Proteomics of Muscle Microdialysates Identifies Potential Circulating Biomarkers in Facioscapulohumeral Muscular Dystrophy
Abstract
:1. Introduction
2. Results
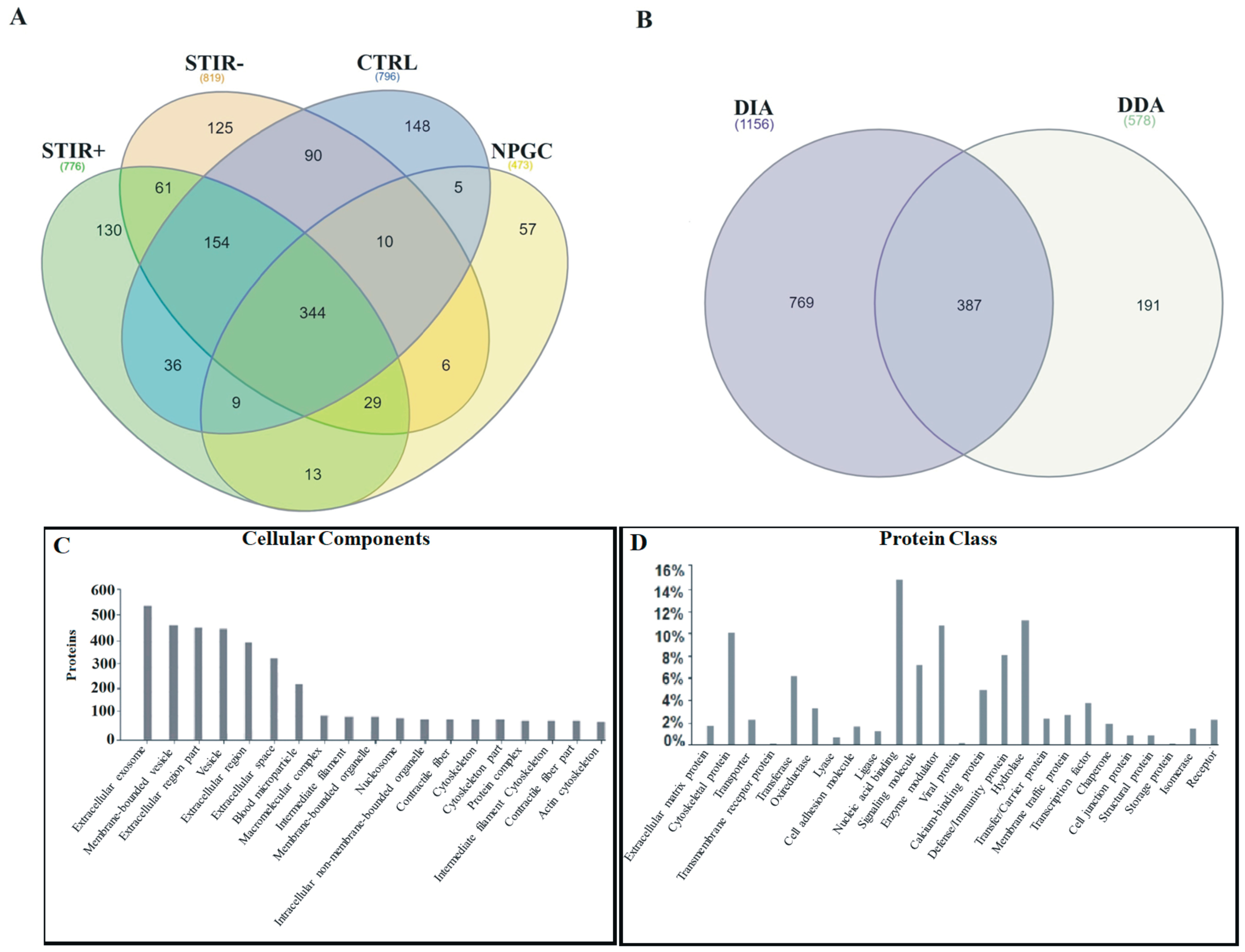
2.1. Combined Proteomic Analysis of Microdialysates
2.2. Comparative Proteomic Analysis Using DIA: STIR+ Versus STIR- Muscles
2.3. Comparison between STIR+ Muscles and Controls
2.4. Comparison between STIR- Muscles and Controls
2.5. DDA Label Free Quantification
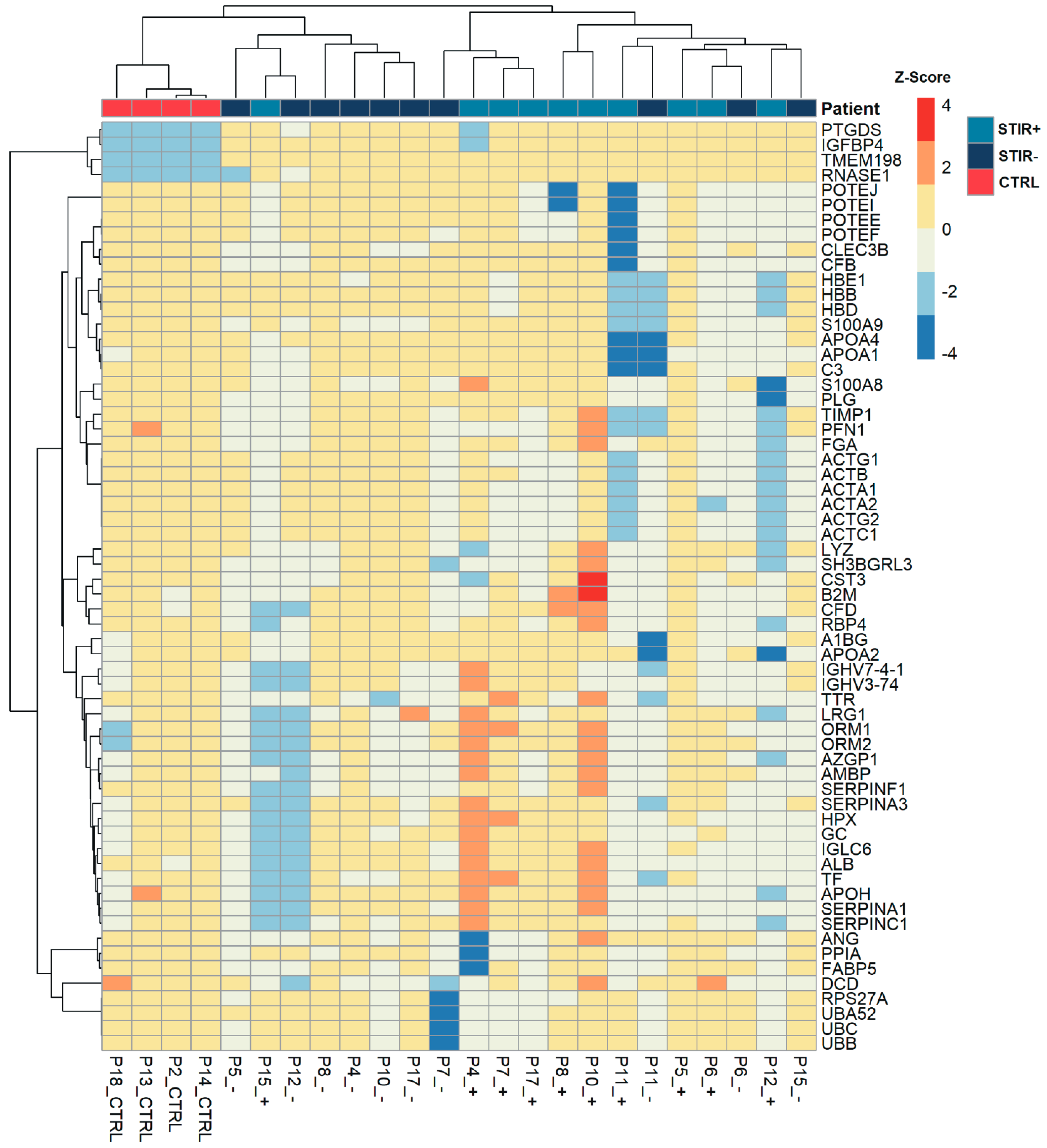
2.6. Targeted Proteomic Approach
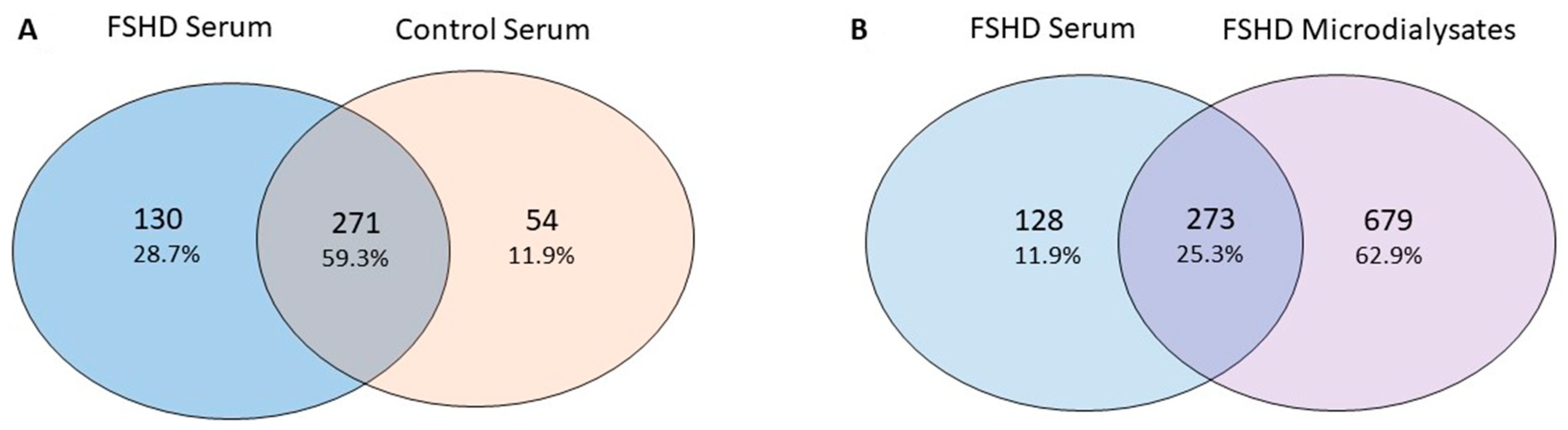
2.7. Serum Analysis
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Mass Spectrometry Based Proteomic Analysis
4.3. Synapt G2Si- Proteomic Analysis
4.4. Orbitrap Elite Proteomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FSHD | Facioscapulohumeral muscular dystrophy |
| MRI | Magnetic Resonance Imaging |
| STIR | Short Tau Inversion Recovery |
| LC-MS | Liquid Chromatography-Mass Spectrometry |
| NPGC | Non-penetrant gene carrier |
| DIA | Data Independent Acquisition |
| DDA | Data Dependent Acquisition |
| GO | Gene Ontology |
| PLGS | ProteinLynx Global Server |
| CTRL | Control |
| PSMs | Peptide Spectrum Matches |
| CSS | Clinical severity scale |
| FASP | Filter-aided sample preparation |
| MRM | Multiple reaction monitoring |
| IPA | Ingenuity Pathway Analysis |
| DAMP | Damage-Associated Molecular Pattern |
| MWCO | Molecular weight cut-off |
References
- Padberg, G.W.; Lunt, P.W.; Koch, M.; Fardeau, M. Diagnostic criteria for facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 1991, 1, 231–234. [Google Scholar] [CrossRef]
- Snider, L.; Geng, L.N.; Lemmers, R.J.L.F.; Kyba, M.; Ware, C.B.; Nelson, A.M.; Tawil, R.; Filippova, G.N.; van der Maarel, S.M.; Tapscott, S.J.; et al. Facioscapulohumeral dystrophy: Incomplete suppression of a retrotransposed gene. PLoS Genet. 2010, 6, e1001181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, L.N.; Yao, Z.; Snider, L.; Fong, A.P.; Cech, J.N.; Young, J.M.; van der Maarel, S.M.; Ruzzo, W.L.; Gentleman, R.C.; Tawil, R.; et al. DUX4 activates germline genes, retroelements, and immune mediators: Implications for facioscapulohumeral dystrophy. Dev. Cell 2012, 22, 38–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kissel, J.T.; McDermott, M.P.; Natarajan, R.; Mendell, J.R.; Pandya, S.; King, W.M.; Griggs, R.C.; Tawil, R.; FSH—Dy Group. Pilot trial of albuterol in facioscapulohumeral muscular dystrophy. Neurology 1998, 50, 1402–1406. [Google Scholar] [CrossRef]
- Statland, J.; Donlin-Smith, C.M.; Tapscott, S.J.; van der Maarel, S.; Tawil, R. Multiplex screen of serum biomarkers in facioscapulohumeral muscular dystrophy. J. Neuromuscul. Dis. 2014, 1, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Petek, L.M.; Rickard, A.M.; Budech, C.; Poliachik, S.L.; Shaw, D.; Ferguson, M.R.; Tawil, R.; Friedman, S.D.; Miller, D.G. A cross sectional study of two independent cohorts identifies serum biomarkers for facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord. 2016, 26, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Heier, C.R.; Zhang, A.; Nguyen, N.Y.; Tully, C.B.; Panigrahi, A.; Gordish-Dressman, H.; Pandey, S.P.; Guglieri, M.; Ryan, M.M.; Clemens, P.R.; et al. Multi-omics identifies circulating miRNA and protein biomarkers for facioscapulohumeral dystrophy. J. Pers. Med. 2020, 10, 236. [Google Scholar] [CrossRef]
- Tasca, G.; Monforte, M.; Ottaviani, P.; Pelliccioni, M.; Frusciante, R.; Laschena, F.; Ricci, E. Magnetic resonance imaging in a large cohort of facioscapulohumeral muscular dystrophy patients: Pattern refinement and implications for clinical trials. Ann. Neurol. 2016, 79, 854–864. [Google Scholar] [CrossRef]
- Monforte, M.; Laschena, F.; Ottaviani, P.; Bagnato, M.R.; Pichiecchio, A.; Tasca, G.; Ricci, E. Tracking muscle wasting and disease activity in facioscapulohumeral muscular dystrophy by qualitative longitudinal imaging. J. Cachexia Sarcopenia Muscle 2019, 10, 1258–1265. [Google Scholar] [CrossRef]
- Tawil, R.; Padberg, G.W.; Shaw, D.W.; van der Maarel, S.M.; Tapscott, S.J.; FSHD Workshop Participants. Clinical trial preparedness in facioscapulohumeral muscular dystrophy: Clinical, tissue, and imaging outcome measures 29–30 May 2015, Rochester, New York. Neuromuscul. Disord. 2016, 26, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Tasca, G.; Monforte, M.; Corbi, M.; Granata, G.; Lucchetti, D.; Sgambato, A.; Ricci, E. Muscle microdialysis to investigate inflammatory biomarkers in facioscapulohumeral muscular dystrophy. Mol. Neurobiol. 2018, 55, 2959–2966. [Google Scholar] [CrossRef] [PubMed]
- Tasca, G.; Pescatori, M.; Monforte, M.; Mirabella, M.; Iannaccone, E.; Frusciante, R.; Cubeddu, T.; Laschena, F.; Ottaviani, P.; Ricci, E. Different molecular signatures in magnetic resonance imaging-staged facioscapulohumeral muscular dystrophy muscles. PLoS ONE 2012, 7, e38779. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Song, E.; Nie, S.; Rodland, K.D.; Liu, T.; Qian, W.-J.; Smith, R.D. Advances in targeted proteomics and applications to biomedical research. Proteomics 2016, 16, 2160–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagannathan, S.; Ogata, Y.; Gafken, P.R.; Tapscott, S.J.; Bradley, R.K. Quantitative proteomics reveals key roles for post-transcriptional gene regulation in the molecular pathology of facioscapulohumeral muscular dystrophy. Elife 2019, 8. [Google Scholar] [CrossRef]
- Carlyle, B.; Trombetta, B.; Arnold, S. Proteomic approaches for the discovery of biofluid biomarkers of neurodegenerative dementias. Proteomes 2018, 6, 32. [Google Scholar] [CrossRef] [Green Version]
- Sandberg, A.; Branca, R.M.M.; Lehtiö, J.; Forshed, J. Quantitative accuracy in mass spectrometry based proteomics of complex samples: The impact of labeling and precursor interference. J. Proteom. 2014, 96, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Vidova, V.; Spacil, Z. A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition. Anal. Chim. Acta 2017, 964, 7–23. [Google Scholar] [CrossRef]
- Takeshita, E.; Komaki, H.; Tachimori, H.; Miyoshi, K.; Yamamiya, I.; Shimizu-Motohashi, Y.; Ishiyama, A.; Saito, T.; Nakagawa, E.; Sugai, K.; et al. Urinary prostaglandin metabolites as Duchenne muscular dystrophy progression markers. Brain Dev. 2018, 40, 918–925. [Google Scholar] [CrossRef]
- Austermann, J.; Spiekermann, C.; Roth, J. S100 proteins in rheumatic diseases. Nat. Rev. Rheumatol. 2018, 14, 528–541. [Google Scholar] [CrossRef]
- Khaki-Khatibi, F.; Qujeq, D.; Kashifard, M.; Moein, S.; Maniati, M.; Vaghari-Tabari, M. Calprotectin in inflammatory bowel disease. Clin. Chim Acta 2020, 510, 556–565. [Google Scholar] [CrossRef]
- Crowe, L.A.N.; McLean, M.; Kitson, S.M.; Melchor, E.G.; Patommel, K.; Cao, H.M.; Reilly, J.H.; Leach, W.J.; Rooney, B.P.; Spencer, S.J.; et al. S100A8 & S100A9: Alarmin mediated inflammation in tendinopathy. Sci. Rep. 2019, 9, 1463. [Google Scholar] [PubMed]
- Ehrchen, J.M.; Sunderkötter, C.; Foell, D.; Vogl, T.; Roth, J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J. Leukoc. Biol. 2009, 86, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Ometto, F.; Friso, L.; Astorri, D.; Botsios, C.; Raffeiner, B.; Punzi, L.; Doria, A. Calprotectin in rheumatic diseases. Exp. Biol. Med. 2017, 242, 859–873. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Schiattarella, G.G.; Perrino, C.; Cattaneo, F.; Pironti, G.; Franzone, A.; Gargiulo, G.; Magliulo, F.; Serino, F.; Carotenuto, G.; et al. Dermcidin: A skeletal muscle myokine modulating cardiomyocyte survival and infarct size after coronary artery ligation. Cardiovasc. Res. 2015, 107, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Stewart, G.D.; Skipworth, R.J.; Ross, J.A.; Fearon, K.C.; Baracos, V.E. The dermcidin gene in cancer: Role in cachexia, carcinogenesis and tumour cell survival. Curr. Opin. Clin. Nutr. Metab. Care 2008, 11, 208–213. [Google Scholar] [CrossRef]
- Núñez-Naveira, L.; Mariñas-Pardo, L.A.; Montero-Martínez, C. Mass spectrometry analysis of the exhaled breath condensate and proposal of dermcidin and S100A9 as possible markers for lung cancer prognosis. Lung 2019, 197, 523–531. [Google Scholar] [CrossRef]
- Wang, L.H.; Friedman, S.D.; Shaw, D.; Snider, L.; Wong, C.-J.; Budech, C.B.; Poliachik, S.L.; Gove, N.E.; Lewis, L.M.; Campbell, A.E.; et al. MRI-informed muscle biopsies correlate MRI with pathology and DUX4 target gene expression in FSHD. Hum. Mol. Genet. 2019, 28, 476–486. [Google Scholar] [CrossRef]
- Vignesh, P.; Rawat, A.; Sharma, M.; Singh, S. Complement in autoimmune diseases. Clin. Chim. Acta 2017, 465, 123–130. [Google Scholar] [CrossRef]
- Suelves, M.; Vidal, B.; Ruiz, V.; Baeza-Raja, B.; Diaz-Ramos, A.; Cuartas, I.; Lluis, F.; Parra, M.; Jardi, M.; Lopez-Alemany, R.; et al. The plasminogen activation system in skeletal muscle regeneration: Antagonistic roles of urokinase-type plasminogen activator (uPA) and its inhibitor (PAI-1). Front. Biosci. 2005, 10, 2978–2985. [Google Scholar] [CrossRef] [Green Version]
- Celegato, B.; Capitanio, D.; Pescatori, M.; Romualdi, C.; Pacchioni, B.; Cagnin, S.; Viganò, A.; Colantoni, L.; Begum, S.; Ricci, E.; et al. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics 2006, 6, 5303–5321. [Google Scholar] [CrossRef] [Green Version]
- Lepedda, A.J.; Deiana, G.A.; Lobina, O.; Nieddu, G.; Baldinu, P.; De Muro, P.; Andreetta, F.; Sechi, E.; Arru, G.; Corda, D.G.; et al. Plasma vitronectin is reduced in patients with myasthenia gravis: Diagnostic and pathophysiological potential. J. Circ. Biomark 2019, 8, 1849454419875912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irigoyen, J.P.; Muñoz-Cánoves, P.; Montero, L.; Koziczak, M.; Nagamine, Y. The plasminogen activator system: Biology and regulation. Cell. Mol. Life Sci. 1999, 56, 104–132. [Google Scholar] [CrossRef] [PubMed]
- Cenik, B.K.; Liu, N.; Chen, B.; Bezprozvannaya, S.; Olson, E.N.; Bassel-Duby, R. Myocardin-related transcription factors are required for skeletal muscle development. Development 2016, 143, 2853–2861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collard, L.; Herledan, G.; Pincini, A.; Guerci, A.; Randrianarison-Huetz, V.; Sotiropoulos, A. Nuclear actin and myocardin-related transcription factors control disuse muscle atrophy through regulation of SRF activity. J. Cell Sci. 2014, 127, 5157–5163. [Google Scholar] [CrossRef] [Green Version]
- Morosetti, R.; Mirabella, M.; Gliubizzi, C.; Broccolini, A.; Sancricca, C.; Pescatori, M.; Gidaro, T.; Tasca, G.; Frusciante, R.; Tonali, P.A.; et al. Isolation and characterization of mesoangioblasts from facioscapulohumeral muscular dystrophy muscle biopsies. Stem Cells 2007, 25, 3173–3182. [Google Scholar] [CrossRef]
- Bárcena, C.; Aran, G.; Perea, L.; Sanjurjo, L.; Téllez, É.; Oncins, A.; Masnou, H.; Serra, I.; García-Gallo, M.; Kremer, L.; et al. CD5L is a pleiotropic player in liver fibrosis controlling damage, fibrosis and immune cell content. EBioMedicine 2019, 43, 513–524. [Google Scholar] [CrossRef] [Green Version]
- Sanjurjo, L.; Amézaga, N.; Aran, G.; Naranjo-Gómez, M.; Arias, L.; Armengol, C.; Borràs, F.E.; Sarrias, M.-R. The human CD5L/AIM-CD36 axis: A novel autophagy inducer in macrophages that modulates inflammatory responses. Autophagy 2015, 11, 487–502. [Google Scholar] [CrossRef] [Green Version]
- Zografos, E.; Anagnostopoulos, A.K.; Papadopoulou, A.; Legaki, E.; Zagouri, F.; Marinos, E.; Tsangaris, G.T.; Gazouli, M. Serum proteomic signatures of male breast cancer. Cancer Genom. Proteom. 2019, 16, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Marini, F.; Carregari, V.C.; Greco, V.; Ronci, M.; Iavarone, F.; Persichilli, S.; Castagnola, M.; Urbani, A.; Pieroni, L. Exploring the hela dark mitochondrial proteome. Front. Cell Dev. Biol 2020, 8, 137. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Tenzer, S. Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 2016, 11, 795–812. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.-Z.; Vissers, J.P.C.; Geromanos, S.J. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteom. 2006, 5, 144–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Zhao, Y.; Angeles, A.; Hamuro, L.L.; Arnold, M.E.; Shen, J.X. A novel and cost effective method of removing excess albumin from plasma/serum samples and its impacts on LC-MS/MS bioanalysis of therapeutic proteins. Anal. Chem. 2014, 86, 8336–8343. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]

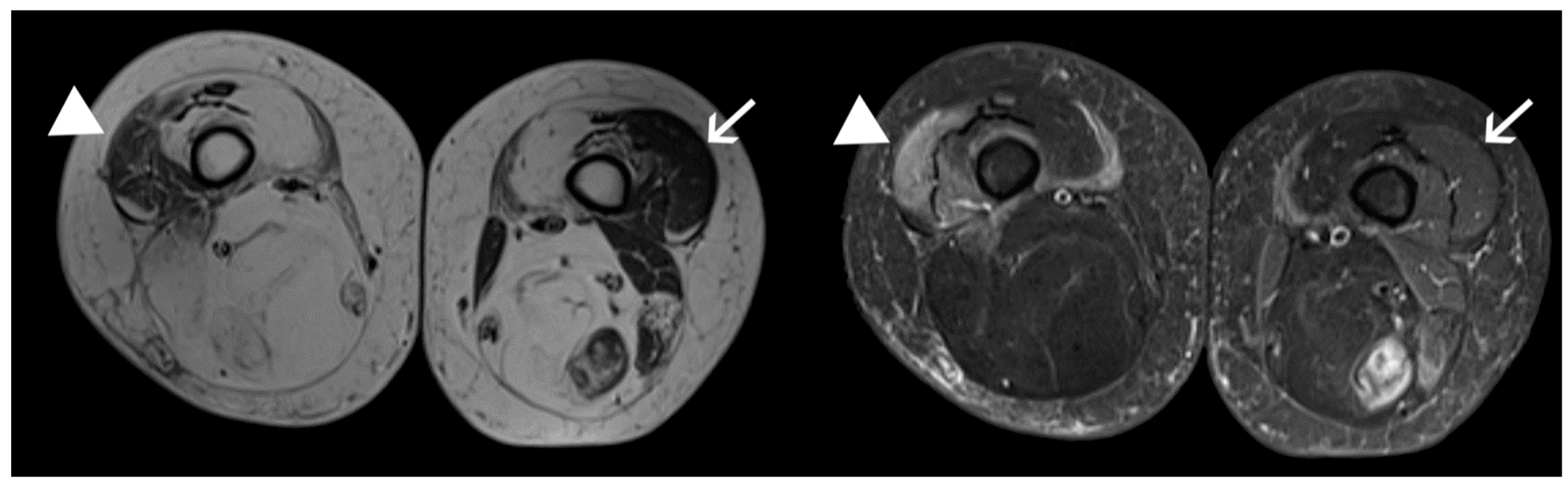
| CATEGORY | n | AGE | SEX | EcoRI | CSS |
|---|---|---|---|---|---|
| FSHD | 10 | 41.7 ± 13.4; range 18–58 | 5 M–5 F | 21.5; range 15–25 | 3; range 1.5–3.5 |
| CONTROLS | 4 | 41.5 ± 16.4; range 24–60 | 3 M–1 F | --- | --- |
| Accession Number | Protein Description | Score | STIR+:STIR- Ratio |
|---|---|---|---|
| P41222 | Prostaglandin-H2 D-isomerase (PTGDS) | 661.92 | 2.89 |
| P06702 | Protein S100-A9 (S100A9) | 4710.25 | 2.23 |
| P22692 | Insulin-like growth factor-binding protein 4 (IGFBP4) | 450.89 | 1.80 |
| P81605 | Dermcidin (DCD) | 3227.52 | 1.77 |
| A2NJV5 | Immunoglobulin kappa variable 2-29 (IGKV2-29) | 2955.26 | 1.67 |
| A0A075B6S2 | Immunoglobulin kappa variable 2D-29 (IGKV2D-29) | 2955.26 | 1.65 |
| A0A075B6P5 | Immunoglobulin kappa variable 2-28 (IGKV2-28) | 2955.26 | 1.65 |
| P06310 | Immunoglobulin kappa variable 2-30 (IGKV2-30) | 2955.26 | 1.63 |
| A0A0A0MRZ7 | Immunoglobulin kappa variable 2D-26 (IGKV2D-26) | 2955.26 | 1.63 |
| A0A075B6S6 | Immunoglobulin kappa variable 2D-30 (IGKV2D-30) | 2955.26 | 1.62 |
| P01615 | Immunoglobulin kappa variable 2D-28 (IGKV2D-28) | 2955.26 | 1.62 |
| P01614 | Immunoglobulin kappa variable 2D-40 (IGKV2D-40) | 3081.8 | 1.62 |
| A0A087WW87 | Immunoglobulin kappa variable 2-40 (IGKV2-40) | 3081.8 | 1.58 |
| P05109 | Protein S100-A8 (S100A8) | 3410.17 | 1.42 |
| P01619 | Immunoglobulin kappa variable 3-20 (IGKV3-20) | 11,910.68 | 1.42 |
| P02766 | Transthyretin (TTR) | 21,752.78 | 1.34 |
| P02790 | Hemopexin (HPX) | 10,037.44 | 1.31 |
| P11217 | Glycogen phosphorylase_ muscle form (PYGM) | 669.66 | 1.26 |
| P07998 | Ribonuclease pancreatic (RNASE1) | 3437.69 | 1.26 |
| P11216 | Glycogen phosphorylase_ brain (PYGB) | 632.03 | 1.25 |
| P02652 | Apolipoprotein A-II (APOA2) | 18,158.41 | 1.23 |
| P59665 | Neutrophil defensin 1 (DEFA1) | 3886.69 | 0.81 |
| P59666 | Neutrophil defensin 3 (DEFA3) | 3886.69 | 0.79 |
| P62937 | Peptidyl-prolyl cis-trans isomerase A (PPIA) | 891.54 | 0.73 |
| Q9H299 | SH3 domain-binding glutamic acid-rich-like protein 3 (SH3BGRL3) | 2135.99 | 0.73 |
| P61626 | Lysozyme C (LYZ) | 6273.03 | 0.71 |
| P02042 | Hemoglobin subunit delta (HBD) | 29,240.84 | 0.71 |
| P07451 | Carbonic anhydrase 3 (CA3) | 1277.63 | 0.70 |
| P0CG47 | Polyubiquitin-B (UBB) | 685.65 | 0.69 |
| P62987 | Ubiquitin-60S ribosomal protein L40 (UBA52) | 685.65 | 0.68 |
| P62979 | Ubiquitin-40S ribosomal protein S27a (RPS27A) | 690.87 | 0.67 |
| P0CG48 | Polyubiquitin-C (UBC) | 685.65 | 0.67 |
| P68871 | Hemoglobin subunit beta (HBB) | 67,024.84 | 0.65 |
| P69905 | Hemoglobin subunit alpha (HBA1) | 28,016.07 | 0.65 |
| P02671 | Fibrinogen alpha chain (FGA) | 3726.94 | 0.63 |
| P06727 | Apolipoprotein A-IV (APOA4) | 3903.4 | 0.59 |
| P00747 | Plasminogen (PLG) | 2244.15 | 0.57 |
| P02100 | Hemoglobin subunit epsilon (HBE1) | 1198.27 | 0.48 |
| P07737 | Profilin-1 (PFN1) | 8113.64 | 0.47 |
| A5A3E0 | POTE ankyrin domain family member F (POTEF) | 354.23 | 0.43 |
| Q02325 | Plasminogen-like protein B (PLGLB1) | 895.85 | 0.42 |
| Q6S8J3 | POTE ankyrin domain family member E (POTEE) | 363.36 | 0.42 |
| P69891 | Hemoglobin subunit gamma-1 (HBG1) | 2857.95 | 0.41 |
| P63261 | Actin_ cytoplasmic 2 (ACTG1) | 2756.19 | 0.40 |
| P60709 | Actin_ cytoplasmic 1 (ACTB) | 2756.19 | 0.40 |
| P69892 | Hemoglobin subunit gamma-2 (HBG2) | 2888.5 | 0.39 |
| P68032 | Actin_ alpha cardiac muscle (ACTC1) | 1401.05 | 0.33 |
| P63267 | Actin_ gamma-enteric smooth muscle (ACTG2) | 1401.05 | 0.32 |
| P62736 | Actin_ aortic smooth muscle (ACTA2) | 1401.05 | 0.32 |
| P68133 | Actin_ alpha skeletal muscle (ACTA1) | 1401.05 | 0.32 |
| Accession Number | Protein Description | PSMs STIR+ | PSMs STIR- | PSMs CTRL | Fold Change STIR+/STIR- | Fold Change STIR+/CTRL | p Value STIR+/STIR- | p Value STIR+/CTRL |
|---|---|---|---|---|---|---|---|---|
| Q8NE71 | ATP-binding cassette sub-family F member 1 (ABCF1) | 3.5 | 1 | 1 | 3.5 | 3.5 | 0.007 | 0.007 |
| P13796 | Plastin-2 (LCP1) | 4 | 1 | 12.5 | 4 | −3.13 | 0.003 | |
| Q14019 | Coactosin-like protein (COTL1) | 4 | 1 | 6.5 | 4 | −1.63 | 0.038 | |
| P61769 | Beta-2-microglobulin (B2M) | 27 | 14 | 11.5 | 1.93 | 2.35 | 0.012 | 0.029 |
| P04040 | Catalase (CAT) | 35.5 | 16 | 8.5 | 2.22 | 4.18 | 0.040 | 0.017 |
| P81605 | Dermcidin (DCD) | 19 | 8.5 | 13 | 2.24 | 1.46 | 0.020 | |
| O75223 | Gamma-glutamylcyclotransferase (GGCT) | 9.5 | 3 | 3 | 3.17 | 3.17 | 0.006 | 0.006 |
| P15924 | Desmoplakin (DSP) | 65.5 | 29.5 | 7.5 | 2.22 | 8.73 | 0.137 | 0.026 |
| P22352 | Glutathione peroxidase 3 (GPX3) | 4.5 | 1 | 1 | 4.5 | 4.5 | 0.003 | 0.003 |
| P04433 | Ig kappa chain V-III region VG (Fragment) | 11 | 5 | 5 | 2.2 | 2.2 | 0.027 | |
| P69905 | Hemoglobin subunit alpha (HBA1) | 182 | 56 | 74 | 3.25 | 2.46 | 0.042 | 0.027 |
| P01743 | Ig heavy chain V-I region HG3 | 9.5 | 4,5 | 7 | 2.11 | 1.36 | 0.019 | 0.038 |
| P01767 | Ig heavy chain V-III region BUT | 10.5 | 6.5 | 14.5 | 1.62 | −1.38 | 0.030 | 0.030 |
| P01597 | Ig kappa chain V-I region DEE | 13.5 | 7.5 | 8 | 1.8 | 1.69 | 0.063 | 0.008 |
| B9A064 | Immunoglobulin lambda-like polypeptide 5 | 143.5 | 87 | 104.5 | 1.65 | 1.37 | 0.091 | 0.008 |
| P02788 | Lactotransferrin (LTF) | 15 | 1 | 3.5 | 15 | 4.29 | <0.001 | 0.009 |
| P61626 | Lysozyme C (LYZ) | 13 | 8.5 | 6.5 | 1.53 | 2 | 0.061 | 0.04 |
| P07737 | Profilin-1 (PFN1) | 29 | 15 | 22 | 1.93 | 1.32 | 0.025 | 0.02 |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein (HSPG2) | 13 | 8 | 6 | 1.63 | 2.17 | 0.155 | 0.038 |
| P04220 | Ig mu heavy chain disease protein | 61.5 | 39 | 16 | 1.58 | 3.84 | 0.118 | 0.03 |
| P01620 | Ig kappa chain V-III region SIE | 36.5 | 23.5 | 27.5 | 1.55 | 1.33 | 0.143 | 0.006 |
| P05109 | Protein S100-A8 (S100A8) | 13.5 | 6.5 | 7.5 | 2.08 | 1.8 | 0.047 | 0.014 |
| P01871 | Ig mu chain C region (IGHM) | 85.5 | 56 | 23.5 | 1.53 | 3.64 | 0.176 | 0.048 |
| P06702 | Protein S100-A9 (S100A9) | 17.5 | 10 | 11.5 | 1.75 | 1.52 | 0.022 | 0.014 |
| P02766 | Transthyretin (TTR) | 126.5 | 77.5 | 101 | 1.63 | 1.25 | 0.004 | 0.005 |
| O75112 | LIM domain-binding protein 3 (LDB3) | 1 | 9.5 | 29 | −9.5 | −29 | 0.02 | <0.001 |
| P05976 | Myosin light chain 1/3, skeletal muscle isoform (MYL1) | 1 | 19 | 19 | −19 | −19 | 0.007 | 0.001 |
| Q96A32 | Myosin regulatory light chain 2, skeletal muscle isoform (MYLPF) | 1 | 23 | 25.5 | −23 | −25.5 | <0.001 | 0.031 |
| Q9UKX2 | Myosin-2 (MYH2) | 8.67 | 143 | 192 | −16.5 | −22.15 | <0.001 | 0.03 |
| P00441 | Superoxide dismutase [Cu-Zn] (SOD1) | 3.5 | 9 | 10 | −2.57 | −2.86 | 0.039 | 0.028 |
| Pt ID | Dx | Age | Sex | EcoRI | CSS | Muscle | Age at Onset | Symptoms at Onset | Treatment | Past Medical History |
|---|---|---|---|---|---|---|---|---|---|---|
| p2 | CTRL | 24 | M | N.P. | 0 | Vastus lateralis | N.A. | N.A. | None | Unremarkable |
| p4 | FSHD | 43 | F | 17 | 3 | Gastrocnemius lateralis | Second decade | Scapular winging | Levothyroxine | Hypothyroidism |
| p5 | FSHD | 53 | F | 24 | 3.5 | Peroneus | Second decade | Scapular winging | Levothyroxine | Hypothyroidism |
| p6 | FSHD | 55 | F | 15 | 3.5 | Vastus lateralis | Second decade | Scapular winging | Amlodipine, bisoprolol, hydrochlorothiazide | Hypertension |
| p7 | FSHD | 29 | M | 20 | 2.5 | Biceps femoris short head | Second decade | Difficulty in raising arms | None | Unremarkable |
| p8 | FSHD | 18 | M | 23 | 1.5 | Semimembranosus | Second decade | Difficulty in raising arms | None | Unremarkable |
| p9 | NPGC | 47 | F | 33 | 0 | Vastus lateralis | N.A. | N.A. | None | Headache |
| p10 | FSHD | 44 | M | 25 | 3.5 | Gastrocnemius lateralis | Fourth decade | Scapular winging | None | Unremarkable |
| p11 | FSHD | 58 | F | 23 | 3 | Gracilis | Second decade | Difficulty in raising arms | Cholecalciferol | Osteoporosis |
| p12 | FSHD | 53 | M | 24 | 3 | Extensor digitorum longus | Fifth decade | Scapular winging | None | Unremarkable |
| p13 | CTRL | 60 | M | >40 | 0 | Vastus lateralis | N.A. | N.A. | None | Unremarkable |
| p14 | CTRL | 50 | M | >40 | 0 | Vastus lateralis | N.A. | N.A. | None | Unremarkable |
| p15 | FSHD | 34 | M | 19 | 3 | Semitendinosus | Third decade | Facial weakness | None | Unremarkable |
| p17 | FSHD | 30 | F | 18 | 3 | Tibialis anterior | Second decade | Scapular winging | None | Unremarkable |
| p18 | CTRL | 32 | F | N.P. | 0 | Vastus lateralis | N.A. | N.A. | None | Unremarkable |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corasolla Carregari, V.; Monforte, M.; Di Maio, G.; Pieroni, L.; Urbani, A.; Ricci, E.; Tasca, G. Proteomics of Muscle Microdialysates Identifies Potential Circulating Biomarkers in Facioscapulohumeral Muscular Dystrophy. Int. J. Mol. Sci. 2021, 22, 290. https://doi.org/10.3390/ijms22010290
Corasolla Carregari V, Monforte M, Di Maio G, Pieroni L, Urbani A, Ricci E, Tasca G. Proteomics of Muscle Microdialysates Identifies Potential Circulating Biomarkers in Facioscapulohumeral Muscular Dystrophy. International Journal of Molecular Sciences. 2021; 22(1):290. https://doi.org/10.3390/ijms22010290
Chicago/Turabian StyleCorasolla Carregari, Victor, Mauro Monforte, Giuseppe Di Maio, Luisa Pieroni, Andrea Urbani, Enzo Ricci, and Giorgio Tasca. 2021. "Proteomics of Muscle Microdialysates Identifies Potential Circulating Biomarkers in Facioscapulohumeral Muscular Dystrophy" International Journal of Molecular Sciences 22, no. 1: 290. https://doi.org/10.3390/ijms22010290
APA StyleCorasolla Carregari, V., Monforte, M., Di Maio, G., Pieroni, L., Urbani, A., Ricci, E., & Tasca, G. (2021). Proteomics of Muscle Microdialysates Identifies Potential Circulating Biomarkers in Facioscapulohumeral Muscular Dystrophy. International Journal of Molecular Sciences, 22(1), 290. https://doi.org/10.3390/ijms22010290







