Reduced Gonadotrophin Receptor Expression Is Associated with a More Aggressive Ovarian Cancer Phenotype
Abstract
:1. Introduction
2. Results
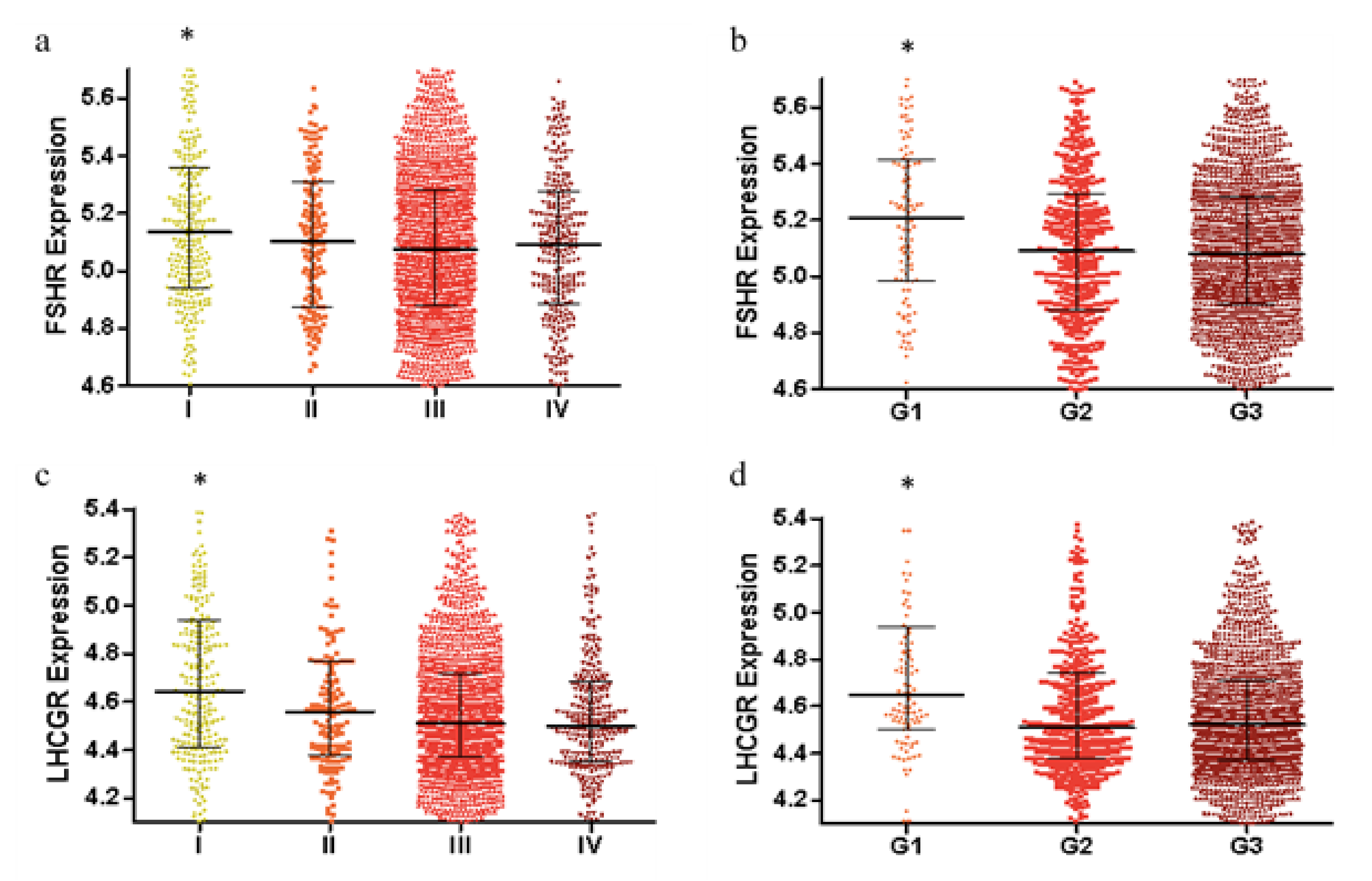
2.1. FSHR and LHCGR Expression Are Associated with Tumor Stage and Tumor Grade
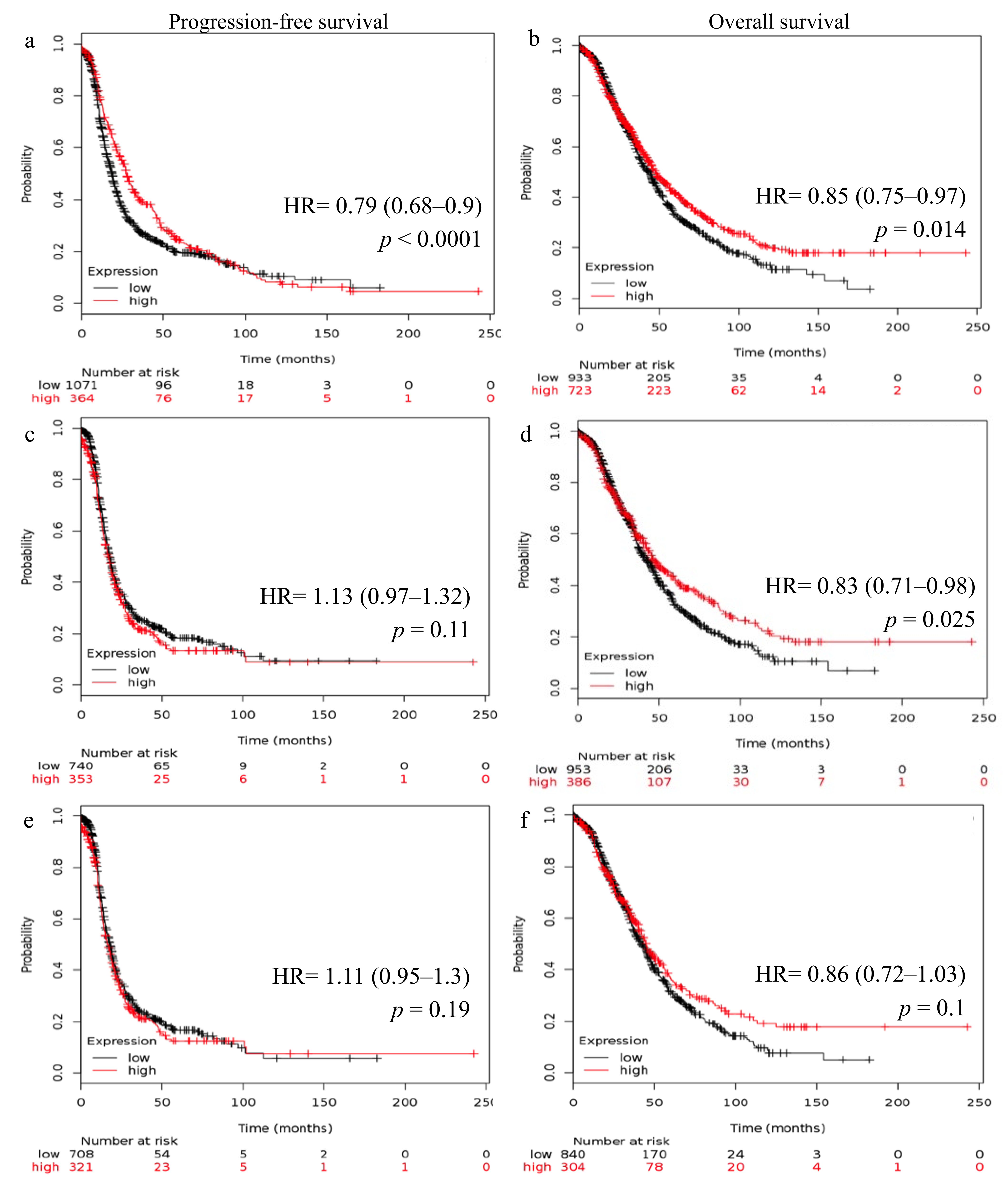
2.2. Reduced FSHR and LHCGR mRNA Expression Is Associated with Poor Patient Outcome
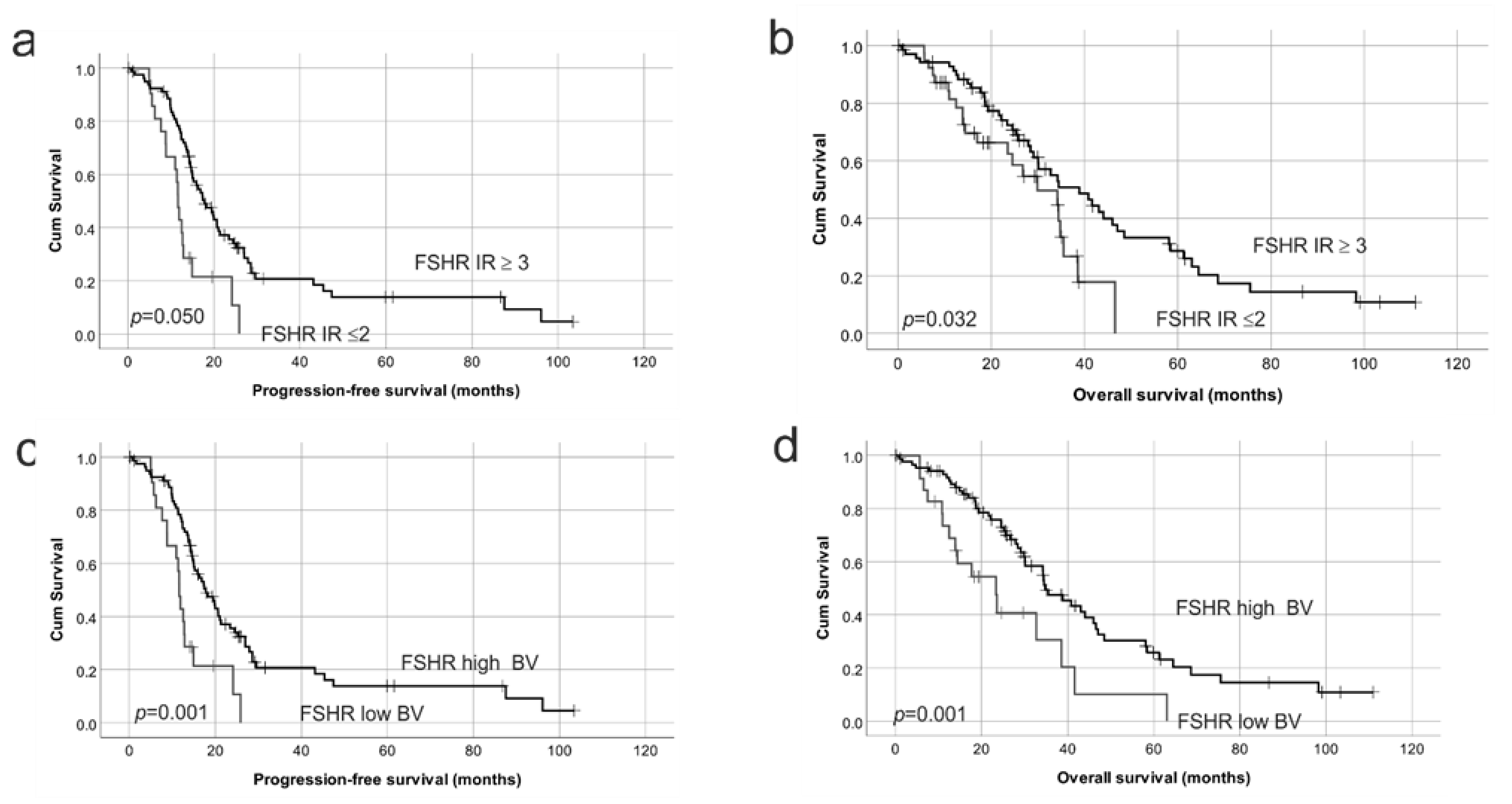
2.3. FSHR Protein Expression Is Associated with Patient Outcome
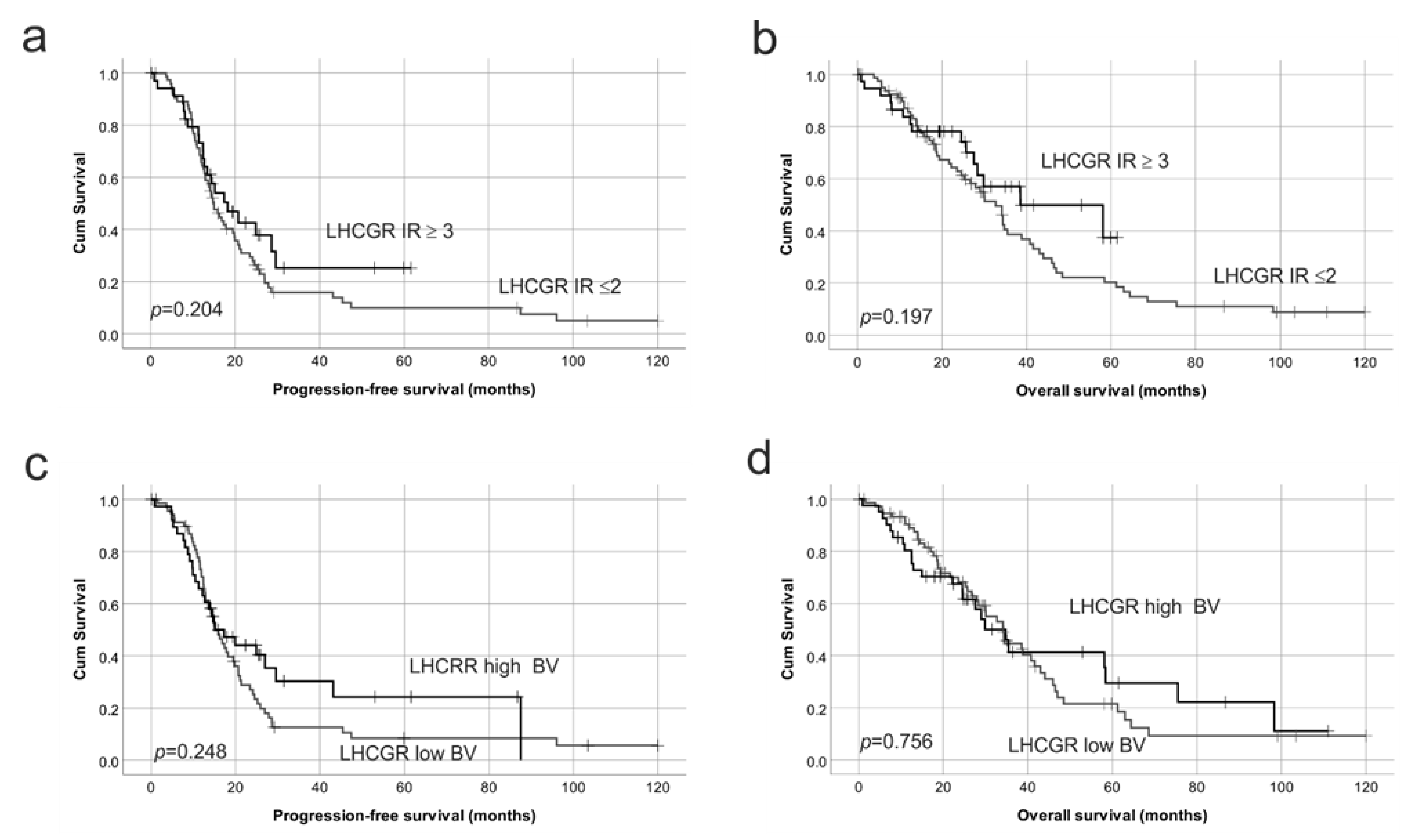
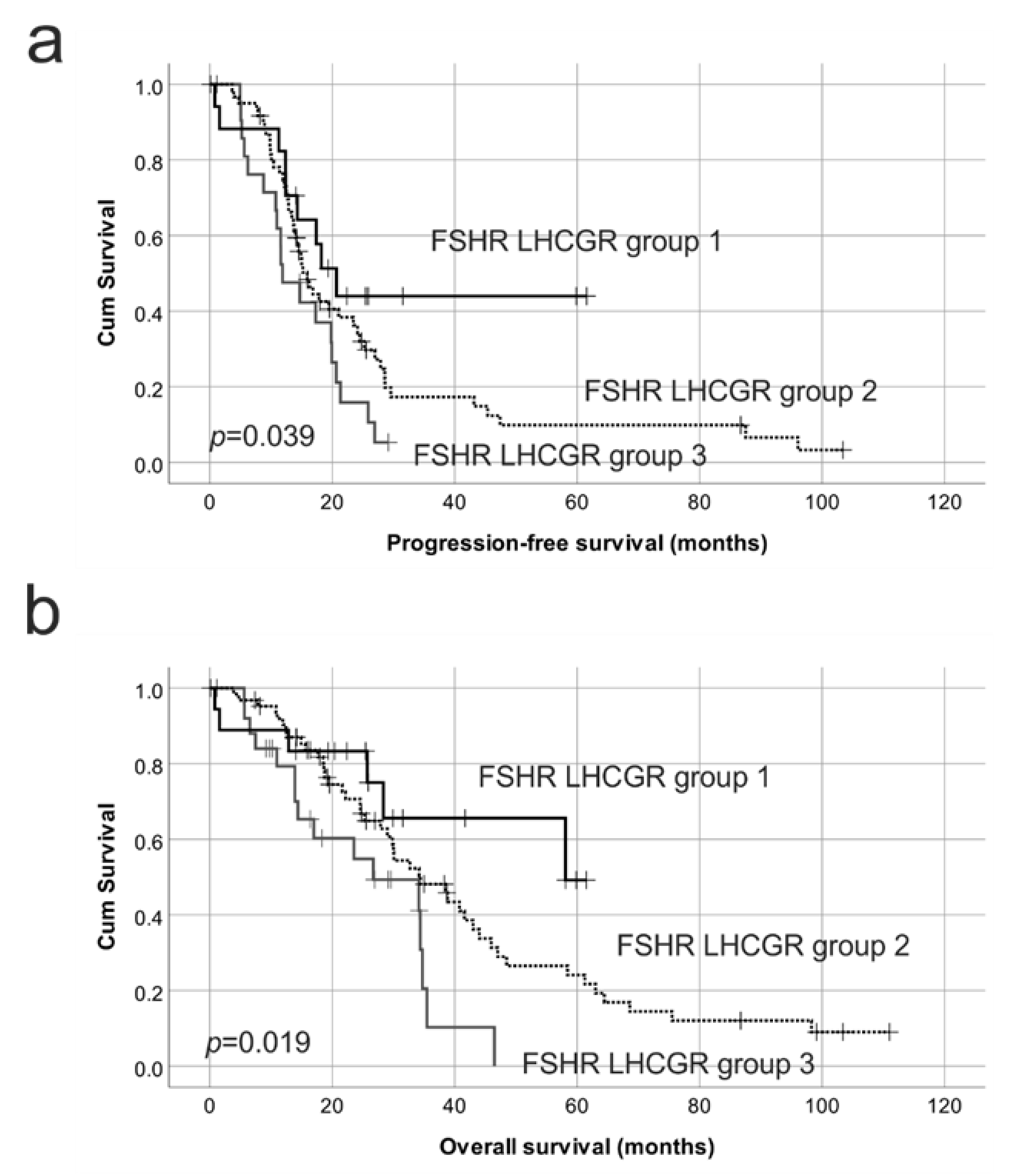
2.4. Reduced Protein Expression of Both FSHR and LHCGR Are Associated with Poor Patient Outcome
2.5. Characterization of Gonadotropin Receptor Expression in Serous Ovarian Cancer Cell Lines
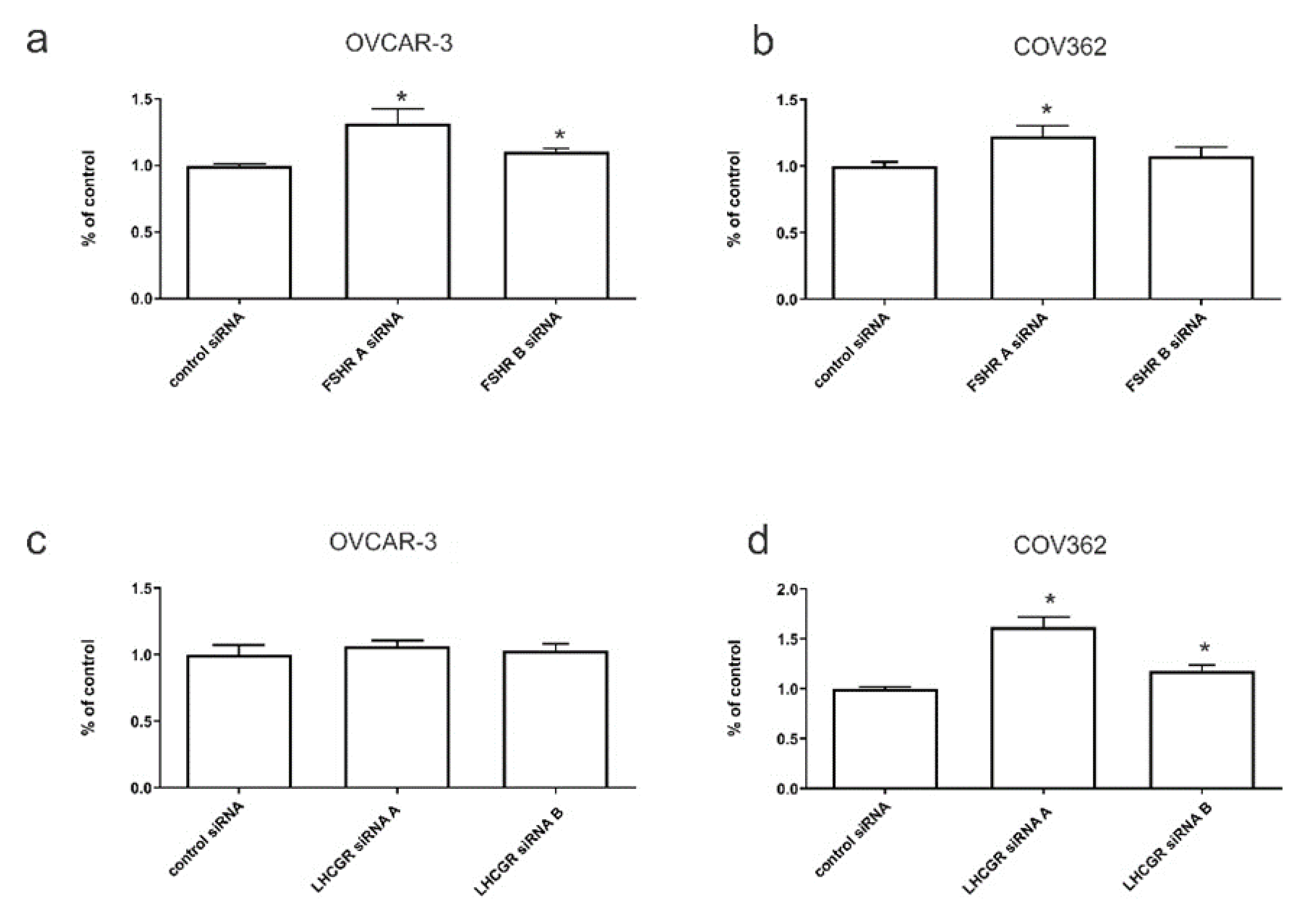
2.6. The Effects of FSHR and LHCGR Knockdown on Serous Ovarian Cancer Invasion In Vitro
3. Discussions
4. Materials and Methods
4.1. Public Ovarian Cancer Transcriptomic Microarray Databases
4.2. Patient Tissue Cohort
4.3. FSHR and LHCGR Immunohistochemistry
4.4. Immunohistochemistry Assessment
4.5. Cell Lines
4.6. Quantitative Real-Time PCR (qRT-PCR)
4.7. Western Blot
4.8. FSHR and LHCGR siRNA Treatment
4.9. Invasion Assays
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Ricciardelli, C.; Oehler, M.K. Diverse molecular pathways in ovarian cancer and their clinical significance. Maturitas 2009, 62, 270–275. [Google Scholar] [CrossRef]
- Colombo, P.E.; Fabbro, M.; Theillet, C.; Bibeau, F.; Rouanet, P.; Ray-Coquard, I. Sensitivity and resistance to treatment in the primary management of epithelial ovarian cancer. Crit. Rev. Oncol. Hematol. 2014, 89, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Pope, C.; McNeilly, J.R.; Coutts, S.; Millar, M.; Anderson, R.A.; McNeilly, A.S. Gonadotrope and thyrotrope development in the human and mouse anterior pituitary gland. Dev. Biol. 2006, 297, 172–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Magid, M.S.; Kramer, E.E.; Chen, Y.T. Follicle-stimulating hormone receptor is expressed in human ovarian surface epithelium and fallopian tube. Am. J. Pathol. 1996, 148, 47–53. [Google Scholar] [PubMed]
- Parrott, J.A.; Doraiswamy, V.; Kim, G.; Mosher, R.; Skinner, M.K. Expression and actions of both the follicle stimulating hormone receptor and the luteinizing hormone receptor in normal ovarian surface epithelium and ovarian cancer. Mol. Cell Endocrinol. 2001, 172, 213–222. [Google Scholar] [CrossRef]
- Choi, J.H.; Wong, A.S.; Huang, H.F.; Leung, P.C. Gonadotropins and ovarian cancer. Endocr. Rev. 2007, 28, 440–461. [Google Scholar] [CrossRef] [Green Version]
- Van Look, P.F.; Baird, D.T. Regulatory mechanisms during the menstrual cycle. Eur. J. Obs. Gynecol. Reprod. Biol. 1980, 11, 121–144. [Google Scholar] [CrossRef]
- Bose, C.K. Follicle stimulating hormone receptor in ovarian surface epithelium and epithelial ovarian cancer. Oncol. Res. 2008, 17, 231–238. [Google Scholar] [CrossRef]
- Chakravarti, S.; Collins, W.P.; Forecast, J.D.; Newton, J.R.; Oram, D.H.; Studd, J.W. Hormonal profiles after the menopause. Br. Med. J. 1976, 2, 784–787. [Google Scholar] [CrossRef] [Green Version]
- Mertens-Walker, I.; Baxter, R.C.; Marsh, D.J. Gonadotropin signalling in epithelial ovarian cancer. Cancer Lett. 2012, 324, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Puett, D.; Li, Y.; Angelova, K.; Demars, G.; Meehan, T.P.; Fanelli, F.; Narayan, P. Structure-function relationships of the luteinizing hormone receptor. Ann. N. Y. Acad. Sci. 2005, 1061, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.R.; Hendrickson, W.A. Structure of human follicle-stimulating hormone in complex with its receptor. Nature 2005, 433, 269–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perales-Puchalt, A.; Svoronos, N.; Rutkowski, M.R.; Allegrezza, M.J.; Tesone, A.J.; Payne, K.K.; Wickramasinghe, J.; Nguyen, J.M.; O’Brien, S.W.; Gumireddy, K.; et al. Follicle-stimulating hormone receptor is expressed by most ovarian cancer subtypes and is a safe and effective immunotherapeutic target. Clin. Cancer Res. 2017, 23, 441–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Lin, L.; Parkash, V.; Schwartz, P.E.; Lauchlan, S.C.; Zheng, W. Quantitative analysis of follicle-stimulating hormone receptor in ovarian epithelial tumors: A novel approach to explain the field effect of ovarian cancer development in secondary mullerian systems. Int. J. Cancer 2003, 103, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Wen, H.; Ju, X.; Bi, R.; Chen, X.; Yang, W.; Wu, X. Expression of hypothalamic-pituitary-gonadal axis-related hormone receptors in low-grade serous ovarian cancer (lgsc). J. Ovarian Res. 2017, 10, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenhard, M.; Lennerova, T.; Ditsch, N.; Kahlert, S.; Friese, K.; Mayr, D.; Jeschke, U. Opposed roles of follicle-stimulating hormone and luteinizing hormone receptors in ovarian cancer survival. Histopathology 2011, 58, 990–994. [Google Scholar] [CrossRef]
- Lu, J.J.; Zheng, Y.; Kang, X.; Yuan, J.M.; Lauchlan, S.C.; Pike, M.C.; Zheng, W. Decreased luteinizing hormone receptor mrna expression in human ovarian epithelial cancer. Gynecol. Oncol. 2000, 79, 158–168. [Google Scholar] [CrossRef]
- Mandai, M.; Konishi, I.; Kuroda, H.; Fukumoto, M.; Komatsu, T.; Yamamoto, S.; Nanbu, K.; Rao, C.V.; Mori, T. Messenger ribonucleic acid expression of lh/hcg receptor gene in human ovarian carcinomas. Eur. J. Cancer 1997, 33, 1501–1507. [Google Scholar] [CrossRef]
- Tan, T.Z.; Yang, H.; Ye, J.; Low, J.; Choolani, M.; Tan, D.S.; Thiery, J.P.; Huang, R.Y. Csiovdb: A microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget 2015, 6, 43843–43852. [Google Scholar] [CrossRef] [Green Version]
- Gyorffy, B.; Lanczky, A.; Szallasi, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Lu, J.J.; Luo, F.; Zheng, Y.; Feng, Y.; Felix, J.C.; Lauchlan, S.C.; Pike, M.C. Ovarian epithelial tumor growth promotion by follicle-stimulating hormone and inhibition of the effect by luteinizing hormone. Gynecol. Oncol. 2000, 76, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Wen, H.; Bi, R.; Ju, X.; Chen, X.; Yang, W.; Wu, X. A clinically applicable molecular classification for high-grade serous ovarian cancer based on hormone receptor expression. Sci. Rep. 2016, 6, 25408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moeker, N.; Peters, S.; Rauchenberger, R.; Ghinea, N.; Kunz, C. Antibody selection for cancer target validation of fsh-receptor in immunohistochemical settings. Antibodies 2017, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Chrusciel, M.; Ponikwicka-Tyszko, D.; Wolczynski, S.; Huhtaniemi, I.; Rahman, N.A. Extragonadal fshr expression and function-is it real? Front. Endocrinol. (Lausanne) 2019, 10, 32. [Google Scholar] [CrossRef] [Green Version]
- Radu, A.; Pichon, C.; Camparo, P.; Antoine, M.; Allory, Y.; Couvelard, A.; Fromont, G.; Hai, M.T.; Ghinea, N. Expression of follicle-stimulating hormone receptor in tumor blood vessels. N. Engl. J. Med. 2010, 363, 1621–1630. [Google Scholar] [CrossRef]
- Vannier, B.; Loosfelt, H.; Meduri, G.; Pichon, C.; Milgrom, E. Anti-human fsh receptor monoclonal antibodies: Immunochemical and immunocytochemical characterization of the receptor. Biochemistry 1996, 35, 1358–1366. [Google Scholar] [CrossRef]
- Ricciardelli, C.; Bianco-Miotto, T.; Jindal, S.; Butler, L.M.; Leung, S.; McNeil, C.M.; O’Toole, S.A.; Ebrahimie, E.; Millar, E.K.A.; Sakko, A.J.; et al. The magnitude of androgen receptor positivity in breast cancer is critical for reliable prediction of disease outcome. Clin. Cancer Res. 2018, 24, 2328–2341. [Google Scholar] [CrossRef] [Green Version]
- Tranchant, T.; Durand, G.; Gauthier, C.; Crepieux, P.; Ulloa-Aguirre, A.; Royere, D.; Reiter, E. Preferential beta-arrestin signalling at low receptor density revealed by functional characterization of the human fsh receptor a189 v mutation. Mol. Cell. Endocrinol. 2011, 331, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Casarini, L.; Reiter, E.; Simoni, M. Beta-arrestins regulate gonadotropin receptor-mediated cell proliferation and apoptosis by controlling different fshr or lhcgr intracellular signaling in the hgl5 cell line. Mol. Cell. Endocrinol. 2016, 437, 11–21. [Google Scholar] [CrossRef]
- Casarini, L.; Lazzaretti, C.; Paradiso, E.; Limoncella, S.; Riccetti, L.; Sperduti, S.; Melli, B.; Marcozzi, S.; Anzivino, C.; Sayers, N.S.; et al. Membrane estrogen receptor (gper) and follicle-stimulating hormone receptor (fshr) heteromeric complexes promote human ovarian follicle survival. iScience 2020, 23, 101812. [Google Scholar] [CrossRef] [PubMed]
- Stelmaszewska, J.; Chrusciel, M.; Doroszko, M.; Akerfelt, M.; Ponikwicka-Tyszko, D.; Nees, M.; Frentsch, M.; Li, X.; Kero, J.; Huhtaniemi, I.; et al. Revisiting the expression and function of follicle-stimulation hormone receptor in human umbilical vein endothelial cells. Sci Rep. 2016, 6, 37095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponikwicka-Tyszko, D.; Chrusciel, M.; Stelmaszewska, J.; Bernaczyk, P.; Sztachelska, M.; Sidorkiewicz, I.; Doroszko, M.; Tomaszewski, J.; Tapanainen, J.S.; Huhtaniemi, I.; et al. Functional expression of fsh receptor in endometriotic lesions. J. Clin. Endocrinol. Metab. 2016, 101, 2905–2914. [Google Scholar] [CrossRef] [PubMed]
- Nouaille, S.; Mondeil, S.; Finoux, A.L.; Moulis, C.; Girbal, L.; Cocaign-Bousquet, M. The stability of an mrna is influenced by its concentration: A potential physical mechanism to regulate gene expression. Nucleic Acids Res. 2017, 45, 11711–11724. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.; Sadygov, R.G. Predicting the protein half-life in tissue from its cellular properties. PLoS ONE 2017, 12, e0180428. [Google Scholar] [CrossRef] [Green Version]
- Sardella, C.; Russo, D.; Raggi, F.; Lombardi, M.; Urbani, C.; Brogioni, S.; Boggi, U.; Funel, N.; Chifenti, B.; Campani, D.; et al. Ectopic expression of fsh receptor isoforms in neoplastic but not in endothelial cells from pancreatic neuroendocrine tumors. J. Endocrinol. Investig. 2013, 36, 174–179. [Google Scholar]
- Tourkova, I.L.; Witt, M.R.; Li, L.; Larrouture, Q.; Liu, L.; Luo, J.; Robinson, L.J.; Blair, H.C. Follicle stimulating hormone receptor in mesenchymal stem cells integrates effects of glycoprotein reproductive hormones. Ann. N. Y. Acad. Sci. 2015, 1335, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.Y.; Chen, J.; Zheng, Y.F.; Gao, X.L.; Kang, Y.; Liu, J.C.; Cheng, M.J.; Sun, H.; Xu, C.J. Follicle-stimulating hormone peptide can facilitate paclitaxel nanoparticles to target ovarian carcinoma in vivo. Cancer Res. 2009, 69, 6506–6514. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Jia, L.; Feng, Y.; Zheng, W. Overexpression of follicle-stimulating hormone receptor facilitates the development of ovarian epithelial cancer. Cancer Lett. 2009, 278, 56–64. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, Y.; Li, L.; Zhu, L.; Zhang, D.; Zhang, S.; Chen, Y. Identification of two novel, alternatively spliced mrna transcripts of the human follicle-stimulating hormone receptor. Mol. Reprod. Dev. 2013, 80, 916–923. [Google Scholar] [CrossRef]
- Robinson, L.J.; Tourkova, I.; Wang, Y.; Sharrow, A.C.; Landau, M.S.; Yaroslavskiy, B.B.; Sun, L.; Zaidi, M.; Blair, H.C. Fsh-receptor isoforms and fsh-dependent gene transcription in human monocytes and osteoclasts. Biochem. Biophys. Res. Commun. 2010, 394, 12–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Giron, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, T.; Delgado, C.; Dufau, M.L. Phosphorylation and glycosylation of the luteinizing hormone receptor. Proc. Natl. Acad. Sci. USA 1989, 86, 1470–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricciardelli, C.; Lokman, N.A.; Ween, M.P.; Oehler, M.K. Women in cancer thematic review: Ovarian cancer-peritoneal cell interactions promote extracellular matrix processing. Endocr. Relat. Cancer 2016, 23, T155–T168. [Google Scholar] [CrossRef] [Green Version]
- Bukovsky, A.; Indrapichate, K.; Fujiwara, H.; Cekanova, M.; Ayala, M.E.; Dominguez, R.; Caudle, M.R.; Wimalsena, J.; Elder, R.F.; Copas, P.; et al. Multiple luteinizing hormone receptor (lhr) protein variants, interspecies reactivity of anti-lhr mab clone 3b5, subcellular localization of lhr in human placenta, pelvic floor and brain, and possible role for lhr in the development of abnormal pregnancy, pelvic floor disorders and alzheimer’s disease. Reprod. Biol. Endocrinol. 2003, 1, 46. [Google Scholar]
- Yarram, S.J.; Perry, M.J.; Christopher, T.J.; Westby, K.; Brown, N.L.; Lamminen, T.; Rulli, S.B.; Zhang, F.P.; Huhtaniemi, I.; Sandy, J.R.; et al. Luteinizing hormone receptor knockout (lurko) mice and transgenic human chorionic gonadotropin (hcg)-overexpressing mice (hcg alphabeta+) have bone phenotypes. Endocrinology 2003, 144, 3555–3564. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Krishna, A. Effects of gnrh agonist treatment on steroidogenesis and folliculogenesis in the ovary of cyclic mice. J. Ovarian Res. 2010, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- Danilovich, N.; Roy, I.; Sairam, M.R. Ovarian pathology and high incidence of sex cord tumors in follitropin receptor knockout (forko) mice. Endocrinology 2001, 142, 3673–3684. [Google Scholar] [CrossRef]
- Chen, X.; Aravindakshan, J.; Yang, Y.; Sairam, M.R. Early alterations in ovarian surface epithelial cells and induction of ovarian epithelial tumors triggered by loss of fsh receptor. Neoplasia 2007, 9, 521–531. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Zhang, J.; Zhu, Y.; Zhang, Z.; Sun, H.; Feng, Y. Follicle-stimulating hormone induced epithelial-mesenchymal transition of epithelial ovarian cancer cells through follicle-stimulating hormone receptor pi3k/akt-snail signaling pathway. Int. J. Gynecol. Cancer 2014, 24, 1564–1574. [Google Scholar] [CrossRef]
- Li, S.; Ma, D.; Zhu, C. Effects of antisense oligodeoxynucleotide to follicle-stimulating hormone receptor on the cell proliferation and apoptosis in cells derived from human ovarian mucinous cystadenocarcinoma in vitro. J. Huazhong Univ. Sci. Technol. Med. Sci. 2007, 27, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Choi, K.C.; Auersperg, N.; Leung, P.C. Overexpression of follicle-stimulating hormone receptor activates oncogenic pathways in preneoplastic ovarian surface epithelial cells. J. Clin. Endocrinol. Metab. 2004, 89, 5508–5516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warrenfeltz, S.W.; Lott, S.A.; Palmer, T.M.; Gray, J.C.; Puett, D. Luteinizing hormone-induced up-regulation of erbb-2 is insufficient stimulant of growth and invasion in ovarian cancer cells. Mol. Cancer Res. 2008, 6, 1775–1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricciardelli, C.; Lokman, N.A.; Cheruvu, S.; Tan, I.A.; Ween, M.P.; Pyragius, C.E.; Ruszkiewicz, A.; Hoffmann, P.; Oehler, M.K. Transketolase is upregulated in metastatic peritoneal implants and promotes ovarian cancer cell proliferation. Clin. Exp. Metastasis 2015, 32, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time pcr data by the comparative c(t) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Ween, M.P.; Lokman, N.A.; Hoffmann, P.; Rodgers, R.J.; Ricciardelli, C.; Oehler, M.K. Transforming growth factor-beta-induced protein secreted by peritoneal cells increases the metastatic potential of ovarian cancer cells. Int. J. Cancer 2011, 128, 1570–1584. [Google Scholar] [CrossRef] [Green Version]
- Ricciardelli, C.; Lokman, N.A.; Pyragius, C.E.; Ween, M.P.; Macpherson, A.M.; Ruszkiewicz, A.; Hoffmann, P.; Oehler, M.K. Keratin 5 overexpression is associated with serous ovarian cancer recurrence and chemotherapy resistance. Oncotarget 2017, 8, 17819–17832. [Google Scholar] [CrossRef] [Green Version]




| a. Univariate Cox Regression Analyses for Progression-Free Survival and Overall Survival. | ||||||
| Progression-Free Survival | Overall Survival | |||||
| Variable | Relative Risk | 95% CI | p-Value | RELATIVE RISK | 95% CI | p-Value |
| Age a | 1.48 | 0.97–2.25 | 0.066 | 1.34 | 0.85–2.10 | 0.204 |
| Tumor stage b | 0.97 | 0.50–1.88 | 0.929 | 0.87 | 0.35–2.17 | 0.764 |
| Tumor grade c | 0.94 | 0.56–1.56 | 0.805 | 0.96 | 0.56–1.65 | 0.888 |
| Residual disease d | 1.72 | 0.90–3.30 | 0.101 | 2.39 | 1.12–5.11 | 0.024 |
| FSHR e | 0.63 | 0.39–1.01 | 0.054 | 0.55 | 0.54–0.96 | 0.035 |
| FSHR BV f | 0.39 | 0.23–0.68 | 0.001 | 0.40 | 0.23–0.73 | 0.001 |
| LHCGR g | 0.85 | 0.66–1.09 | 0.206 | 0.83 | 0.62–1.11 | 0.199 |
| LHCGR BV h | 0.76 | 0.47–1.21 | 0.248 | 0.93 | 0.57–1.51 | 0.756 |
| FSHR and LHCGR groups i | 1.00 | 1.00 | ||||
| 3 vs. 2 | 0.62 | 0.36–1.06 | 0.081 | 0.52 | 0.28–0.94 | 0.032 |
| 3 vs. 1 | 0.37 | 0.17–0.83 | 0.016 | 0.30 | 0.11–0.77 | 0.013 |
| b. Multivariate Cox Regression Analyses for Progression-Free Survival and Overall Survival. All Variables Are Significant in Univariate Analysis | ||||||
| Progression-Free Survival (n = 79) | Overall Survival (n = 84) | |||||
| Variable | RELATIVE RISK | 95% CI | p-Value | Relative Risk | 95% CI | p-Value |
| Residual disease d | 1.52 | 0.75–3.09 | 0.248 | 3.06 | 1.19–7.89 | 0.021 |
| FSHR BV f | 0.37 | 0.19–0.70 | 0.002 | 0.48 | 0.25–0.89 | 0.02 |
| FSHR and LHCGR groups i | 1.00 | 1.00 | ||||
| 3 vs. 2 | 0.76 | 0.41–1.40 | 0.381 | 0.40 | 0.14–1.09 | 0.073 |
| 3 vs. 1 | 0.58 | 0.23–1.48 | 0.255 | 0.50 | 0.25–0.98 | 0.044 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheung, J.; Lokman, N.A.; Abraham, R.D.; Macpherson, A.M.; Lee, E.; Grutzner, F.; Ghinea, N.; Oehler, M.K.; Ricciardelli, C. Reduced Gonadotrophin Receptor Expression Is Associated with a More Aggressive Ovarian Cancer Phenotype. Int. J. Mol. Sci. 2021, 22, 71. https://doi.org/10.3390/ijms22010071
Cheung J, Lokman NA, Abraham RD, Macpherson AM, Lee E, Grutzner F, Ghinea N, Oehler MK, Ricciardelli C. Reduced Gonadotrophin Receptor Expression Is Associated with a More Aggressive Ovarian Cancer Phenotype. International Journal of Molecular Sciences. 2021; 22(1):71. https://doi.org/10.3390/ijms22010071
Chicago/Turabian StyleCheung, Janelle, Noor A. Lokman, Riya D. Abraham, Anne M. Macpherson, Eunice Lee, Frank Grutzner, Nicolae Ghinea, Martin K. Oehler, and Carmela Ricciardelli. 2021. "Reduced Gonadotrophin Receptor Expression Is Associated with a More Aggressive Ovarian Cancer Phenotype" International Journal of Molecular Sciences 22, no. 1: 71. https://doi.org/10.3390/ijms22010071





