The Evolving Landscape of Exosomes in Neurodegenerative Diseases: Exosomes Characteristics and a Promising Role in Early Diagnosis
Abstract
:1. Introduction
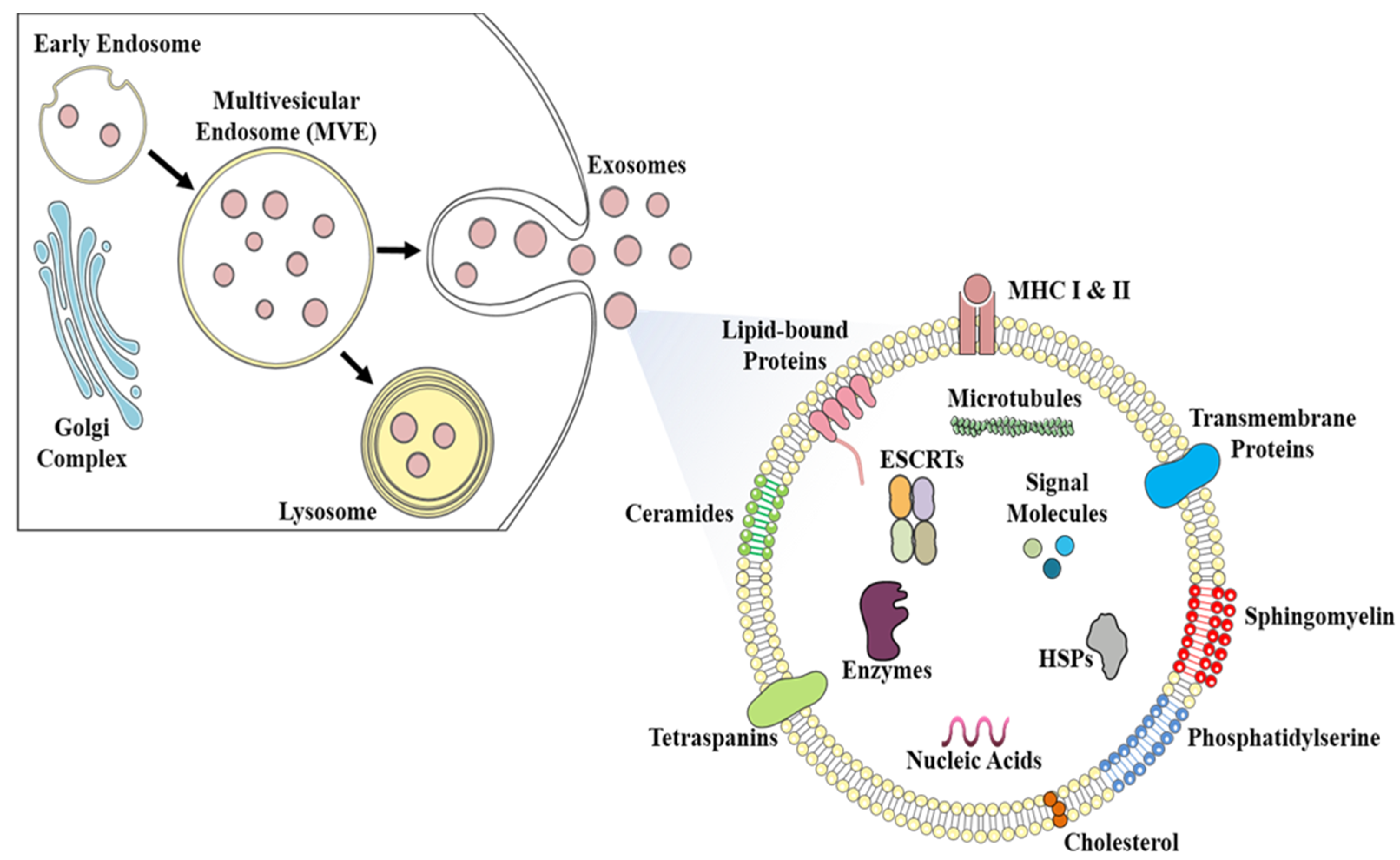
2. Exosomes: Biogenesis, Composition, and Their Diverse Functions
3. Ultrastructural, Biochemical, and Size Based Characterization of Exosomes
3.1. Physical Analysis of Exosomes
3.2. Biochemical Analysis of Exosomes
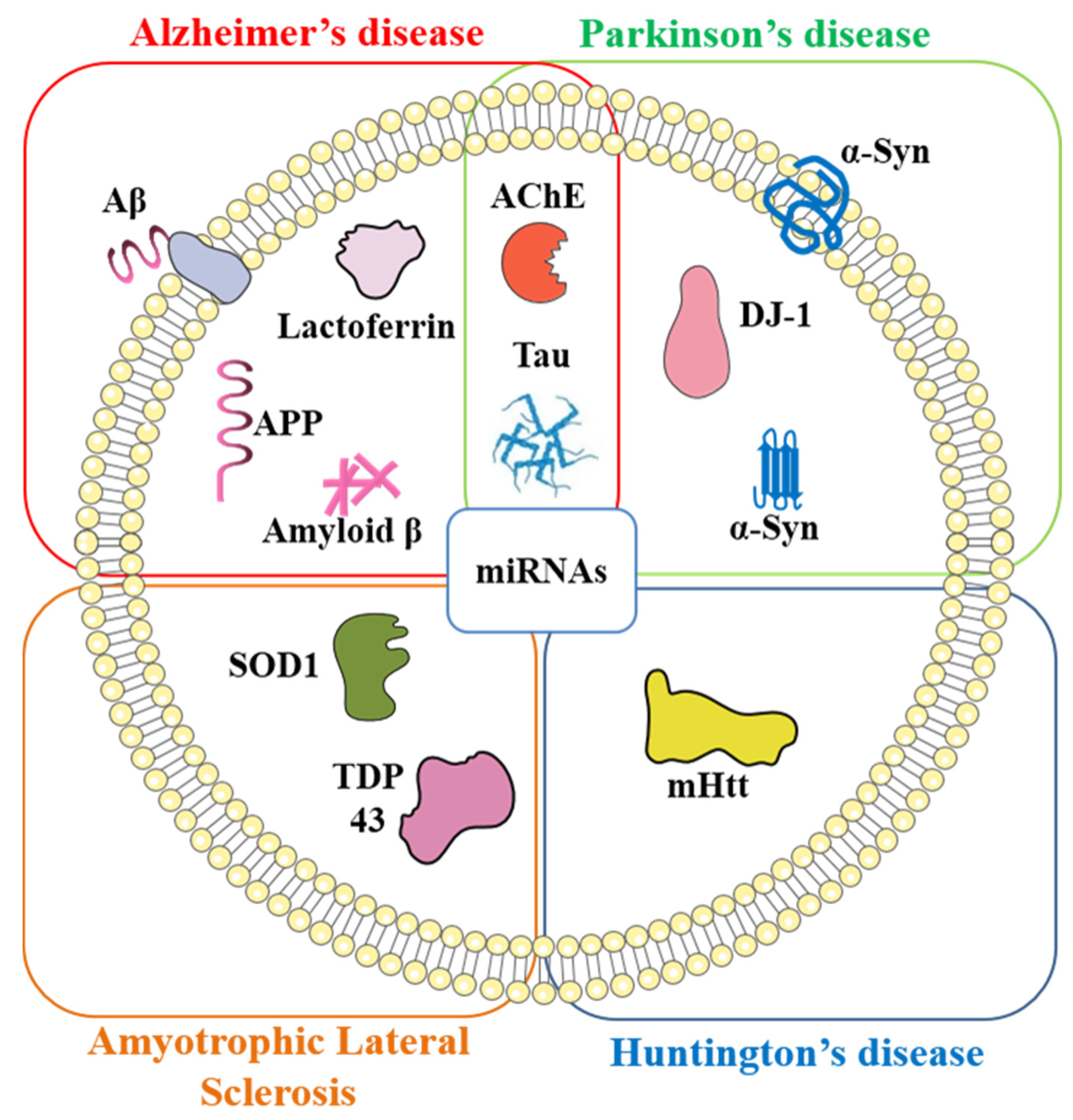
4. Exosomal Biomarkers and Their Role in Neurodegenerative Diseases
4.1. Alzheimer’s Disease
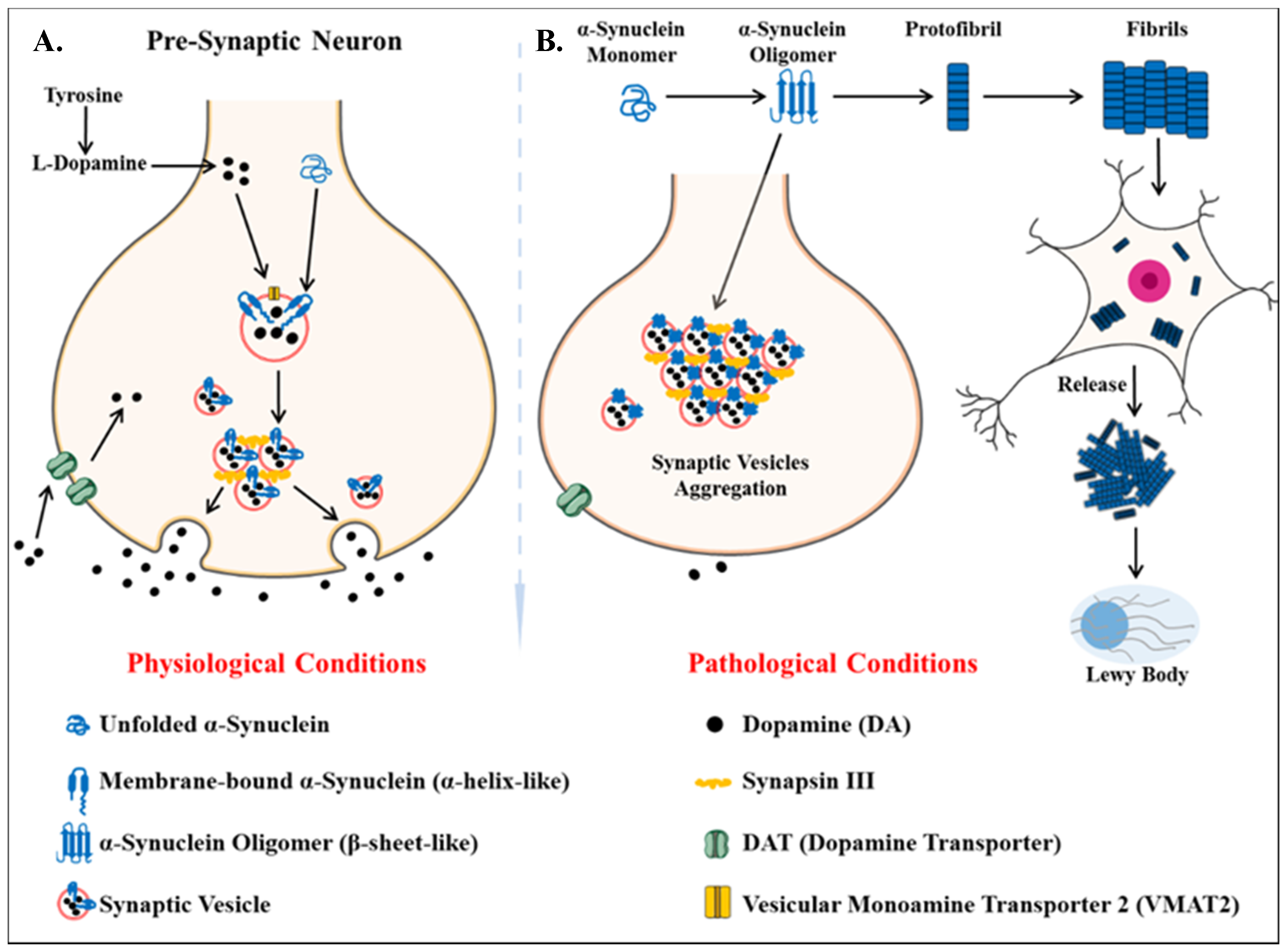
4.2. Parkinson’s Disease
4.3. Other Neurodegenerative Disorders
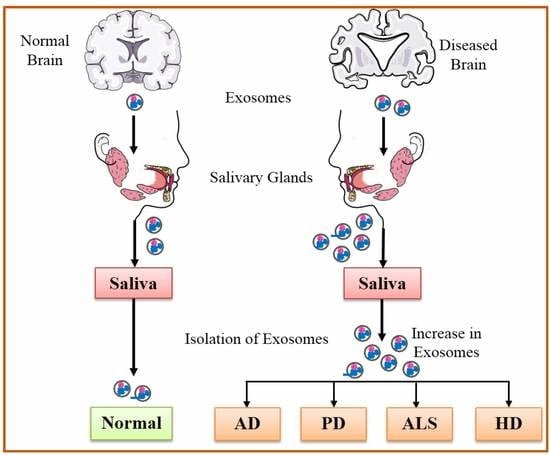
5. Potential of Salivary Exosomes Based Biomarkers for Early Diagnosis of Neurodegenerative Diseases
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Frost, B.; Diamond, M.I. Prion-like mechanisms in neurodegenerative diseases. Nat. Rev. Neurosci. 2010, 11, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Jeromin, A.; Bowser, R. Biomarkers in Neurodegenerative Diseases. Adv. Neurobiol. 2017, 15, 491–528. [Google Scholar] [CrossRef] [PubMed]
- Obrocki, P.; Khatun, A.; Ness, D.; Senkevich, K.; Hanrieder, J.; Capraro, F.; Mattsson, N.; Andreasson, U.; Portelius, E.; Ashton, N.J.; et al. Perspectives in fluid biomarkers in neurodegeneration from the 2019 biomarkers in neurodegenerative diseases course—a joint PhD student course at University College London and University of Gothenburg. Alzheimer’s Res. Ther. 2020, 12, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlyle, B.C.; Trombetta, B.A.; Arnold, S.E. Proteomic Approaches for the Discovery of Biofluid Biomarkers of Neurodegenerative Dementias. Proteomes 2018, 6, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, S.R.L.; Toffoli, M.; Campbell, P.; Schapira, A.H.V. Biofluid Biomarkers in Parkinson’s Disease: Clarity Amid Controversy. Mov. Disord. 2020, 35, 1128–1133. [Google Scholar] [CrossRef] [PubMed]
- Galasko, D. Expanding the Repertoire of Biomarkers for Alzheimer’s Disease: Targeted and Non-targeted Approaches. Front. Neurol. 2015. [Google Scholar] [CrossRef] [Green Version]
- Khan, T.K.; Alkon, D.L. Alzheimer’s Disease Cerebrospinal Fluid and Neuroimaging Biomarkers: Diagnostic Accuracy and Relationship to Drug Efficacy. J. Alzheimer’s Dis. 2015, 46, 817–836. [Google Scholar] [CrossRef]
- Greenberg, N.; Grassano, A.; Thambisetty, M.; Lovestone, S.; Legido-Quigley, C. A proposed metabolic strategy for monitoring disease progression in Alzheimer’s disease. Electrophoresis 2009, 30, 1235–1239. [Google Scholar] [CrossRef]
- Orešič, M.; Hyötyläinen, T.; Herukka, S.K.; Sysi-Aho, M.; Mattila, I.; Seppänan-Laakso, T.; Julkunen, V.; Gopalacharyulu, P.V.; Hallikainen, M.; Koikkalainen, J.; et al. Metabolome in progression to Alzheimer’s disease. Transl. Psychiatry 2011, 1, e57. [Google Scholar] [CrossRef]
- Trushina, E.; Mielke, M.M. Recent advances in the application of metabolomics to Alzheimer’s Disease. Biochim. Biophys. Acta 2014, 1842, 1232–1239. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.; Wang, M. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Saeedi, S.; Israel, S.; Nagy, C.; Turecki, G. The emerging role of exosomes in mental disorders. Transl. Psychiatry 2019, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Duban, L.; Segura, E.; Véron, P.; Lantz, O.; Amigorena, S. Indirect activation of naïve CD4+ T cells by dendritic cell-derived exosomes. Nat. Immunol. 2002, 3, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Mesci, P.; Carromeu, C.; McClatchy, D.R.; Schiapparelli, L.; Yates, J.R., 3rd; Muotri, A.R.; Cline, H.T. Exosomes regulate neurogenesis and circuit assembly. Proc. Natl. Acad. Sci. USA 2019, 116, 16086–16094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- Ching, R.C.; Kingham, P.J. The role of exosomes in peripheral nerve regeneration. Neural Regen. Res. 2015, 10, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Bang, C.; Thum, T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ. Cardiovasc. Genet. 2010, 3, 484–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fröhlich, D.; Kuo, W.P.; Frühbeis, C.; Sun, J.-J.; Zehendner, C.M.; Luhmann, H.J.; Pinto, S.; Toedling, J.; Trotter, J.; Krämer-Albers, E.-M. Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130510. [Google Scholar] [CrossRef]
- Frühbeis, C.; Fröhlich, D.; Kuo, W.P.; Amphornrat, J.; Thilemann, S.; Saab, A.S.; Kirchhoff, F.; Möbius, W.; Goebbels, S.; Nave, K.-A.; et al. Neurotransmitter-Triggered Transfer of Exosomes Mediates Oligodendrocyte–Neuron Communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef] [Green Version]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef] [Green Version]
- Février, B.; Raposo, G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Wang, B.; Kodali, M.C.; Chen, C.; Kim, E.; Patters, B.J.; Lan, L.; Kumar, S.; Wang, X.; Yue, J.; et al. In vivo evidence for the contribution of peripheral circulating inflammatory exosomes to neuroinflammation. J. Neuroinflamm. 2018, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Bakhti, M.; Winter, C.; Simons, M. Inhibition of myelin membrane sheath formation by oligodendrocyte-derived exosome-like vesicles. J. Biol. Chem. 2011, 286, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krämer-Albers, E.M.; Bretz, N.; Tenzer, S.; Winterstein, C.; Möbius, W.; Berger, H.; Nave, K.A.; Schild, H.; Trotter, J. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: Trophic support for axons? In Proteomics—Clinical Applications; Wiley: Weinheim, Germany, 2007; Volume 1, pp. 1446–1461. [Google Scholar]
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Ségaliny, A.; et al. Elucidation of Exosome Migration Across the Blood–Brain Barrier Model In Vitro. Cell. Mol. Bioeng. 2016, 9, 509–529. [Google Scholar] [CrossRef]
- Zhuang, X.; Xiang, X.; Grizzle, W.; Sun, D.; Zhang, S.; Axtell, R.C.; Ju, S.; Mu, J.; Zhang, L.; Steinman, L.; et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011, 19, 1769–1779. [Google Scholar] [CrossRef]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef] [Green Version]
- Malhotra, H.; Sheokand, N.; Kumar, S.; Chauhan, A.S.; Kumar, M.; Jakhar, P.; Boradia, V.M.; Raje, C.I.; Raje, M. Exosomes: Tunable Nano Vehicles for Macromolecular Delivery of Transferrin and Lactoferrin to Specific Intracellular Compartment. J. Biomed. Nanotechnol. 2016, 12, 1101–1114. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Ratajczak, J. Extracellular microvesicles/exosomes: Discovery, disbelief, acceptance, and the future? Leukemia 2020. [Google Scholar] [CrossRef]
- Chung, I.-M.; Rajakumar, G.; Venkidasamy, B.; Subramanian, U.; Thiruvengadam, M. Exosomes: Current use and future applications. Clin. Chim. Acta 2020, 500, 226–232. [Google Scholar] [CrossRef]
- Cheshmi, B.; Cheshomi, H. Salivary exosomes: Properties, medical applications, and isolation methods. Mol. Biol. Rep. 2020, 47, 6295–6307. [Google Scholar] [CrossRef]
- Conigliaro, A.; Corrado, C.; Fontana, S.; Alessandro, R. Chapter 1—Exosome basic mechanisms. In Exosomes; Edelstein, L., Smythies, J., Quesenberry, P., Noble, D., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 1–21. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Norman, S., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta (BBA) Biomembr. 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015, 1295, 179–209. [Google Scholar] [CrossRef]
- Sokolova, V.; Ludwig, A.K.; Hornung, S.; Rotan, O.; Horn, P.A.; Epple, M.; Giebel, B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids and surfaces. Colloids Surf. B Biointerfaces 2011, 87, 146–150. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, E.; Coumans, F.A.; Grootemaat, A.E.; Gardiner, C.; Sargent, I.L.; Harrison, P.; Sturk, A.; van Leeuwen, T.G.; Nieuwland, R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thromb. Haemost. 2014, 12, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, H.F.G.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated Platelets Release Two Types of Membrane Vesicles: Microvesicles by Surface Shedding and Exosomes Derived From Exocytosis of Multivesicular Bodies and alpha-Granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT Pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radulovic, M.; Stenmark, H. ESCRTs in membrane sealing. Biochem. Soc. Trans. 2018, 46, 773–778. [Google Scholar] [CrossRef] [Green Version]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klumperman, J.; Raposo, G. The complex ultrastructure of the endolysosomal system. Cold Spring Harbor Perspect. Biol. 2014, 6, a016857. [Google Scholar] [CrossRef] [Green Version]
- Anand, S.; Samuel, M.; Kumar, S.; Mathivanan, S. Ticket to a bubble ride: Cargo sorting into exosomes and extracellular vesicles. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2019, 1867, 140203. [Google Scholar] [CrossRef]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-K.; Kang, B.; Kim, O.Y.; Choi, D.-S.; Lee, J.; Kim, S.R.; Go, G.; Yoon, Y.J.; Kim, J.H.; Jang, S.C.; et al. EVpedia: An integrated database of high-throughput data for systemic analyses of extracellular vesicles. J. Extracell. Vesicles 2013, 2, 20384. [Google Scholar] [CrossRef]
- Hemler, M.E. Tetraspanin Proteins Mediate Cellular Penetration, Invasion, and Fusion Events and Define a Novel Type of Membrane Microdomain. Annu. Rev. Cell Dev. Biol. 2003, 19, 397–422. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.; Morohashi, Y.; Yoshimura, S.-i.; Manrique-Hoyos, N.; Jung, S.; Lauterbach, M.A.; Bakhti, M.; Grønborg, M.; Möbius, W.; Rhee, J.; et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A–C. J. Cell Biol. 2010, 189, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [Green Version]
- Fan, Q.; Yang, L.; Zhang, X.; Peng, X.; Wei, S.; Su, D.; Zhai, Z.; Hua, X.; Li, H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018, 414, 107–115. [Google Scholar] [CrossRef]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.T.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New technologies for analysis of extracellular vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Gandham, S.; Su, X.; Wood, J.; Nocera, A.L.; Alli, S.C.; Milane, L.; Zimmerman, A.; Amiji, M.; Ivanov, A.R. Technologies and standardization in research on extracellular vesicles. Trends Biotechnol. 2020. [Google Scholar] [CrossRef]
- Hartjes, T.A.; Mytnyk, S.; Jenster, G.W.; van Steijn, V.; van Royen, M.E. Extracellular vesicle quantification and characterization: Common methods and emerging approaches. Bioengineering 2019, 6, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cizmar, P.; Yuana, Y. Detection and characterization of extracellular vesicles by transmission and cryo-transmission electron microscopy. In Extracellular Vesicles; Springer: Berlin/Heidelberg, Germany, 2017; pp. 221–232. [Google Scholar]
- Rikkert, L.G.; Nieuwland, R.; Terstappen, L.; Coumans, F. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracell. Vesicles 2019, 8, 1555419. [Google Scholar] [CrossRef] [PubMed]
- Milne, J.L.; Borgnia, M.J.; Bartesaghi, A.; Tran, E.E.; Earl, L.A.; Schauder, D.M.; Lengyel, J.; Pierson, J.; Patwardhan, A.; Subramaniam, S. Cryo-electron microscopy–a primer for the non-microscopist. FEBS J. 2013, 280, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Koifman, N.a.; Biran, I.; Aharon, A.; Brenner, B.; Talmon, Y. A direct-imaging cryo-EM study of shedding extracellular vesicles from leukemic monocytes. J. Struct. Biol. 2017, 198, 177–185. [Google Scholar] [CrossRef]
- Yuana, Y.; Koning, R.I.; Kuil, M.E.; Rensen, P.C.; Koster, A.J.; Bertina, R.M.; Osanto, S. Cryo-electron microscopy of extracellular vesicles in fresh plasma. J. Extracell. Vesicles 2013, 2, 21494. [Google Scholar] [CrossRef]
- Yuana, Y.; Oosterkamp, T.H.; Bahatyrova, S.; Ashcroft, B.; Garcia Rodriguez, P.; Bertina, R.M.; Osanto, S. Atomic force microscopy: A novel approach to the detection of nanosized blood microparticles. J. Thromb. Haemost. 2010, 8, 315–323. [Google Scholar] [CrossRef]
- Sharma, S.; Rasool, H.I.; Palanisamy, V.; Mathisen, C.; Schmidt, M.; Wong, D.T.; Gimzewski, J.K. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 2010, 4, 1921–1926. [Google Scholar] [CrossRef] [Green Version]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.L.; Dobson, P.J. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 780–788. [Google Scholar] [CrossRef] [Green Version]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2, 19671. [Google Scholar] [CrossRef]
- Gercel-Taylor, C.; Atay, S.; Tullis, R.H.; Kesimer, M.; Taylor, D.D. Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients. Anal. Biochem. 2012, 428, 44–53. [Google Scholar] [CrossRef]
- Bachurski, D.; Schuldner, M.; Nguyen, P.-H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M. Extracellular vesicle measurements with nanoparticle tracking analysis–An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 2019, 8, 1596016. [Google Scholar] [CrossRef] [PubMed]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef]
- Lawrie, A.; Albanyan, A.; Cardigan, R.; Mackie, I.; Harrison, P. Microparticle sizing by dynamic light scattering in fresh-frozen plasma. Vox Sang. 2009, 96, 206–212. [Google Scholar] [CrossRef]
- Xu, Y.; Nakane, N.; Maurer-Spurej, E. Novel test for microparticles in platelet-rich plasma and platelet concentrates using dynamic light scattering. Transfusion 2011, 51, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.; Broekman, M.L.; de Vrij, J. Tunable resistive pulse sensing for the characterization of extracellular vesicles. In Exosomes and Microvesicles; Springer: Berlin/Heidelberg, Germany, 2017; pp. 21–33. [Google Scholar]
- Vogel, R.; Coumans, F.A.; Maltesen, R.G.; Böing, A.N.; Bonnington, K.E.; Broekman, M.L.; Broom, M.F.; Buzás, E.I.; Christiansen, G.; Hajji, N. A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing. J. Extracell. Vesicles 2016, 5, 31242. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.; Pal, A.K.; Jambhrunkar, S.; Patel, P.; Thakur, S.S.; Reátegui, E.; Parekh, H.S.; Saá, P.; Stassinopoulos, A.; Broom, M.F. High-resolution single particle zeta potential characterisation of biological nanoparticles using tunable resistive pulse sensing. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coumans, F.A.; van der Pol, E.; Böing, A.N.; Hajji, N.; Sturk, G.; van Leeuwen, T.G.; Nieuwland, R. Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J. Extracell. Vesicles 2014, 3, 25922. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.A.; Gool, E.L.; Nieuwland, R. Bulk immunoassays for analysis of extracellular vesicles. Platelets 2017, 28, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Kowal, E.J.; Ter-Ovanesyan, D.; Regev, A.; Church, G.M. Extracellular vesicle isolation and analysis by western blotting. In Extracellular Vesicles; Springer: Berlin/Heidelberg, Germany, 2017; pp. 143–152. [Google Scholar]
- Logozzi, M.; Di Raimo, R.; Mizzoni, D.; Fais, S. Immunocapture-based ELISA to characterize and quantify exosomes in both cell culture supernatants and body fluids. Methods Enzymol. 2020. [Google Scholar] [CrossRef]
- Serrano-Pertierra, E.; Oliveira-Rodríguez, M.; Matos, M.; Gutiérrez, G.; Moyano, A.; Salvador, M.; Rivas, M.; Blanco-López, M.C. Extracellular Vesicles: Current Analytical Techniques for Detection and Quantification. Biomolecules 2020, 10, 824. [Google Scholar] [CrossRef]
- Ueda, K.; Ishikawa, N.; Tatsuguchi, A.; Saichi, N.; Fujii, R.; Nakagawa, H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014, 4, 6232. [Google Scholar] [CrossRef] [PubMed]
- Schageman, J.; Zeringer, E.; Li, M.; Barta, T.; Lea, K.; Gu, J.; Magdaleno, S.; Setterquist, R.; Vlassov, A.V. The complete exosome workflow solution: From isolation to characterization of RNA cargo. BioMed Res. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Yip, K.W.; Spence, T.; Liu, F.-F. MicroRNAs in extracellular vesicles: Potential cancer biomarkers. J. Hum. Genet. 2017, 62, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poncelet, P.; Robert, S.; Bailly, N.; Garnache-Ottou, F.; Bouriche, T.; Devalet, B.; Segatchian, J.H.; Saas, P.; Mullier, F. Tips and tricks for flow cytometry-based analysis and counting of microparticles. Transfus. Apher. Sci. 2015, 53, 110–126. [Google Scholar] [CrossRef] [PubMed]
- Orozco, A.F.; Lewis, D.E. Flow cytometric analysis of circulating microparticles in plasma. Cytom. Part A 2010, 77, 502–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pospichalova, V.; Svoboda, J.; Dave, Z.; Kotrbova, A.; Kaiser, K.; Klemova, D.; Ilkovics, L.; Hampl, A.; Crha, I.; Jandakova, E. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [Google Scholar] [CrossRef] [PubMed]
- Kormelink, T.G.; Arkesteijn, G.J.; Nauwelaers, F.A.; van den Engh, G.; Nolte-’t Hoen, E.N.; Wauben, M.H. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry. Cytom. Part A 2016, 89, 135–147. [Google Scholar] [CrossRef]
- Stoner, S.A.; Duggan, E.; Condello, D.; Guerrero, A.; Turk, J.R.; Narayanan, P.K.; Nolan, J.P. High sensitivity flow cytometry of membrane vesicles. Cytom. Part A 2016, 89, 196–206. [Google Scholar] [CrossRef] [Green Version]
- Van der Vlist, E.; Aalberts, M.; Mertens, H.; Bosch, B.; Bartelink, W.; Mastrobattista, E.; van Gaal, E.; Stoorvogel, W.; Arkesteijn, G.; Wauben, M. Quantitative and qualitative flow cytometric analysis of nanosized cell-derived membrane vesicles. Nanomed. Nanotechnol. Biol. Med. 2011, 8, 712–720. [Google Scholar]
- Choi, D.; Montermini, L.; Jeong, H.; Sharma, S.; Meehan, B.; Rak, J. Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by nano-flow cytometry. ACS Nano 2019, 13, 10499–10511. [Google Scholar] [CrossRef] [PubMed]
- Ferri, C.P.; Prince, M.; Brayne, C.; Brodaty, H.; Fratiglioni, L.; Ganguli, M.; Hall, K.; Hasegawa, K.; Hendrie, H.; Huang, Y.; et al. Global prevalence of dementia: A Delphi consensus study. Lancet 2005, 366, 2112–2117. [Google Scholar] [CrossRef]
- Elahi, F.M.; Miller, B.L. A clinicopathological approach to the diagnosis of dementia. Nat. Rev. Neurol. 2017, 13, 457–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morley, J.E.; Morris, J.C.; Berg-Weger, M.; Borson, S.; Carpenter, B.D.; Del Campo, N.; Dubois, B.; Fargo, K.; Fitten, L.J.; Flaherty, J.H.; et al. Brain health: The importance of recognizing cognitive impairment: An IAGG consensus conference. J. Am. Med. Dir. Assoc. 2015, 16, 731–739. [Google Scholar] [CrossRef] [Green Version]
- van der Flier, W.M.; Scheltens, P. Epidemiology and risk factors of dementia. J. Neurol. Neurosurg. Psychiatry 2005, 76 (Suppl. 5), v2–v7. [Google Scholar] [CrossRef] [Green Version]
- Perl, D.P. Neuropathology of Alzheimer’s disease. Mt. Sinai J. Med. 2010, 77, 32–42. [Google Scholar] [CrossRef]
- Kumar, S.; Rezaei-Ghaleh, N.; Terwel, D.; Thal, D.R.; Richard, M.; Hoch, M.; Mc Donald, J.M.; Wüllner, U.; Glebov, K.; Heneka, M.T. Extracellular phosphorylation of the amyloid β-peptide promotes formation of toxic aggregates during the pathogenesis of Alzheimer’s disease. EMBO J. 2011, 30, 2255–2265. [Google Scholar] [CrossRef] [Green Version]
- Goate, A.; Chartier-Harlin, M.-C.; Mullan, M.; Brown, J.; Crawford, F.; Fidani, L.; Giuffra, L.; Haynes, A.; Irving, N.; James, L. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 1991, 349, 704–706. [Google Scholar] [CrossRef]
- Kang, J.; Lemaire, H.; Unterbeck, A.; Salbaum, J.; Masters, C.; Grzeschik, K.; Multihaup, G.; Beyreuther, K.; Muller-Hill, B. Amyloid production secretase. Nature 1987, 325, 733–736. [Google Scholar] [CrossRef]
- Weidemann, A.; König, G.; Bunke, D.; Fischer, P.; Salbaum, J.; Masters, C.L.; Beyreuther, K. Cell 1989, 57, 115–126. [CrossRef]
- Sisodia, S.; Koo, E.; Beyreuther, K.; Unterbeck, A.; Price, D. Evidence that beta-amyloid protein in Alzheimer’s disease is not derived by normal processing. Science 1990, 248, 492–495. [Google Scholar] [CrossRef] [PubMed]
- Esch, F.S.; Keim, P.S.; Beattie, E.C.; Blacher, R.W.; Culwell, A.R.; Oltersdorf, T.; McClure, D.; Ward, P.J. Cleavage of amyloid beta peptide during constitutive processing of its precursor. Science 1990, 248, 1122–1124. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B. Proteases and proteolysis in Alzheimer disease: A multifactorial view on the disease process. Physiol. Rev. 2010, 90, 465–494. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.-C.; Heath, S.; Even, G.; Campion, D.; Sleegers, K.; Hiltunen, M.; Combarros, O.; Zelenika, D.; Bullido, M.J.; Tavernier, B. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009, 41, 1094–1099. [Google Scholar] [CrossRef]
- Chartier-Hariln, M.-C.; Parfitt, M.; Legrain, S.; Pérez-Tur, J.; Brousseau, T.; Evans, A.; Berr, C.; Vldal, O.; Roques, P.; Gourlet, V. Apolipoprotein E, ɛ4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: Analysis of the 19q13. 2 chromosomal region. Hum. Mol. Genet. 1994, 3, 569–574. [Google Scholar] [CrossRef]
- Houlden, H.; Crook, R.; Backhovens, H.; Prihar, G.; Baker, M.; Hutton, M.; Rossor, M.; Martin, J.J.; Van Broeckhoven, C.; Hardy, J. ApoE genotype is a risk factor in nonpresenilin early-onset alzheimer’s disease families. Am. J. Med. Genet. 1998, 81, 117–121. [Google Scholar] [CrossRef]
- Rebeck, G.W.; Reiter, J.S.; Strickland, D.K.; Hyman, B.T. Apolipoprotein E in sporadic Alzheimer’s disease: Allelic variation and receptor interactions. Neuron 1993, 11, 575–580. [Google Scholar] [CrossRef]
- Shi, J.; Han, P.; Kuniyoshi, S.M. Cognitive impairment in neurological diseases: Lessons from apolipoprotein E. J. Alzheimer’s Dis. 2014, 38, 1–9. [Google Scholar] [CrossRef]
- Liu, C.-C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [Green Version]
- Jarrett, J.T.; Berger, E.P.; Lansbury, P.T., Jr. The carboxy terminus of the. beta. amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry 1993, 32, 4693–4697. [Google Scholar] [CrossRef]
- McLean, C.A.; Cherny, R.A.; Fraser, F.W.; Fuller, S.J.; Smith, M.J.; Vbeyreuther, K.; Bush, A.I.; Masters, C.L. Soluble pool of Aβ amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann. Neurol. 1999, 46, 860–866. [Google Scholar] [CrossRef]
- Mc Donald, J.M.; Savva, G.M.; Brayne, C.; Welzel, A.T.; Forster, G.; Shankar, G.M.; Selkoe, D.J.; Ince, P.G.; Walsh, D.M. The presence of sodium dodecyl sulphate-stable Aβ dimers is strongly associated with Alzheimer-type dementia. Brain 2010, 133, 1328–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samanta, S.; Rajasingh, S.; Drosos, N.; Zhou, Z.; Dawn, B.; Rajasingh, J. Exosomes: New molecular targets of diseases. Acta Pharmacol. Sin. 2018, 39, 501–513. [Google Scholar] [CrossRef] [PubMed]
- D’anca, M.; Fenoglio, C.; Serpente, M.; Arosio, B.; Cesari, M.; Scarpini, E.A.; Galimberti, D. Exosome determinants of physiological aging and age related neurodegenerative diseases. Front. Aging Neurosci. 2019, 11, 232. [Google Scholar] [CrossRef] [Green Version]
- Raber, J.; Wong, D.; Buttini, M.; Orth, M.; Bellosta, S.; Pitas, R.E.; Mahley, R.W.; Mucke, L. Isoform-specific effects of human apolipoprotein E on brain function revealed in ApoE knockout mice: Increased susceptibility of females. Proc. Natl. Acad. Sci. USA 1998, 95, 10914–10919. [Google Scholar] [CrossRef] [Green Version]
- Holtzman, D.M.; Bales, K.R.; Tenkova, T.; Fagan, A.M.; Parsadanian, M.; Sartorius, L.J.; Mackey, B.; Olney, J.; McKeel, D.; Wozniak, D. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2000, 97, 2892–2897. [Google Scholar] [CrossRef] [Green Version]
- Caselli, R.J.; Dueck, A.C.; Osborne, D.; Sabbagh, M.N.; Connor, D.J.; Ahern, G.L.; Baxter, L.C.; Rapcsak, S.Z.; Shi, J.; Woodruff, B.K. Longitudinal modeling of age-related memory decline and the APOE ε4 effect. N. Engl. J. Med. 2009, 361, 255–263. [Google Scholar] [CrossRef] [Green Version]
- Selwood, S.P.; Parvathy, S.; Cordell, B.; Ryan, H.S.; Oshidari, F.; Vincent, V.; Yesavage, J.; Lazzeroni, L.C.; Murphy, G.M., Jr. Gene expression profile of the PDAPP mouse model for Alzheimer’s disease with and without Apolipoprotein, E. Neurobiol. Aging 2009, 30, 574–590. [Google Scholar] [CrossRef]
- Trommer, B.L.; Shah, C.; Yun, S.H.; Gamkrelidze, G.; Pasternak, E.S.; Ye, G.L.; Sotak, M.; Sullivan, P.M.; Pasternak, J.F.; LaDu, M.J. ApoE isoform affects LTP in human targeted replacement mice. Neuroreport 2004, 15, 2655–2658. [Google Scholar] [CrossRef]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease β-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [Green Version]
- Sharples, R.A.; Vella, L.J.; Nisbet, R.M.; Naylor, R.; Perez, K.; Barnham, K.J.; Masters, C.L.; Hill, A.F. Inhibition of γ-secretase causes increased secretion of amyloid precursor protein C-terminal fragments in association with exosomes. FASEB J. 2008, 22, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.; Duran-Aniotz, C.; Castilla, J.; Estrada, L.; Soto, C. De novo induction of amyloid-β deposition in vivo. Mol. Psychiatry 2012, 17, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.; Benussi, L.; Furlan, R.; Ghidoni, R.; Verderio, C. Extracellular vesicles in Alzheimer’s disease: Friends or foes? Focus on aβ-vesicle interaction. Int. J. Mol. Sci. 2015, 16, 4800–4813. [Google Scholar] [CrossRef] [PubMed]
- Ghate, P.S.; Sidhar, H.; Carlson, G.A.; Giri, R.K. Development of a novel cellular model of Alzheimer’s disease utilizing neurosphere cultures derived from B6C3-Tg(APPswe, PSEN1dE9)85Dbo/J embryonic mouse brain. SpringerPlus 2014, 3, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, E.H.; Squazzo, S.L. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J. Biol. Chem. 1994, 269, 17386–17389. [Google Scholar]
- Shankar, G.M.; Leissring, M.A.; Adame, A.; Sun, X.; Spooner, E.; Masliah, E.; Selkoe, D.J.; Lemere, C.A.; Walsh, D.M. Biochemical and immunohistochemical analysis of an Alzheimer’s disease mouse model reveals the presence of multiple cerebral Aβ assembly forms throughout life. Neurobiol. Dis. 2009, 36, 293–302. [Google Scholar] [CrossRef] [Green Version]
- Lesné, S.; Koh, M.T.; Kotilinek, L.; Kayed, R.; Glabe, C.G.; Yang, A.; Gallagher, M.; Ashe, K.H. A specific amyloid-β protein assembly in the brain impairs memory. Nature 2006, 440, 352–357. [Google Scholar] [CrossRef]
- Simonsen, A.H.; Herukka, S.-K.; Andreasen, N.; Baldeiras, I.; Bjerke, M.; Blennow, K.; Engelborghs, S.; Frisoni, G.B.; Gabryelewicz, T.; Galluzzi, S. Recommendations for CSF AD biomarkers in the diagnostic evaluation of dementia. Alzheimer’s Dement. 2017, 13, 274–284. [Google Scholar] [CrossRef] [Green Version]
- Hornung, S.; Dutta, S.; Bitan, G. CNS-Derived Blood Exosomes as a Promising Source of Biomarkers: Opportunities and Challenges. Front. Mol. Neurosci. 2020, 13, 38. [Google Scholar] [CrossRef] [Green Version]
- Goetzl, E.J.; Mustapic, M.; Kapogiannis, D.; Eitan, E.; Lobach, I.V.; Goetzl, L.; Schwartz, J.B.; Miller, B.L. Cargo proteins of plasma astrocyte-derived exosomes in Alzheimer’s disease. Faseb J. 2016, 30, 3853–3859. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Sui, Y.-T.; Peskind, E.R.; Li, G.; Hwang, H.; Devic, I.; Ginghina, C.; Edgar, J.S.; Pan, C.; Goodlett, D.R.; et al. Salivary Tau Species are Potential Biomarkers of Alzheimer’s Disease. J. Alzheimer’s Dis. 2011, 27, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Pareja, F.; del Ser, T.; Valentí, M.; de la Fuente, M.; Bartolome, F.; Carro, E. Salivary lactoferrin as biomarker for Alzheimer’s disease: Brain-immunity interactions. Alzheimer’s Dement. 2020, 16, 1196–1204. [Google Scholar] [CrossRef]
- Ahmadi-Motamayel, F.; Goodarzi, M.T.; Tarazi, S.; Vahabian, M. Evaluation of salivary acetylcholinesterase and pseudocholinesterase in patients with Alzheimer’s disease: A case–control study. Spec. Care Dent. 2019, 39, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Lebouvier, T.; Chaumette, T.; Paillusson, S.; Duyckaerts, C.; Bruley des Varannes, S.; Neunlist, M.; Derkinderen, P. The second brain and Parkinson’s disease. Eur. J. Neurosci. 2009, 30, 735–741. [Google Scholar] [CrossRef]
- Poewe, W.; Seppi, K.; Tanner, C.; Halliday, G.; Brundin, P.; Volkmann, J.; Schrag, A.; Lang, A. Parkinson disease. Nat. Rev. Dis. Primers 2017, 3, 17013. [Google Scholar] [CrossRef] [PubMed]
- Twelves, D.; Perkins, K.S.; Counsell, C. Systematic review of incidence studies of Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2003, 18, 19–31. [Google Scholar] [CrossRef]
- Pringsheim, T.; Jette, N.; Frolkis, A.; Steeves, T.D. The prevalence of Parkinson’s disease: A systematic review and meta-analysis. Mov. Disord. 2014, 29, 1583–1590. [Google Scholar] [CrossRef] [PubMed]
- Baldereschi, M.; Di Carlo, A.; Rocca, W.; Vanni, P.; Maggi, S.; Perissinotto, E.; Grigoletto, F.; Amaducci, L.; Inzitari, D. Parkinson’s disease and parkinsonism in a longitudinal study: Two-fold higher incidence in men. Neurology 2000, 55, 1358–1363. [Google Scholar] [CrossRef]
- Kusumi, M.; Nakashima, K.; Harada, H.; Nakayama, H.; Takahashi, K. Epidemiology of Parkinson’s disease in Yonago City, Japan: Comparison with a study carried out 12 years ago. Neuroepidemiology 1996, 15, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Cerri, S.; Mus, L.; Blandini, F. Parkinson’s Disease in Women and Men: What’s the Difference? J. Parkinson’s Dis. 2019, 9, 501–515. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Gao, J.; Wang, J.; Xie, A. Prion-like mechanisms in Parkinson’s disease. Front. Neurosci. 2019, 13, 552. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, J.M.; Lees, A.J. Ageing and Parkinson’s disease: Substantia nigra regional selectivity. Brain 1991, 114, 2283–2301. [Google Scholar] [CrossRef] [PubMed]
- Damier, P.; Hirsch, E.; Agid, Y.; Graybiel, A. The substantia nigra of the human brain: II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain 1999, 122, 1437–1448. [Google Scholar] [CrossRef]
- Dickson, D.W. Parkinson’s disease and parkinsonism: Neuropathology. Cold Spring Harbor Perspect. Med. 2012, 2, a009258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irizarry, M.C.; Growdon, W.; Gomez-Isla, T.; Newell, K.; George, J.M.; Clayton, D.F.; Hyman, B.T. Nigral and cortical Lewy bodies and dystrophic nigral neurites in Parkinson’s disease and cortical Lewy body disease contain α-synuclein immunoreactivity. J. Neuropathol. Exp. Neurol. 1998, 57, 334–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vekrellis, K.; Xilouri, M.; Emmanouilidou, E.; Rideout, H.J.; Stefanis, L. Pathological roles of α-synuclein in neurological disorders. Lancet Neurol. 2011, 10, 1015–1025. [Google Scholar] [CrossRef]
- Burré, J. The synaptic function of α-synuclein. J. Parkinson’s Dis. 2015, 5, 699–713. [Google Scholar]
- Alvarez-Erviti, L.; Rodriguez-Oroz, M.C.; Cooper, J.M.; Caballero, C.; Ferrer, I.; Obeso, J.A.; Schapira, A.H. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch. Neurol. 2010, 67, 1464–1472. [Google Scholar] [CrossRef] [Green Version]
- Halliday, G.M.; McCann, H. The progression of pathology in Parkinson’s disease. Ann. N. Y. Acad. Sci. 2010, 1184, 188–195. [Google Scholar] [CrossRef]
- Tolosa, E.; Wenning, G.; Poewe, W. The diagnosis of Parkinson’s disease. Lancet Neurol. 2006, 5, 75–86. [Google Scholar] [CrossRef]
- Chaudhuri, K.R.; Schapira, A.H. Non-motor symptoms of Parkinson’s disease: Dopaminergic pathophysiology and treatment. Lancet Neurol. 2009, 8, 464–474. [Google Scholar] [CrossRef]
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 2015, 30, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Burke, R.E.; Dauer, W.T.; Vonsattel, J.P.G. A critical evaluation of the Braak staging scheme for Parkinson’s disease. Ann. Neurol. Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 2008, 64, 485–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stayte, S.; Vissel, B. Advances in non-dopaminergic treatments for Parkinson’s disease. Front. Neurosci. 2014, 8, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bronstein, J.M.; Tagliati, M.; Alterman, R.L.; Lozano, A.M.; Volkmann, J.; Stefani, A.; Horak, F.B.; Okun, M.S.; Foote, K.D.; Krack, P. Deep brain stimulation for Parkinson disease: An expert consensus and review of key issues. Arch. Neurol. 2011, 68, 165. [Google Scholar] [CrossRef]
- Stern, M.B.; Follett, K.A.; Weaver, F.M. Randomized trial of deep brain stimulation for Parkinson disease: Thirty-six-month outcomes; turning tables: Should GPi become the preferred DBS target for Parkinson disease? Author response. Neurology 2013, 80, 225. [Google Scholar]
- Deuschl, G.; Agid, Y. Subthalamic neurostimulation for Parkinson’s disease with early fluctuations: Balancing the risks and benefits. Lancet Neurol. 2013, 12, 1025–1034. [Google Scholar] [CrossRef]
- Bengoa-Vergniory, N.; Roberts, R.F.; Wade-Martins, R.; Alegre-Abarrategui, J. Alpha-synuclein oligomers: A new hope. Acta Neuropathol. 2017, 134, 819–838. [Google Scholar] [CrossRef] [Green Version]
- Luth, E.S.; Stavrovskaya, I.G.; Bartels, T.; Kristal, B.S.; Selkoe, D.J. Soluble, prefibrillar α-synuclein oligomers promote complex I-dependent, Ca2+-induced mitochondrial dysfunction. J. Biol. Chem. 2014, 289, 21490–21507. [Google Scholar] [CrossRef] [Green Version]
- Lindström, V.; Gustafsson, G.; Sanders, L.H.; Howlett, E.H.; Sigvardson, J.; Kasrayan, A.; Ingelsson, M.; Bergström, J.; Erlandsson, A. Extensive uptake of α-synuclein oligomers in astrocytes results in sustained intracellular deposits and mitochondrial damage. Mol. Cell. Neurosci. 2017, 82, 143–156. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xilouri, M.; Brekk, O.R.; Stefanis, L. Alpha-synuclein and protein degradation systems: A reciprocal relationship. Mol. Neurobiol. 2013, 47, 537–551. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, G.A.; Silva, J.L. Alpha-synuclein stepwise aggregation reveals features of an early onset mutation in Parkinson’s disease. Commun. Biol. 2019, 2, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brundin, P.; Li, J.-Y.; Holton, J.L.; Lindvall, O.; Revesz, T. Research in motion: The enigma of Parkinson’s disease pathology spread. Nat. Rev. Neurosci. 2008, 9, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Sun, T.; An, J.; Wen, L.; Liu, F.; Bu, Z.; Cui, Y.; Feng, J. Potential Roles of Exosomes in Parkinson’s Disease: From Pathogenesis, Diagnosis, and Treatment to Prognosis. Front. Cell Dev. Biol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Paisan-Ruiz, C.; Lewis, P.A.; Singleton, A.B. LRRK2: Cause, risk, and mechanism. J. Parkinson’s Dis. 2013, 3, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Ben Gedalya, T.; Loeb, V.; Israeli, E.; Altschuler, Y.; Selkoe, D.J.; Sharon, R. α-Synuclein and Polyunsaturated Fatty Acids Promote Clathrin-Mediated Endocytosis and Synaptic Vesicle Recycling. Traffic 2009, 10, 218–234. [Google Scholar] [CrossRef] [Green Version]
- Cabin, D.E.; Shimazu, K.; Murphy, D.; Cole, N.B.; Gottschalk, W.; McIlwain, K.L.; Orrison, B.; Chen, A.; Ellis, C.E.; Paylor, R. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking α-synuclein. J. Neurosci. 2002, 22, 8797–8807. [Google Scholar] [CrossRef] [Green Version]
- Emmanouilidou, E.; Melachroinou, K.; Roumeliotis, T.; Garbis, S.D.; Ntzouni, M.; Margaritis, L.H.; Stefanis, L.; Vekrellis, K. Cell-produced α-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J. Neurosci. 2010, 30, 6838–6851. [Google Scholar] [CrossRef] [Green Version]
- Grey, M.; Dunning, C.J.; Gaspar, R.; Grey, C.; Brundin, P.; Sparr, E.; Linse, S. Acceleration of α-synuclein aggregation by exosomes. J. Biol. Chem. 2015, 290, 2969–2982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.; Lang, H.; Geng, N.; Wang, J.; Li, N.; Wang, X. Exosomes of BV-2 cells induced by alpha-synuclein: Important mediator of neurodegeneration in PD. Neurosci. Lett. 2013, 548, 190–195. [Google Scholar] [CrossRef]
- Hall, S.; Surova, Y.; Öhrfelt, A.; Zetterberg, H.; Lindqvist, D.; Hansson, O. CSF biomarkers and clinical progression of Parkinson disease. Neurology 2015, 84, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.-W.; Yang, S.-Y.; Yang, C.-C.; Chang, C.-W.; Wu, Y.-R. Plasma and Serum Alpha-Synuclein as a Biomarker of Diagnosis in Patients with Parkinson’s Disease. Front. Neurol. 2020, 10, 1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohmichi, T.; Mitsuhashi, M.; Tatebe, H.; Kasai, T.; El-Agnaf, O.M.A.; Tokuda, T. Quantification of brain-derived extracellular vesicles in plasma as a biomarker to diagnose Parkinson’s and related diseases. Parkinsonism Relat. Disord. 2019, 61, 82–87. [Google Scholar] [CrossRef]
- Kitamura, Y.; Kojima, M.; Kurosawa, T.; Sasaki, R.; Ichihara, S.; Hiraku, Y.; Tomimoto, H.; Murata, M.; Oikawa, S. Proteomic profiling of exosomal proteins for blood-based biomarkers in Parkinson’s Disease. Neuroscience 2018, 392, 121–128. [Google Scholar] [CrossRef]
- Cao, Z.; Wu, Y.; Liu, G.; Jiang, Y.; Wang, X.; Wang, Z.; Feng, T. α-Synuclein in salivary extracellular vesicles as a potential biomarker of Parkinson’s disease. Neurosci. Lett. 2019, 696, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Masters, J.M.; Noyce, A.J.; Warner, T.T.; Giovannoni, G.; Proctor, G.B. Elevated salivary protein in Parkinson’s disease and salivary DJ-1 as a potential marker of disease severity. Parkinsonism Relat. Disord. 2015, 21, 1251–1255. [Google Scholar] [CrossRef] [Green Version]
- Rani, K.; Rastogi, S.; Vishwakarma, P.; Bharti, P.S.; Sharma, V.; Renu, K.; Modi, G.P.; Vishnu, V.Y.; Chatterjee, P.; Dey, A.B. A novel approach to correlate the salivary exosomes and their protein cargo in the progression of cognitive impairment into Alzheimer’s disease. J. Neurosci. Methods 2020. [Google Scholar] [CrossRef]
- McKeever, P.M.; Schneider, R.; Taghdiri, F.; Weichert, A.; Multani, N.; Brown, R.A.; Boxer, A.L.; Karydas, A.; Miller, B.; Robertson, J. MicroRNA expression levels are altered in the cerebrospinal fluid of patients with young-onset Alzheimer’s disease. Mol. Neurobiol. 2018, 55, 8826–8841. [Google Scholar] [CrossRef] [Green Version]
- Jain, G.; Stuendl, A.; Rao, P.; Berulava, T.; Centeno, T.P.; Kaurani, L.; Burkhardt, S.; Delalle, I.; Kornhuber, J.; Hüll, M. A combined miRNA–piRNA signature to detect Alzheimer’s disease. Transl. Psychiatry 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, L.; Qiu, Q.; Zhang, H.; Chu, L.; Du, Y.; Zhang, J.; Zhou, C.; Liang, F.; Shi, S.; Wang, S. Concordance between the assessment of Aβ42, T-tau, and P-T181-tau in peripheral blood neuronal-derived exosomes and cerebrospinal fluid. Alzheimer’s Dement. 2019, 15, 1071–1080. [Google Scholar] [CrossRef]
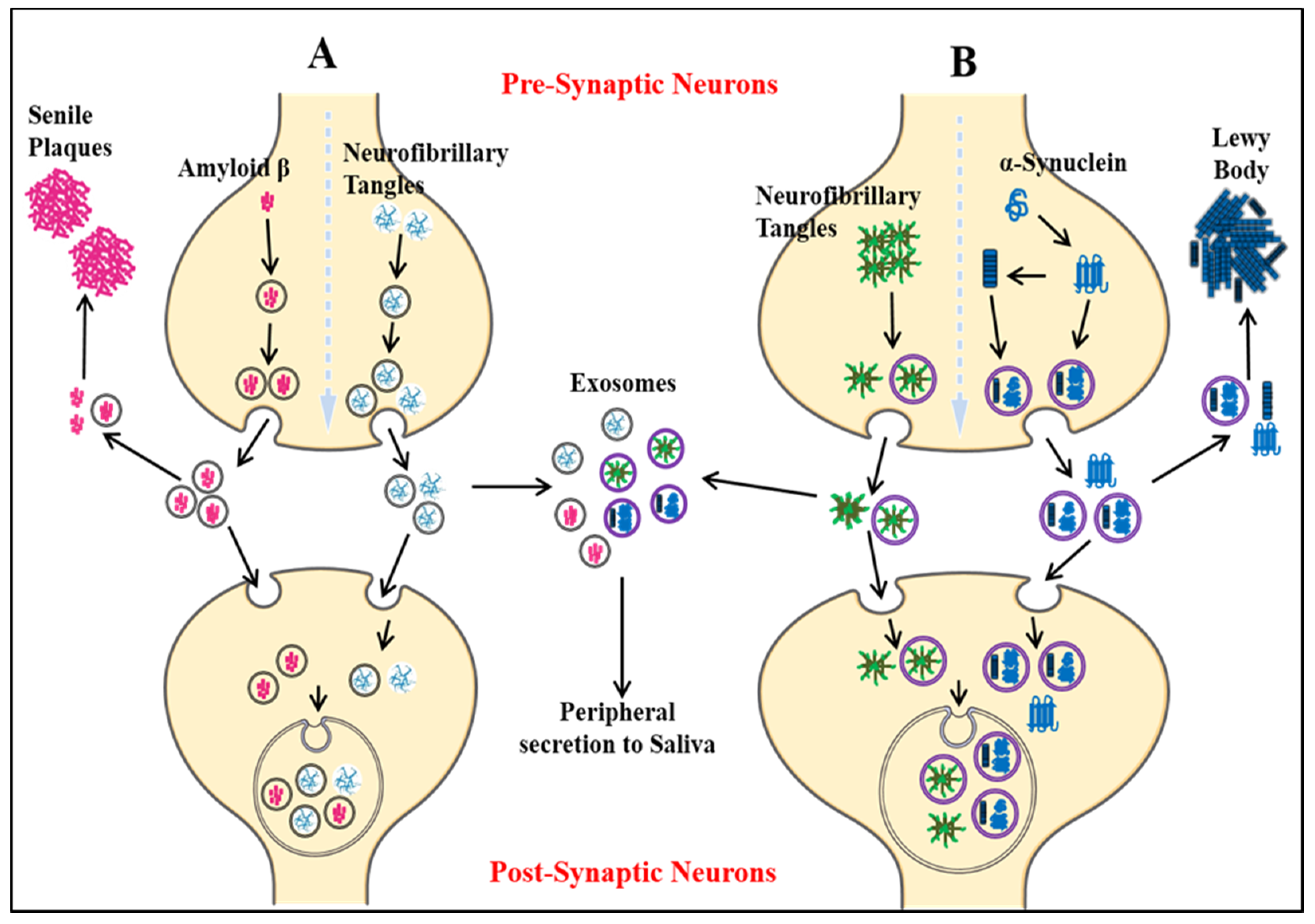
- Xiao, T.; Zhang, W.; Jiao, B.; Pan, C.-Z.; Liu, X.; Shen, L. The role of exosomes in the pathogenesis of Alzheimer’disease. Transl. Neurodegener. 2017, 6, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.K.; Langfelder, P.; Horvath, S.; Palazzolo, M.J. Exosomes and homeostatic synaptic plasticity are linked to each other and to Huntington’s, Parkinson’s, and other neurodegenerative diseases by database-enabled analyses of comprehensively curated datasets. Front. Neurosci. 2017, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, S.; Jedrychowski, M.P.; Yanamandra, K.; Ikezu, S.; Gygi, S.P.; Ikezu, T. Proteomic Profiling of Extracellular Vesicles Derived from Cerebrospinal Fluid of Alzheimer’s Disease Patients: A Pilot Study. Cells 2020, 9, 1959. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-G.; Song, J.; Zhang, Y.-Q.; Wang, P.-C. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol. Med. Rep. 2014, 10, 2395–2400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043. [Google Scholar] [CrossRef] [Green Version]
- Goetzl, E.J.; Kapogiannis, D.; Schwartz, J.B.; Lobach, I.V.; Goetzl, L.; Abner, E.L.; Jicha, G.A.; Karydas, A.M.; Boxer, A.; Miller, B.L. Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer’s disease. FASEB J. 2016, 30, 4141–4148. [Google Scholar] [CrossRef] [Green Version]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Kapogiannis, D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 2015, 85, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Winston, C.N.; Goetzl, E.J.; Schwartz, J.B.; Elahi, F.M.; Rissman, R.A. Complement protein levels in plasma astrocyte-derived exosomes are abnormal in conversion from mild cognitive impairment to Alzheimer’s disease dementia. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2019, 11, 61–66. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Gu, D.; Meng, M.; Gordon, M.L. TDP-43 Is Elevated in Plasma Neuronal-Derived Exosomes of Patients With Alzheimer’s Disease. Front. Aging Neurosci. 2020, 12, 166. [Google Scholar] [CrossRef] [PubMed]
- Winston, C.N.; Goetzl, E.J.; Akers, J.C.; Carter, B.S.; Rockenstein, E.M.; Galasko, D.; Masliah, E.; Rissman, R.A. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2016, 3, 63–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Sun, L.; Cao, W.; Yin, C.; Sun, W.; Liu, P.; Tan, L.; Xu, Z.; Zhao, W. Association between plasma exosome neurogranin and brain structure in patients with Alzheimer’s disease: A protocol study. BMJ Open 2020, 10, e036990. [Google Scholar] [CrossRef] [PubMed]
- Guix, F.X.; Corbett, G.T.; Cha, D.J.; Mustapic, M.; Liu, W.; Mengel, D.; Chen, Z.; Aikawa, E.; Young-Pearse, T.; Kapogiannis, D. Detection of aggregation-competent tau in neuron-derived extracellular vesicles. Int. J. Mol. Sci. 2018, 19, 663. [Google Scholar] [CrossRef] [Green Version]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Carlson, O.D.; Mustapic, M.; Kapogiannis, D. Low neural exosomal levels of cellular survival factors in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 769–773. [Google Scholar] [CrossRef]
- Sun, R.; Wang, H.; Shi, Y.; Gao, D.; Sun, Z.; Chen, Z.; Jiang, H.; Zhang, J. A Pilot Study of Urinary Exosomes in Alzheimer’s Disease. Neurodegener. Dis. 2019, 19, 184–191. [Google Scholar] [CrossRef]
- Song, Z.; Xu, Y.; Zhang, L.; Zhou, L.; Zhang, Y.; Han, Y.; Li, X.; Yu, P.; Qu, Y.; Zhao, W. Comprehensive Proteomic Profiling of Urinary Exosomes and Identification of Potential Non-invasive Early Biomarkers of Alzheimer’s Disease in 5XFAD Mouse Model. Front. Genet. 2020. [Google Scholar] [CrossRef]
- Rani, K.; Mukherjee, R.; Singh, E.; Kumar, S.; Sharma, V.; Vishwakarma, P.; Bharti, P.S.; Nikolajeff, F.; Dinda, A.K.; Goyal, V.; et al. Neuronal exosomes in saliva of Parkinson’s disease patients: A pilot study. Parkinsonism Relat. Disord. 2019, 67, 21–23. [Google Scholar] [CrossRef]
- Guo, M.; Wang, J.; Zhao, Y.; Feng, Y.; Han, S.; Dong, Q.; Cui, M.; Tieu, K. Microglial exosomes facilitate α-synuclein transmission in Parkinson’s disease. Brain 2020, 143, 1476–1497. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, X.; Chen, X.; Wang, L.; Yang, G. Exosome mediated delivery of miR-124 promotes neurogenesis after ischemia. Mol. Ther. Nucleic Acids 2017, 7, 278–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lööv, C.; Scherzer, C.R.; Hyman, B.T.; Breakefield, X.O.; Ingelsson, M. α-Synuclein in extracellular vesicles: Functional implications and diagnostic opportunities. Cell. Mol. Neurobiol. 2016, 36, 437–448. [Google Scholar] [CrossRef]
- Zhao, Z.-H.; Chen, Z.-T.; Zhou, R.-L.; Zhang, X.; Ye, Q.-Y.; Wang, Y.-Z. Increased DJ-1 and α-synuclein in plasma neural-derived exosomes as potential markers for Parkinson’s disease. Front. Aging Neurosci. 2019, 10, 438. [Google Scholar] [CrossRef]
- Shi, M.; Liu, C.; Cook, T.J.; Bullock, K.M.; Zhao, Y.; Ginghina, C.; Li, Y.; Aro, P.; Dator, R.; He, C. Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol. 2014, 128, 639–650. [Google Scholar] [CrossRef]
- Shi, M.; Kovac, A.; Korff, A.; Cook, T.J.; Ginghina, C.; Bullock, K.M.; Yang, L.; Stewart, T.; Zheng, D.; Aro, P. CNS tau efflux via exosomes is likely increased in Parkinson’s disease but not in Alzheimer’s disease. Alzheimer’s Dement. 2016, 12, 1125–1131. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Guo, Y.; Wei, L.; Yu, F.; Yu, B.; Xu, A. Long Noncoding RNA POU3F3 and α-Synuclein in Plasma L1CAM Exosomes Combined with β-Glucocerebrosidase Activity: Potential Predictors of Parkinson’s Disease. Neurother. J. Am. Soc. Exp. Neurother. 2020, 17, 1104–1119. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Z.; Ye, T.; Mabrouk, O.S.; Maltbie, T.; Aasly, J.; West, A.B. Elevated LRRK2 autophosphorylation in brain-derived and peripheral exosomes in LRRK2 mutation carriers. Acta Neuropathol. Commun. 2017, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Fraser, K.B.; Moehle, M.S.; Daher, J.P.; Webber, P.J.; Williams, J.Y.; Stewart, C.A.; Yacoubian, T.A.; Cowell, R.M.; Dokland, T.; Ye, T. LRRK2 secretion in exosomes is regulated by 14-3-3. Hum. Mol. Genet. 2013, 22, 4988–5000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, E.R.; Latourelle, J.C.; Bockholt, J.H.; Bregu, J.; Smock, J.; Paulsen, J.S.; Myers, R.H.; PREDICT-HD CSF Ancillary Study Investigators. MicroRNAs in CSF as prodromal biomarkers for Huntington disease in the PREDICT-HD study. Neurology 2018, 90, e264–e272. [Google Scholar] [CrossRef]
- Denis, H.L.; Lamontagne-Proulx, J.; St-Amour, I.; Mason, S.L.; Weiss, A.; Chouinard, S.; Barker, R.A.; Boilard, E.; Cicchetti, F. Platelet-derived extracellular vesicles in Huntington’s disease. J. Neurol. 2018, 265, 2704–2712. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Vijayan, M.; Bhatti, J.; Reddy, P.H. MicroRNAs as peripheral biomarkers in aging and age-related diseases. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2017; Volume 146, pp. 47–94. [Google Scholar]
- Gaughwin, P.M.; Ciesla, M.; Lahiri, N.; Tabrizi, S.J.; Brundin, P.; Björkqvist, M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington’s disease. Hum. Mol. Genet. 2011, 20, 2225–2237. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhao, Y.; Zhou, X.; Luan, J.; Cui, Y.; Han, J. Comparison of the extraction and determination of serum exosome and miRNA in serum and the detection of miR-27a-3p in serum exosome of ALS patients. Intractable Rare Dis. Res. 2018, 7, 13–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feneberg, E.; Steinacker, P.; Lehnert, S.; Schneider, A.; Walther, P.; Thal, D.R.; Linsenmeier, M.; Ludolph, A.C.; Otto, M. Limited role of free TDP-43 as a diagnostic tool in neurodegenerative diseases. Amyotroph. Lateral Scler. Front. Degener. 2014, 15, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Otake, K.; Kamiguchi, H.; Hirozane, Y. Identification of biomarkers for amyotrophic lateral sclerosis by comprehensive analysis of exosomal mRNAs in human cerebrospinal fluid. BMC Med. Genom. 2019, 12, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banack, S.A.; Dunlop, R.A.; Cox, P.A. An miRNA fingerprint using neural-enriched extracellular vesicles from blood plasma: Towards a biomarker for amyotrophic lateral sclerosis/motor neuron disease. Open Biol. 2020, 10, 200116. [Google Scholar] [CrossRef]
- Bates, G.P.; Dorsey, R.; Gusella, J.F.; Hayden, M.R.; Kay, C.; Leavitt, B.R.; Nance, M.; Ross, C.A.; Scahill, R.I.; Wetzel, R.; et al. Huntington disease. Nat. Rev. Dis. Primers 2015, 1, 15005. [Google Scholar] [CrossRef]
- Jeon, I.; Cicchetti, F.; Cisbani, G.; Lee, S.; Li, E.; Bae, J.; Lee, N.; Li, L.; Im, W.; Kim, M. Human-to-mouse prion-like propagation of mutant huntingtin protein. Acta Neuropathol. 2016, 132, 577–592. [Google Scholar] [CrossRef] [Green Version]
- Fisher, E.R.; Hayden, M.R. Multisource ascertainment of Huntington disease in Canada: Prevalence and population at risk. Mov. Disord. 2014, 29, 105–114. [Google Scholar] [CrossRef]
- Morrison, P.; Harding-Lester, S.; Bradley, A. Uptake of Huntington disease predictive testing in a complete population. Clin. Genet. 2011, 80, 281–286. [Google Scholar] [CrossRef]
- Zhang, X.; Abels, E.R.; Redzic, J.S.; Margulis, J.; Finkbeiner, S.; Breakefield, X.O. Potential transfer of polyglutamine and CAG-repeat RNA in extracellular vesicles in Huntington’s disease: Background and evaluation in cell culture. Cell. Mol. Neurobiol. 2016, 36, 459–470. [Google Scholar] [CrossRef]
- Lee, M.; Liu, T.; Im, W.; Kim, M. Exosomes from adipose-derived stem cells ameliorate phenotype of Huntington’s disease in vitro model. Eur. J. Neurosci. 2016, 44, 2114–2119. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Zhao, T.; Li, X.-J.; Li, S. Mutant huntingtin inhibits αB-crystallin expression and impairs exosome secretion from astrocytes. J. Neurosci. 2017, 37, 9550–9563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corey-Bloom, J.; Haque, A.S.; Park, S.; Nathan, A.S.; Baker, R.W.; Thomas, E.A. Salivary levels of total huntingtin are elevated in Huntington’s disease patients. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Coleman, B.M.; Hill, A.F. Extracellular vesicles–Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin. Cell Dev. Biol. 2015, 40, 89–96. [Google Scholar] [CrossRef]
- Hardiman, O.; Al-Chalabi, A.; Chio, A.; Corr, E.M.; Logroscino, G.; Robberecht, W.; Shaw, P.J.; Simmons, Z.; Van Den Berg, L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 2017, 3, 1–19. [Google Scholar] [CrossRef]
- Phukan, J.; Elamin, M.; Bede, P.; Jordan, N.; Gallagher, L.; Byrne, S.; Lynch, C.; Pender, N.; Hardiman, O. The syndrome of cognitive impairment in amyotrophic lateral sclerosis: A population-based study. J. Neurol. Neurosurg. Psychiatry 2012, 83, 102–108. [Google Scholar] [CrossRef]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.-X. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef]
- Maruyama, H.; Morino, H.; Ito, H.; Izumi, Y.; Kato, H.; Watanabe, Y.; Kinoshita, Y.; Kamada, M.; Nodera, H.; Suzuki, H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010, 465, 223–226. [Google Scholar] [CrossRef]
- Deng, H.X.; Chen, W.; Hong, S.T.; Boycott, K.M.; Gorrie, G.H.; Siddique, N.; Yang, Y.; Fecto, F.; Shi, Y.; Zhai, H.; et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 2011, 477, 211–215. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.O.; Mandrioli, J.; Benatar, M.; Abramzon, Y.; Van Deerlin, V.M.; Trojanowski, J.Q.; Gibbs, J.R.; Brunetti, M.; Gronka, S.; Wuu, J. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010, 68, 857–864. [Google Scholar] [CrossRef] [Green Version]
- Renton, A.E.; Majounie, E.; Waite, A.; Simón-Sánchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; Van Swieten, J.C.; Myllykangas, L. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Kim, N.C.; Wang, Y.-D.; Scarborough, E.A.; Moore, J.; Diaz, Z.; MacLea, K.S.; Freibaum, B.; Li, S.; Molliex, A. Prion-like domain mutations in hnRNPs cause multisystem proteinopathy and ALS. Nature 2013, 495, 467. [Google Scholar] [CrossRef] [PubMed]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farg, M.A.; Sundaramoorthy, V.; Sultana, J.M.; Yang, S.; Atkinson, R.A.; Levina, V.; Halloran, M.A.; Gleeson, P.A.; Blair, I.P.; Soo, K.Y. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 2014, 23, 3579–3595. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Manzano, R.; Lee, Y.; Dafinca, R.; Aoki, M.; Douglas, A.G.L.; Varela, M.A.; Sathyaprakash, C.; Scaber, J.; Barbagallo, P.; et al. C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. Brain 2017, 140, 887–897. [Google Scholar] [CrossRef]
- Gomes, C.; Keller, S.; Altevogt, P.; Costa, J. Evidence for secretion of Cu, Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci. Lett. 2007, 428, 43–46. [Google Scholar] [CrossRef]
- Iguchi, Y.; Eid, L.; Parent, M.; Soucy, G.; Bareil, C.; Riku, Y.; Kawai, K.; Takagi, S.; Yoshida, M.; Katsuno, M.; et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 2016, 139, 3187–3201. [Google Scholar] [CrossRef] [Green Version]
- Maguire, G. Amyotrophic lateral sclerosis as a protein level, non-genomic disease: Therapy with S2RM exosome released molecules. World J. Stem Cells 2017, 9, 187. [Google Scholar] [CrossRef]
- Han, Y.; Jia, L.; Zheng, Y.; Li, W. Salivary exosomes: Emerging roles in systemic disease. Int. J. Biol. Sci. 2018, 14, 633. [Google Scholar] [CrossRef]
- Cheng, J.; Nonaka, T.; Wong, D.T. Salivary exosomes as nanocarriers for cancer biomarker delivery. Materials 2019, 12, 654. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, Y.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yanoshita, R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol. Pharm. Bull. 2008, 31, 1059–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, S.; Munro, C.; Pickler, R.; Grap, M.J.; Elswick, R. Comparison of biomarkers in blood and saliva in healthy adults. Nurs. Res. Pract. 2012. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, M.N.; Shi, J.; Lee, M.; Arnold, L.; Al-Hasan, Y.; Heim, J.; McGeer, P. Salivary beta amyloid protein levels are detectable and differentiate patients with Alzheimer’s disease dementia from normal controls: Preliminary findings. BMC Neurol. 2018, 18, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermejo-Pareja, F.; Antequera, D.; Vargas, T.; Molina, J.A.; Carro, E. Saliva levels of Abeta1-42 as potential biomarker of Alzheimer’s disease: A pilot study. BMC Neurol. 2010, 10, 108. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.; Guo, J.-P.; Kennedy, K.; McGeer, E.G.; McGeer, P.L. A method for diagnosing Alzheimer’s disease based on salivary amyloid-β protein 42 levels. J. Alzheimer’s Dis. 2017, 55, 1175–1182. [Google Scholar] [CrossRef]
- Kim, C.-B.; Song, K.-B. Antibody-Based Magnetic Nanoparticle Immunoassay for Quantification of Salivary Beta-Amyloid Peptides. Biophys. J. 2014, 106, 616a. [Google Scholar] [CrossRef] [Green Version]
- Chan, H.-N.; Xu, D.; Ho, S.-L.; Wong, M.S.; Li, H.-W. Ultra-sensitive detection of protein biomarkers for diagnosis of Alzheimer’s disease. Chem. Sci. 2017, 8, 4012–4018. [Google Scholar] [CrossRef] [Green Version]
- Santos, G.A.; Olave, E.; Pardi, P.C. Salivary Biomarkers in Alzheimer’s Disease. Int. J. Morphol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Tvarijonaviciute, A.; Zamora, C.; Ceron, J.J.; Bravo-Cantero, A.F.; Pardo-Marin, L.; Valverde, S.; Lopez-Jornet, P. Salivary biomarkers in Alzheimer’s disease. Clin. Oral Investig. 2020, 24, 3437–3444. [Google Scholar] [CrossRef]
- Pekeles, H.; Qureshi, H.Y.; Paudel, H.K.; Schipper, H.M.; Gornistky, M.; Chertkow, H. Development and validation of a salivary tau biomarker in Alzheimer’s disease. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2019, 11, 53–60. [Google Scholar] [CrossRef]
- Lau, H.-C.; Lee, I.-K.; Ko, P.-W.; Lee, H.-W.; Huh, J.-S.; Cho, W.-J.; Lim, J.-O. Non-invasive screening for Alzheimer’s disease by sensing salivary sugar using Drosophila cells expressing gustatory receptor (Gr5a) immobilized on an extended gate ion-sensitive field-effect transistor (EG-ISFET) biosensor. PLoS ONE 2015, 10, e0117810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashton, N.J.; Ide, M.; Schöll, M.; Blennow, K.; Lovestone, S.; Hye, A.; Zetterberg, H. No association of salivary total tau concentration with Alzheimer’s disease. Neurobiol. Aging 2018, 70, 125–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carro, E.; Bartolomé, F.; Bermejo-Pareja, F.; Villarejo-Galende, A.; Molina, J.A.; Ortiz, P.; Calero, M.; Rabano, A.; Cantero, J.L.; Orive, G. Early diagnosis of mild cognitive impairment and Alzheimer’s disease based on salivary lactoferrin. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2017, 8, 131–138. [Google Scholar] [CrossRef] [PubMed]
- González-Sánchez, M.; Bartolome, F.; Antequera, D.; Puertas-Martín, V.; González, P.; Gómez-Grande, A.; Llamas-Velasco, S.; Herrero-San Martín, A.; Pérez-Martínez, D.; Villarejo-Galende, A.; et al. Decreased salivary lactoferrin levels are specific to Alzheimer’s disease. EBioMedicine 2020, 57, 102834. [Google Scholar] [CrossRef] [PubMed]
- Sayer, R.; Law, E.; Connelly, P.J.; Breen, K.C. Association of a salivary acetylcholinesterase with Alzheimer’s disease and response to cholinesterase inhibitors. Clin. Biochem. 2004, 37, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiari, S.; Moghadam, N.B.; Ehsani, M.; Mortazavi, H.; Sabour, S.; Bakhshi, M. Can salivary acetylcholinesterase be a diagnostic biomarker for Alzheimer? J. Clin. Diagn. Res. 2017, 11, ZC58. [Google Scholar] [CrossRef]
- Boston, P.F.; Gopalkaje, K.; Manning, L.; Middleton, L.; Loxley, M. Developing a simple laboratory test for Alzheimer’s disease: Measuring acetylcholinesterase in saliva a pilot study. Int. J. Geriatr. Psychiatry 2008, 23, 439–440. [Google Scholar] [CrossRef]
- Liang, Q.; Liu, H.; Zhang, T.; Jiang, Y.; Xing, H.; Zhang, A.-H. Metabolomics-based screening of salivary biomarkers for early diagnosis of Alzheimer’s disease. RSC Adv. 2015, 5, 96074–96079. [Google Scholar] [CrossRef]
- Huan, T.; Tran, T.; Zheng, J.; Sapkota, S.; MacDonald, S.W.; Camicioli, R.; Dixon, R.A.; Li, L. Metabolomics analyses of saliva detect novel biomarkers of Alzheimer’s disease. J. Alzheimer’s Dis. 2018, 65, 1401–1416. [Google Scholar] [CrossRef]
- Yilmaz, A.; Geddes, T.; Han, B.; Bahado-Singh, R.O.; Wilson, G.D.; Imam, K.; Maddens, M.; Graham, S.F. Diagnostic biomarkers of Alzheimer’s disease as identified in saliva using 1H NMR-based metabolomics. J. Alzheimer’s Dis. 2017, 58, 355–359. [Google Scholar] [CrossRef]
- Vivacqua, G.; Latorre, A.; Suppa, A.; Nardi, M.; Pietracupa, S.; Mancinelli, R.; Fabbrini, G.; Colosimo, C.; Gaudio, E.; Berardelli, A. Abnormal salivary total and oligomeric alpha-synuclein in Parkinson’s disease. PLoS ONE 2016, 11, e0151156. [Google Scholar] [CrossRef] [Green Version]
- Vivacqua, G.; Suppa, A.; Mancinelli, R.; Belvisi, D.; Fabbrini, A.; Costanzo, M.; Formica, A.; Onori, P.; Fabbrini, G.; Berardelli, A. Salivary alpha-synuclein in the diagnosis of Parkinson’s disease and Progressive Supranuclear Palsy. Parkinsonism Relat. Disord. 2019, 63, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, H.; Sobhy, S.; El Mously, S.; Abuomira, M.; Mansour, M. Salivary alpha-synuclein (total and oligomeric form): Potential biomarkers in parkinson’s disease. Egypt. J. Neurol. Psychiatry Neurosurg. 2020, 56, 1–6. [Google Scholar] [CrossRef]
- Al-Nimer, M.S.; Mshatat, S.F.; Abdulla, H.I. Saliva α-synuclein and a high extinction coefficient protein: A novel approach in assessment biomarkers of Parkinson’s disease. N. Am. J. Med Sci. 2014, 6, 633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devic, I.; Hwang, H.; Edgar, J.S.; Izutsu, K.; Presland, R.; Pan, C.; Goodlett, D.R.; Wang, Y.; Armaly, J.; Tumas, V. Salivary α-synuclein and DJ-1: Potential biomarkers for Parkinson’s disease. Brain 2011, 134, e178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldman, J.G.; Andrews, H.; Amara, A.; Naito, A.; Alcalay, R.N.; Shaw, L.M.; Taylor, P.; Xie, T.; Tuite, P.; Henchcliffe, C. Cerebrospinal fluid, plasma, and saliva in the BioFIND study: Relationships among biomarkers and Parkinson’s disease features. Mov. Disord. 2018, 33, 282–288. [Google Scholar] [CrossRef] [Green Version]
- Kang, W.; Chen, W.; Yang, Q.; Zhang, L.; Zhang, L.; Wang, X.; Dong, F.; Zhao, Y.; Chen, S.; Quinn, T.J. Salivary total α-synuclein, oligomeric α-synuclein and SNCA variants in Parkinson’s disease patients. Sci. Rep. 2016, 6, 28143. [Google Scholar] [CrossRef] [Green Version]
- Kang, W.-Y.; Yang, Q.; Jiang, X.-F.; Chen, W.; Zhang, L.-Y.; Wang, X.-Y.; Zhang, L.-N.; Quinn, T.J.; Liu, J.; Chen, S.-D. Salivary DJ-1 could be an indicator of Parkinson’s disease progression. Front. Aging Neurosci. 2014, 6, 102. [Google Scholar] [CrossRef] [Green Version]
- Fedorova, T.; Knudsen, C.S.; Mouridsen, K.; Nexo, E.; Borghammer, P. Salivary acetylcholinesterase activity is increased in Parkinson’s disease: A potential marker of parasympathetic dysfunction. Parkinson’s Dis. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Kothari, V.; Velly, A.M.; Cressatti, M.; Liberman, A.; Gornitsky, M.; Schipper, H.M. Evaluation of salivary heme oxygenase-1 as a potential biomarker of early Parkinson’s disease. Mov. Disord. 2018, 33, 583–591. [Google Scholar] [CrossRef]
- Cressatti, M.; Juwara, L.; Galindez, J.M.; Velly, A.M.; Nkurunziza, E.S.; Marier, S.; Canie, O.; Gornistky, M.; Schipper, H.M. Salivary microR-153 and microR-223 Levels as Potential Diagnostic Biomarkers of Idiopathic Parkinson’s Disease. Mov. Disord. 2020, 35, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zheng, J.; Su, L.; Chen, F.; Zhu, R.; Chen, X.; Ye, Q. Increased Salivary microRNAs That Regulate DJ-1 Gene Expression as Potential Markers for Parkinson’s Disease. Front. Aging Neurosci. 2020, 12, 210. [Google Scholar] [CrossRef] [PubMed]
- Bahn, J.H.; Zhang, Q.; Li, F.; Chan, T.-M.; Lin, X.; Kim, Y.; Wong, D.T.; Xiao, X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015, 61, 221–230. [Google Scholar] [CrossRef] [Green Version]
- Corey-Bloom, J.; Fischer, R.S.; Kim, A.; Snell, C.; Parkin, G.M.; Granger, D.A.; Granger, S.W.; Thomas, E.A. Levels of Interleukin-6 in Saliva, but Not Plasma, Correlate with Clinical Metrics in Huntington’s Disease Patients and Healthy Control Subjects. Int. J. Mol. Sci. 2020, 21, 6363. [Google Scholar] [CrossRef] [PubMed]
- Obayashi, K.; Sato, K.; Shimazaki, R.; Ishikawa, T.; Goto, K.; Ueyama, H.; Mori, T.; Ando, Y.; Kumamoto, T. Salivary chromogranin A: Useful and quantitative biochemical marker of affective state in patients with amyotrophic lateral sclerosis. Intern. Med. 2008, 47, 1875–1879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlomagno, C.; Banfi, P.I.; Gualerzi, A.; Picciolini, S.; Volpato, E.; Meloni, M.; Lax, A.; Colombo, E.; Ticozzi, N.; Verde, F.; et al. Human salivary Raman fingerprint as biomarker for the diagnosis of Amyotrophic Lateral Sclerosis. Sci. Rep. 2020, 10, 10175. [Google Scholar] [CrossRef]

| Exosomes | Microvesicles | |
|---|---|---|
| Size | 30–150 nm | 50–1000 nm |
| Morphology | Cup-shaped | Heterogeneous |
| Density | 1.1–1.2 g/mL | 1.08–1.19 g/mL |
| Origin | Multivesicular Endosomes (MVEs) | Plasma Membrane |
| Contents | Protein, miRNA, mRNA | Protein, miRNA, mRNA |
| Protein markers | Alix, Tsg101, CD 81, CD 82, CD63, CD 37, CD9 | Selectins, Integrins, CD40 |
| Technique | Principle | Advantages | Disadvantages |
|---|---|---|---|
| Electron Microscopy (EM) | Electron radiation | Direct imaging of exosomes, higher resolution, ultrastructure, surface topography | Expensive, cumbersome processing and preparation, qualitative, low throughput |
| Atomic Force Microscopy (AFM) | Hooke’s law | Higher resolution, sample processing, surface topography, and substructure | Expensive, Low throughput, qualitative |
| Nanoparticle Tracking Analysis (NTA) | Brownian motion and Stokes–Einstein equation | Size and concentration simultaneously | Better for smaller particles |
| Dynamic Light Scattering (DLS) | Brownian motion | Simple and fast | Not for heterogeneous populations, lower resolution |
| Tunable Resistive Pulse Sensing (TRPS) | Coulter principle | Size, concentration, and zeta potential simultaneously | Better for larger particles, cannot differentiate between exosomes and other particles |
| Western Blotting and ELISA | Antigen-antibody interactions(immunoaffinity) | Detection of exosome-specific proteins, size, and abundance of proteins | Low specificity and quality, higher cost, cross-reactivity |
| qRT-PCR | Amplification using primers and PCR | Quantitative, low sample volume, higher resolution, high throughput | Limited to the analysis of known target RNA sequences |
| Flow Cytometry | Coulter principle, light scattering, fluorescence tags/antibodies | Sample processing, fast, specific, reproducible, quantitative, low sample volume | Size standards do not correlate correctly, limitation on detection of lower sized particles |
| Neurodegenerative Disease | Source of Exosome | Studied Exosome Cargo Content | References |
|---|---|---|---|
| Alzheimer’s disease | Saliva |
| [184] |
| CSF |
| [185,186,187,188,189,190,191,192] | |
| Plasma |
| [135,193,194,195,196,197,198,199,200] | |
| Serum |
| [201] | |
| Urine |
| [202,203] | |
| Parkinson’s disease | Saliva |
| [182,204] |
| CSF |
| [128,205,206] | |
| Plasma |
| [180,207,208,209,210,211] | |
| Urine |
| [212,213] | |
| Huntington Disease | Saliva | ___ | ___ |
| CSF |
| [214] | |
| Plasma |
| [189,215,216,217] | |
| Urine | ____ | ___ | |
| Amyotrophic Lateral Sclerosis | Saliva | ____ | ___ |
| CSF |
| [218,219,220] | |
| Plasma |
| [218,221] | |
| Urine | ____ | ____ |
| Disease | Biomarkers | Outcomes | Methods | References |
|---|---|---|---|---|
| Alzheimer’s disease | Aβ-42 | -Increased in saliva Aβ-42 in MCI, AD patients in comparison to HC -No differences in Aβ-42 in PD and healthy. | ELISA, Sandwich Immunoassay on Magnetic Nanoparticles ELISA kits, detection assay is based on the immunoreaction between the target proteins and their corresponding pair of antibodies followed by fluorescence labeling with a newly developed indolium-based turn-on fluorophore, namely SIM | [249,250,251,252,253,254] |
| Aβ-42 | -The association between saliva Aβ-42 levels and AD was independent - Decrease in Aβ42 | ELISA Magnetic Bead Panel—Multiplex Assay kits | [204,250,255] | |
| Aβ40 | -Aβ40 expression was unchanged within the entire studied sample | ELISA, | [250,253,255] | |
| Complement C4 | -Increase in complement C4 | Magnetic Bead Panel—Multiplex Assay kits, | [255] | |
| t-Tau protein, p-Tau/t-Tau (S396), p-Tau/t-Tau ratio, p-tau | -t-Tau expression in AD patients is significantly lower. -(S396) p-tau/t-tau ratio was significantly elevated in patients with CSF in AD -Higher abundance of p-tau in MCI in comparison to AD -Higher protein abundance in AD and MCI in comparison to healthy controls | Western blotting, Immunoprecipitation, Mass spectrometry, Luminex assays, | [136,256,257] | |
| total tau (t-tau) tau441, and p-tau181 | -No difference in salivary t-tau concentration found between AD and MCI or healthy elderly control -No association of salivary t-tau concentration with neurophysiological assessment or structural magnetic resonance imaging -No significant change | Human Total Tau assay Ultrasensitive single-particle molecule array technology ELISA kits, detection assay is based on the immunoreaction between the target proteins and their corresponding pair of antibodies followed by fluorescence labeling with a newly developed indolium-based turn-on fluorophore, namely SIM | [253,258] | |
| Lactoferrin | -Salivary lactoferrin shows a very high correlation with all MCI and AD patients | MALDI-TOF/TOF mass spectrometer | [259] | |
| Lactoferrin | -Reduced in patients suffering MCI and sporadic AD -Decreased levels in PD | Meta-analysis, ELISA, Amyloid-PET scan | [137,260] | |
| Acetylcholinesterase (AChE) | -The activity of the enzyme was significantly lower in people with AD. Significant age-related decrease in the enzyme | Ellman colorimetric method | [261,262] | |
| Acetylcholinesterase (AChE) | -No statistically significant -Activity of AChE and PChE significantly increased in the group with AD | Ellman colorimetric method | [138,263] | |
| Sphinganine-1-phosphate, ornithine, phenyllactic acid, alpha-amyloid protein | -Sphinganine-1-phosphate, ornithine, phenyllactic acid, inosine, 3-dehydrocarnitine, hypoxanthine in the saliva, of the AD subjects were significantly different from the control sphinganine-1-phosphate, which was upregulated in AD | Metabolomics, faster ultra-performance liquid chromatography (FUPLC) mass spectrometry (MS) | [264] | |
| Higher metabolites level may distinguish AD from CN and MCI with good diagnostic ability | -Increased—Methylguanosine, Histidinyl-Phenylalanine, Choline-cytidine, Glucosylgalactosyl Hydroxylysine, Glutamine-carnitines | Metabolomics, liquid chromatography, mass spectrometry | [265] | |
| Different markers | -Increased—Imidazole, Acetone, Creatine, 5-Aminopentanoate, Propionate, and Acetone -Decreased—Galactose. Altered metabolites may predict early AD and MCI | 1-H NMR | [266] | |
| Exosomal Aβ oligomer and p –tau | -Increased—Aβ oligomer abundance and phospho-tau in AD and CI | Fluoroscence-NTA, western blotting, TEM | [184] | |
| Parkinson’s disease | α-syn levels, a ratio of phospho α-syn/total α-syn, total α-syn, α-syn (Oligo and total) a-syn total/oligo α-syn and the extinction coefficient of the saliva protein α-syn (CSF, Plasma, Saliva) | -Increased α-synolig, α-synolig/α-syntotal in PD significantly higher -The α-synolig/α-syntotal ratio was also higher in patients Increase in α-synolig level and α-synolig/α-syn total ratio and a decrease in α-syntotal level among PD patients -α-syn level is significantly less in PD Total saliva protein and uncontaminated protein with nucleic acids are significantly higher in PD -In CSF, α-syn was lower in PD but Plasma and saliva α-syn did not differ between PD and controls | Unstimulated whole saliva, XYCQ EV enrichment Kit, Western blotting, Luminex multiplex assays ELISA, Western blotting, NTA, Sandwich ELISA BioFIND, ELISA | [182,204,210,267,268,269,270,271] |
| α-syn (CSF, Plasma, Saliva); total α-syn, oligo α-syn and α-syn SNP variants levels | -Plasma and saliva α-syn do not differ between PD and controls. Saliva α-syn neither correlate with CSF α-syn nor distinguish PD from controls | BioFIND, ELISA | [272] | |
| -No difference in salivary total α-syn levels was found between PD patients and HC, it decreased with age in PD patients and was closely associated with the genotypic distribution of rs11931074 and rs894278 in PD | Luminex assay, Gel filtration chromatography and Western blot; PCR and sequencing | [273] | ||
| DJ-1 | -DJ-1 levels tended to increase in Parkinson’s disease, DJ-1 levels correlated with disease severity | Unstimulated whole saliva, Western blotting, Luminex multiplex assays | [183,213,271] | |
| DJ-1 | -DJ-1 levels in the saliva were not changed significantly in PD patients -A significant difference in salivary DJ-1 levels in PD patients based on different disease stages and clinical subtypes -Salivary DJ-1 levels in the advanced stage of PD were significantly higher than those in the early stage of PD | Luminex assay | [274], | |
| Acetylcholinesterase (AChE) | -Increased salivary AChE activity -AChE activity/total protein ratio was significantly increased in PD patients | colorimetric method | [275] | |
| Heme oxygenase-1 | -Significantly higher mean salivary heme oxygenase-1 concentrations in patients with H and Y stage 1 PD (early) than control subjects and stage 2 and stage 3 PD patients | ELISA, Western Blotting | [276] | |
| miRNA-153, mi-RNA-223, miR-7a and miR-7b, mi-RNA 874, mi-RNA 145-3p | -mi-RNA levels decreased in PD patients but did not correlate to disease progression. miR-7a and miR-7b did not correlate with PD or patients. -Ratio of oligomeric α-syn/miR-153 was significantly increased among PD patients, ratio of oligomeric α-syn/miR-223 was not significantly different in the PD patients | RT-qPCR, ELISA | [277,278] | |
| Exosomal L1CAM and α-syn protein abundance | -Increased α-syn protein in PD patient | NTA, Western blotting | [204] | |
| α-synOlig and α-synOlig/α-synTotal | -α-synOlig and α-synOlig/α-synTotal significantly higher in PD patients than healthy controls | NTA, Western blotting | ||
| Healthy | MicroRNA, Piwi-Interacting RNA, and Circular RNA | -miR-223–3p, miR-148a-3p | Unstimulated whole saliva, Bioinformatics analysis | [279] |
| Huntington disease | Interleukin-6 | -The increased level of IL-6 in HD patients in comparison to healthy controls | ELISA | [280] |
| Amyotropic lateral Sclerosis | Chromogranin A | -Higher in terminal ALS patient in comparison to moderately suffering ALS patient | YK070 chromogranin A EIA kit | [281] |
| -Correlation of Raman data of ALS patient’s saliva revealed a direct | Raman Spectroscopy | [282] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rastogi, S.; Sharma, V.; Bharti, P.S.; Rani, K.; Modi, G.P.; Nikolajeff, F.; Kumar, S. The Evolving Landscape of Exosomes in Neurodegenerative Diseases: Exosomes Characteristics and a Promising Role in Early Diagnosis. Int. J. Mol. Sci. 2021, 22, 440. https://doi.org/10.3390/ijms22010440
Rastogi S, Sharma V, Bharti PS, Rani K, Modi GP, Nikolajeff F, Kumar S. The Evolving Landscape of Exosomes in Neurodegenerative Diseases: Exosomes Characteristics and a Promising Role in Early Diagnosis. International Journal of Molecular Sciences. 2021; 22(1):440. https://doi.org/10.3390/ijms22010440
Chicago/Turabian StyleRastogi, Simran, Vaibhav Sharma, Prahalad Singh Bharti, Komal Rani, Gyan P. Modi, Fredrik Nikolajeff, and Saroj Kumar. 2021. "The Evolving Landscape of Exosomes in Neurodegenerative Diseases: Exosomes Characteristics and a Promising Role in Early Diagnosis" International Journal of Molecular Sciences 22, no. 1: 440. https://doi.org/10.3390/ijms22010440
APA StyleRastogi, S., Sharma, V., Bharti, P. S., Rani, K., Modi, G. P., Nikolajeff, F., & Kumar, S. (2021). The Evolving Landscape of Exosomes in Neurodegenerative Diseases: Exosomes Characteristics and a Promising Role in Early Diagnosis. International Journal of Molecular Sciences, 22(1), 440. https://doi.org/10.3390/ijms22010440