | 470 | ||
| CG 3 | 238 | ||
| CG 4 | 286 | ||
| CG 5 | 315 | ||
| CG6 | 261 |
| Comparison | Proteins Downregulated Compared with Controls | Proteins Upregulated Compared with Controls |
|---|---|---|
| Control vs. Untreated | 91 | 73 |
| Control vs. ERT-a | 64 | 55 |
| Control vs. ERT-b | 49 | 56 |
| Comparison | Proteins down regulated compared with untreated group | Proteins upregulated compared with untreated group |
| Untreated vs. ERT-a | 22 | 10 |
| Untreated vs. ERT-b | 10 | 23 |
| Comparison | Proteins down regulated compared with ERT-a group | Proteins upregulated compared with ERT-b group |
| ERT-a vs. ERT-b | 4 | 12 |
| UniProt Code | UniProt Name | Protein Name | Fold Change Relative to Healthy Controls | ||
|---|---|---|---|---|---|
| Untreated | ERT-a | ERT-b | |||
| P14618 | KPYM | Pyruvate kinase PKM | 0.0537 | 0.0770 | 0.1210 |
| P04406 | G3P | Glyceraldehyde-3-phosphate dehydrogenase | 0.0539 | 0.0729 | 0.0632 |
| P00338 | LDHA | l-lactate dehydrogenase A chain | 0.0629 | 0.1039 | 0.1176 |
| P00558 | PGK1 | Phosphoglycerate kinase 1 | 0.0902 | 0.1124 | 0.1742 |
| P07195 | LDHB | l-lactate dehydrogenase B chain | 0.2261 | 0.2049 | 0.2358 |
| P11177 | ODPB | Pyruvate dehydrogenase E1 component subunit beta. mitochondrial | 0.3005 | 0.2520 | 0.3246 |
| P04075 | ALDOA | Fructose-bisphosphatealdolase A | 0.4012 | 0.5957 | 0.6128 |
| P06744 | G6PI | Glucose-6-phosphate isomerase | 0.1645 | 0.3401 | 0.3358 |
| P36871 | PGM1 | Phosphoglucomutase-1 | 0.3284 | 0.2524 | 0.5816 |
| P11413 | G6PD | Glucose-6-phosphate 1-dehydrogenase | 0.3951 | 0.4585 | 0.5714 |
| P26038 | MOES | Moesin | 0.1282 | 0.1284 | 0.2848 |
| P60981 | DEST | Destrin | 0.3604 | 0.2756 | 0.4310 |
| O15145 | ARPC3 | Actin-related protein 2/3 complex subunit 3 | 0.5094 | 0.4480 | 0.5276 |
| P31146 | CORO1A | Coronin-1A | 0.1283 | 0.1418 | 0.2020 |
| P09493 | TPM1 | Tropomyosin alpha-1 chain | 0.2997 | 0.6517 | 0.7882 |
| P35527 | K1C9 | Keratin. type I cytoskeletal 9 | 0.3019 | 0.5949 | 0.6857 |
| P35908 | K22E | Keratin. type II cytoskeletal 2 epidermal | 0.2940 | 0.6149 | 0.8652 |
| O15143 | ARC1B | Actin-related protein 2/3 complex subunit 1B | 0.3132 | 0.4472 | 0.4072 |
| P04264 | K2C1 | Keratin. type II cytoskeletal 1 | 0.3301 | 0.6055 | 0.7358 |
| P02538 | K2C6A | Keratin. type II cytoskeletal 6A | 0.4352 | 1.0918 | 2.2909 |
| P67936 | TPM4 | Tropomyosin alpha-4 chain | 0.4602 | 0.8777 | 0.5522 |
| P02533 | K1C14 | Keratin. type I cytoskeletal 14 | 0.5186 | 0.8316 | 0.7752 |
| P61160 | ARP2 | Actin-related protein 2 | 0.3608 | 0.3315 | 0.3566 |
| O15511 | ARPC5 | Actin-related protein 2/3 complex subunit 5 | 0.4050 | 0.5206 | 0.4899 |
| P60709 | ACTB | Actin. cytoplasmic 1 | 0.3010 | 0.2164 | 0.2500 |
| Q99439 | CNN2 | Calponin-2 | 0.5653 | 1.2121 | 0.9130 |
| P35612 | ADDB | Beta-adducin | 0.4505 | 0.5356 | 0.7874 |
| Q13813 | SPTN1 | Spectrin alpha chain. non-erythrocytic 1 | 0.6223 | 0.9501 | 0.9308 |
| P50552 | VASP | Vasodilator-stimulated phosphoprotein | 0.4809 | 0.7258 | 0.6589 |
| O75083 | WDR1 | WD repeat-containing protein 1 | 0.6081 | 0.5059 | 0.4914 |
| Q16181 | SEPT7 | Septin-7 | 0.3998 | 0.2507 | 0.3119 |
| P17931 | LEG3 | Galectin-3 | 0.3903 | 0.4969 | 0.6100 |
| Q96QK1 | VPS35 | Vacuolar protein sorting-associated protein 35 | 0.4053 | 0.3453 | 0.7819 |
| Q15833 | STXB2 | Syntaxin-binding protein 2 | 0.4043 | 0.5382 | 0.6145 |
| P68036 | UB2L3 | Ubiquitin-conjugating enzyme E2 L3 | 0.3696 | 0.6336 | 0.5254 |
| P30040 | ERP29 | Endoplasmic reticulum resident protein 29 | 0.5751 | 0.8206 | 0.6180 |
| Q13492 | PICAL | Phosphatidylinositol-binding clathrin assembly protein | 0.6298 | 0.6427 | 0.6980 |
| P30049 | ATPD | ATP synthasesubunit delta. mitochondrial | 0.5906 | 0.8645 | 0.9240 |
| P25705 | ATPA | ATP synthasesubunit alpha. mitochondrial | 0.3060 | 0.2776 | 0.5251 |
| O75390 | CISY | Citrate synthase. mitochondrial | 0.6794 | 0.5843 | 0.6636 |
| P30044 | PRDX5 | Peroxiredoxin-5. mitochondrial | 0.4486 | 0.6043 | 0.5455 |
| P06576 | ATPB | ATP synthase subunit beta. mitochondrial | 0.4890 | 0.7480 | 0.7076 |
| Q99798 | ACON | Aconitatehydratase. mitochondrial | 0.5100 | 0.4129 | 0.6735 |
| P13804 | ETFA | Electron transfer flavoprotein subunit alpha. mitochondrial | 0.2472 | 0.1565 | 0.1542 |
| P10809 | CH60 | 60 kDa heat shock protein. mitochondrial | 0.1905 | 0.4708 | 0.3593 |
| P61604 | CH10 | 10 kDa heat shock protein. mitochondrial | 0.3779 | 0.3537 | 0.3176 |
| P00505 | AATM | Aspartate aminotransferase. mitochondrial | 0.5693 | 0.4255 | 0.3930 |
| P40926 | MDHM | Malate dehydrogenase. mitochondrial | 0.6669 | 0.5397 | 0.6540 |
| P69905 | HBA | Hemoglobin subunit alpha | 0.2772 | 0.6311 | 0.4201 |
| P68871 | HBB | Hemoglobin subunit beta | 0.2063 | 0.6678 | 0.3896 |
| P55072 | TERA | Transitional endoplasmic reticulum ATPase | 0.2347 | 0.1977 | 0.3911 |
| Q14697 | GANAB | Neutral alpha-glucosidase AB | 0.5454 | 0.4364 | 0.4613 |
| P08133 | ANXA6 | Annexin A6 | 0.4899 | 0.7086 | 0.9723 |
| P09525 | ANXA4 | Annexin A4 | 0.4646 | 0.6825 | 0.7587 |
| P04083 | ANXA1 | Annexin A1 | 0.2010 | 0.5650 | 0.6752 |
| P52209 | 6PGD | 6-phosphogluconate dehydrogenase decarboxylating | 0.1136 | 0.1570 | 0.2218 |
| P05089 | ARGI1 | Arginase-1 | 0.2834 | 0.4069 | 0.4421 |
| P29401 | TKT | Transketolase | 0.3432 | 0.3358 | 0.2993 |
| Q16762 | THTR | Thiosulfate sulfurtransferase | 0.4924 | 0.7856 | 0.8081 |
| P30566 | PUR8 | Adenylosuccinatelyase | 0.2364 | 0.1361 | 0.7371 |
| Q00013 | EM55 | 55 kDa erythrocyte membrane protein | 0.4414 | 0.4275 | 0.6262 |
| P02766 | TTHY | Transthyretin | 0.4808 | 0.4167 | 0.5573 |
| Q9H2U2 | IPYR2 | Inorganic pyrophosphatase2.mitochondrial | 0.5533 | 0.6667 | 0.7373 |
| Q7L5Y6 | DET1 | DET1 homolog | 0.3518 | 0.5613 | 0.6315 |
| P19971 | TYPH | Thymidine phosphorylase | 0.2521 | 0.2706 | 0.4983 |
| P00488 | F13A | Coagulation factor XIII A chain | 0.2992 | 0.3087 | 0.3043 |
| P06737 | PYGL | Glycogen phosphorylase. liver form | 0.1167 | 0.1441 | 0.2094 |
| P62136 | PP1A | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 0.5690 | 0.4578 | 0.5951 |
| P30101 | PDIA3 | Protein disulfide-isomerase A3 | 0.5598 | 1.0088 | 1.1788 |
| P55786 | PSA | Puromycin-sensitive aminopeptidase | 0.4389 | 0.6256 | 0.6560 |
| P07741 | APT | Adenine phosphoribosyltransferase | 0.2659 | 0.2516 | 0.3201 |
| Q06323 | PSME1 | Proteasome activator complexsubunit 1 | 0.3035 | 0.4744 | 0.4342 |
| P08571 | CD14 | Monocyte differentiation antigen CD14 | 0.4434 | 0.3594 | 1.0551 |
| P02652 | APOA2 | Apolipoprotein A-II | 0.3696 | 0.2974 | 0.8084 |
| P30086 | PEBP1 | Phosphatidylethanolamine-binding protein 1 | 0.5324 | 0.6353 | 0.8049 |
| P17612 | KAPCA | cAMP-dependent protein kinase catalytic subunit alpha | 0.2921 | 0.2831 | 0.4253 |
| P01860 | IGHG3 | Immunoglobulin heavy constant gamma 3 | 0.5132 | 0.4999 | 0.4892 |
| Q5VTE0 | EF1A3 | Putative elongation factor 1-alpha-like 3 | 0.3466 | 0.3686 | 0.4249 |
| Q9NTK5 | OLA1 | Obg-like ATPase 1 | 0.1099 | 0.3431 | 0.2186 |
| P62826 | RAN | GTP-binding nuclear protein Ran | 0.1277 | 0.1178 | 0.3363 |
| O00299 | CLIC1 | Chloride intracellular channel protein 1 | 0.2541 | 0.4800 | 0.6103 |
| P38606 | VATA | V-type proton ATPase catalytic subunit A | 0.3386 | 0.4063 | 0.3411 |
| P31948 | STIP1 | Stress-induced-phosphoprotein 1 | 0.3071 | 0.5992 | 0.5741 |
| Q15366 | PCBP2 | Poly(rC)-binding protein 2 | 0.4584 | 0.3282 | 0.4541 |
| P61978 | HNRPK | Heterogeneous nuclear ribonucleoprotein K | 0.5087 | 0.5807 | 0.6304 |
| P09651 | ROA1 | Heterogeneous nuclear ribonucleoprotein A1 | 0.5798 | 0.6174 | 0.6532 |
| P26583 | HMGB2 | High mobility group protein B2 | 0.1873 | 0.4159 | 0.6264 |
| P16402 | H13 | Histone H1.3 | 0.0761 | 0.2883 | 0.2277 |
| P16401 | H15 | Histone H1.5 | 0.0896 | 0.7384 | 1.0462 |
| P40199 | CEAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 | 0.2537 | 0.4984 | 0.7378 |
| Q92882 | OSTF1 | Osteoclast-stimulating factor 1 | 0.2963 | 0.6296 | 0.9094 |
| UniProt Code | UniProt Name | Protein Name | Fold change Relative to Healthy Controls | ||
|---|---|---|---|---|---|
| Untreated | ERT-a | ERT-b | |||
| P30405 | PPIF | Peptidyl-prolylcis-transisomerase F. mitochondrial | 1.8656 | 0.9764 | 1.2728 |
| P62318 | SMD3 | Small nuclear ribonucleoproteinSm D3 | 2.1112 | 1.8227 | 1.5860 |
| Q9H4B7 | TBB1 | Tubulin beta-1 chain | 2.6765 | 1.4412 | 1.5400 |
| P61224 | RAP1B | Ras-related protein Rap-1b | 2.8033 | 1.2182 | 1.1940 |
| P14780 | MMP9 | Matrix metalloproteinase-9 | 3.1051 | 1.7732 | 1.5370 |
| P08567 | PLEK | Pleckstrin | 1.4669 | 1.0482 | 1.0934 |
| Q6DRA6 | H2B2D | Putative histone H2B type 2-D | 8.3208 | 2.3169 | 1.3557 |
| Q9BTM1 | H2AJ | Histone H2A.J | 8.8293 | 2.3197 | 1.3487 |
| Q99879 | H2B1M | Histone H2B type 1-M | 9.9891 | 2.1137 | 1.4663 |
| P62805 | H4 | Histone H4 | 12.0044 | 3.0822 | 2.1018 |
| Q6UX71 | PXDC2 | Plexin domain-containing protein 2 | 2.0905 | 1.0293 | 1.0988 |
| Q8WWA1 | TMM40 | Transmembrane protein 40 | 4.9507 | 1.5721 | 1.2002 |
| P62314 | SMD1 | Small nuclear ribonucleo protein Sm D1 | 2.3589 | 1.9495 | 1.7400 |
| P20338 | RAB4A | Ras -related protein Rab-4A | 1.5322 | 2.1434 | 1.0633 |
| Q96P48 | ARAP1 | Arf-GAP with Rho-GAP domain. ANK repeat and PH domain-containing protein 1 | 2.036 | 1.8559 | 1.3848 |
| P01137 | TGFB1 | Transforming growth factor beta-1 proprotein | 2.336 | 1.0245 | 1.2298 |
| P05106 | ITB3 | Integrin beta-3 | 1.6777 | 1.0676 | 0.9028 |
| P02775 | CXCL7 | Platelet basic protein | 3.1669 | 1.0947 | 1.0450 |
| P41218 | MNDA | Myeloid cell nuclear differentiation antigen | 3.2656 | 1.7116 | 0.7412 |
| P02776 | PLF4 | Platelet factor 4 | 3.9596 | 1.3654 | 1.3027 |
| P11234 | RALB | Ras-related protein Ral-B | 2.004 | 1.1184 | 1.0789 |
| P12838 | DEF4 | Neutrophil defensin 4 | 2.4608 | 2.3711 | 1.2386 |
| P59666 | DEF3 | Neutrophil defensin 3 | 8.9484 | 3.7253 | 1.5188 |
| O14773 | TPP1 | Tripeptidyl-peptidase 1 | 1.4441 | 1.1461 | 1.0245 |
| Q8NBS9 | TXND5 | Thioredoxin domain-containing protein 5 | 1.5722 | 1.0267 | 1.2345 |
| P24158 | PRTN3 | Myeloblastin | 2.0115 | 1.8480 | 1.4986 |
| P50990 | TCPQ | T-complexprotein 1 subunittheta | 2.0416 | 1.2148 | 1.2408 |
| P12724 | ECP | Eosinophil cationic protein | 2.4191 | 1.0259 | 0.5594 |
| P17213 | BPI | Bactericidal permeability-increasing protein | 3.3966 | 1.5635 | 1.2036 |
| P05164 | PERM | Myeloperoxidase | 5.511 | 2.0122 | 1.2871 |
| P20160 | CAP7 | Azurocidin | 7.2103 | 2.0481 | 1.1568 |
| Q13231 | CHIT1 | Chitotriosidase-1 | 2.1411 | 2.0308 | 1.7411 |
| P00387 | NB5R3 | NADH-cytochrome b5 reductase 3 | 2.4463 | 1.7933 | 1.2666 |
| P23229 | ITA6 | Integrin alpha-6 | 1.8854 | 1.0913 | 0.6926 |
| P21926 | CD9 | CD9 antigen | 1.9864 | 1.0081 | 0.9969 |
| Q9Y6C2 | EMIL1 | EMILIN-1 | 2.4765 | 1.1540 | 1.2063 |
| P04004 | VTNC | Vitronectin | 4.0358 | 3.1389 | 4.1505 |
| P07996 | TSP1 | Thrombospondin-1 | 2.0331 | 1.0455 | 1.0711 |
| Q6UX06 | OLFM4 | Olfactomedin-4 | 2.8132 | 2.4717 | 2.1722 |
| Q15084 | PDIA6 | Protein disulfide-isomerase A6 | 1.4635 | 1.0387 | 1.1286 |
| P04839 | CY24B | Cytochrome b-245 heavy chain | 1.5946 | 1.6427 | 1.2274 |
| Q9HDC9 | APMAP | Adipocyte plasma membrane-associated protein | 1.615 | 1.3920 | 1.3258 |
| P61769 | B2MG | Beta-2-microglobulin | 1.6224 | 1.1067 | 0.8221 |
| P24557 | THAS | Thromboxane-A synthase | 1.7477 | 1.6320 | 1.4601 |
| P04844 | RPN2 | Dolichyl-diphospho-oligosaccharide—proteinglycosyltransferasesubunit 2 | 1.8642 | 1.3634 | 1.2661 |
| Q9NQC3 | RTN4 | Reticulon-4 | 1.9867 | 1.5387 | 1.0570 |
| Q14165 | MLEC | Malectin | 2.1586 | 1.0898 | 0.9708 |
| Q9BSJ8 | ESYT1 | Extended synaptotagmin-1 | 2.4649 | 1.2970 | 1.2265 |
| Q8TC12 | RDH11 | Retinol dehydrogenase 11 | 3.061 | 1.3400 | 0.8831 |
| P23219 | PGH1 | Prostaglandin G/H synthase 1 | 3.6929 | 1.5669 | 1.3489 |
| P02774 | VTDB | Vitamin D-binding protein | 2.4327 | 1.4855 | 1.6653 |
| P41240 | CSK | Tyrosine-protein kinase CSK | 3.4288 | 1.8739 | 3.5935 |
| P02749 | APOH | Beta-2-glycoprotein 1 | 5.374 | 2.4436 | 3.0788 |
| Q00325 | MPCP | Phosphate carrier protein. mitochondrial | 2.2758 | 2.5252 | 1.3470 |
| Q9UFN0 | NPS3A | ProteinNip Snap homolog 3ª | 1.6409 | 1.3436 | 1.3143 |
| Q96P48 | ARAP1 | Arf-GAP with Rho-GAP domain. ANK repeat and PH domain-containing protein 1 | 2.036 | 1.8559 | 1.3848 |
| P20061 | TCO1 | Transcobalamin-1 | 1.7894 | 1.2442 | 1.2795 |
| P80188 | NGAL | Neutrophil gelatinase-associated lipocalin | 1.983 | 1.5798 | 1.2060 |
| P08246 | ELNE | Neutrophil elastase | 2.9474 | 2.0215 | 0.9159 |
| P00747 | PLMN | Plasminogen | 3.3382 | 1.2731 | 1.4806 |
| Q8NBM8 | PCYXL | Prenylcysteine oxidase-like | 3.8514 | 1.0675 | 1.1120 |
| P02788 | TRFL | Lactotransferrin | 7.6776 | 2.2862 | 1.4302 |
| Q5SQ64 | LY66F | Lymphocyte antigen 6 complex locus protein G6f | 2.1107 | 1.3705 | 1.0059 |
| P54108 | CRIS3 | Cysteine-rich secretory protein 3 | 1.7124 | 1.7709 | 1.2384 |
| P54578 | UBP14 | Ubiquitin carboxyl-terminal hydrolase 14 | 1.6928 | 0.8851 | 1.3075 |
| Q7L5Y6 | DET1 | DET1 homolog | 2.8429 | 0.6266 | 0.5570 |
| P19971 | TYPH | Thymidine phosphorylase | 3.9674 | 3.6955 | 0.5058 |
| P00488 | F13A | Coagulation factor XIII A chain | 3.3421 | 3.2396 | 0.9832 |
| P06737 | PYGL | Glycogen phosphorylase. liver form | 8.5721 | 6.9374 | 0.5571 |
| P62136 | PP1A | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 1.7576 | 2.1843 | 0.9561 |
| P30101 | PDIA3 | Protein disulfide-isomerase A3 | 1.7864 | 0.5549 | 0.4749 |
| P55786 | PSA | Puromycin-sensitive aminopeptidase | 2.2783 | 0.7016 | 0.6691 |
| P07741 | APT | Adenine phosphoribosyltransferase | 3.7614 | 3.9742 | 0.8305 |
| Q99623 | PHB2 | Prohibitin-2 | 1.7628 | 2.1771 | 1.6235 |
| UniProtCode | UniProt Name | Protein Name | CG | Fold Change Relative to Untreated Group | |
|---|---|---|---|---|---|
| ERT-a | ERT-b | ||||
| Q96AG4 | LRC59 | Leucine-rich repeat-containing protein 59 | ND | 0.5690 | 0.6684 |
| Q12913 | PTPRJ | Receptor-type tyrosine-protein phosphatase eta | ND | 0.4456 | 0.5427 |
| P10153 | RNAS2 | Non-secretory ribonuclease | ND | 0.4025 | 0.1628 |
| Q06323 | PSME1 | Proteasome activator complex subunit 1 | ND | 0.6398 | 0.6990 |
| P27695 | APEX1 | DNA-(apurinic or apyrimidinic site) endonuclease | ND | 0.7912 | 0.5368 |
| P02042 | HBD | Hemoglobin subunit delta | ND | 0.3031 | 0.5340 |
| O43684 | BUB3 | Mitotic check point protein BUB3 | ND | 0.5946 | 0.4602 |
| Q9Y2Y8 | PRG3 | Proteoglycan 3 | ND | 0.5416 | 0.3759 |
| UG | EA nM/h/mg | Before ERT | EA nM/h/mg | After ERT | EA nM/h/mg | Healthy Controls | EA nM/h/mg |
|---|---|---|---|---|---|---|---|
| 1 | 0.2 | ERT-a 1 | 0.7 | ERT-b 1 | 1.8 | CG 1 | 4.8 |
| 2 | 0.2 | ERT-a 2 | 0.6 | ERT-b 2 | 2.1 | CG 2 | 14.2 |
| 3 | 0.2 | ERT-a 3 | 0.7 | ERT-b 3 | 2.2 | CG 3 | 2.7 |
| 4 | 0.0 | ERT-a 4 | 1.0 | ERT-b 4 | 2.8 | CG 4 | 3.6 |
| 5 | 0.1 | ERT-a 5 | 1.6 | ERT-b 5 | 6.7 | CG 5 | 3.1 |
| 6 | 0.2 | - | - | - | - | CG 6 | 4.8 |
| 7 | 0.1 | - | - | - | - | - | - |
| 8 | 0.2 | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez, V.J.; Bravo, S.B.; Chantada-Vazquez, M.P.; Colón, C.; De Castro, M.J.; Morales, M.; Vitoria, I.; Tomatsu, S.; Otero-Espinar, F.J.; Couce, M.L. Characterization of New Proteomic Biomarker Candidates in Mucopolysaccharidosis Type IVA. Int. J. Mol. Sci. 2021, 22, 226. https://doi.org/10.3390/ijms22010226
Álvarez VJ, Bravo SB, Chantada-Vazquez MP, Colón C, De Castro MJ, Morales M, Vitoria I, Tomatsu S, Otero-Espinar FJ, Couce ML. Characterization of New Proteomic Biomarker Candidates in Mucopolysaccharidosis Type IVA. International Journal of Molecular Sciences. 2021; 22(1):226. https://doi.org/10.3390/ijms22010226
Chicago/Turabian StyleÁlvarez, Víctor J., Susana B. Bravo, Maria Pilar Chantada-Vazquez, Cristóbal Colón, María J. De Castro, Montserrat Morales, Isidro Vitoria, Shunji Tomatsu, Francisco J. Otero-Espinar, and María L. Couce. 2021. "Characterization of New Proteomic Biomarker Candidates in Mucopolysaccharidosis Type IVA" International Journal of Molecular Sciences 22, no. 1: 226. https://doi.org/10.3390/ijms22010226
APA StyleÁlvarez, V. J., Bravo, S. B., Chantada-Vazquez, M. P., Colón, C., De Castro, M. J., Morales, M., Vitoria, I., Tomatsu, S., Otero-Espinar, F. J., & Couce, M. L. (2021). Characterization of New Proteomic Biomarker Candidates in Mucopolysaccharidosis Type IVA. International Journal of Molecular Sciences, 22(1), 226. https://doi.org/10.3390/ijms22010226









