Ovule Development and in Planta Transformation of Paphiopedilum Maudiae by Agrobacterium-Mediated Ovary-Injection
Abstract
:1. Introduction
2. Results
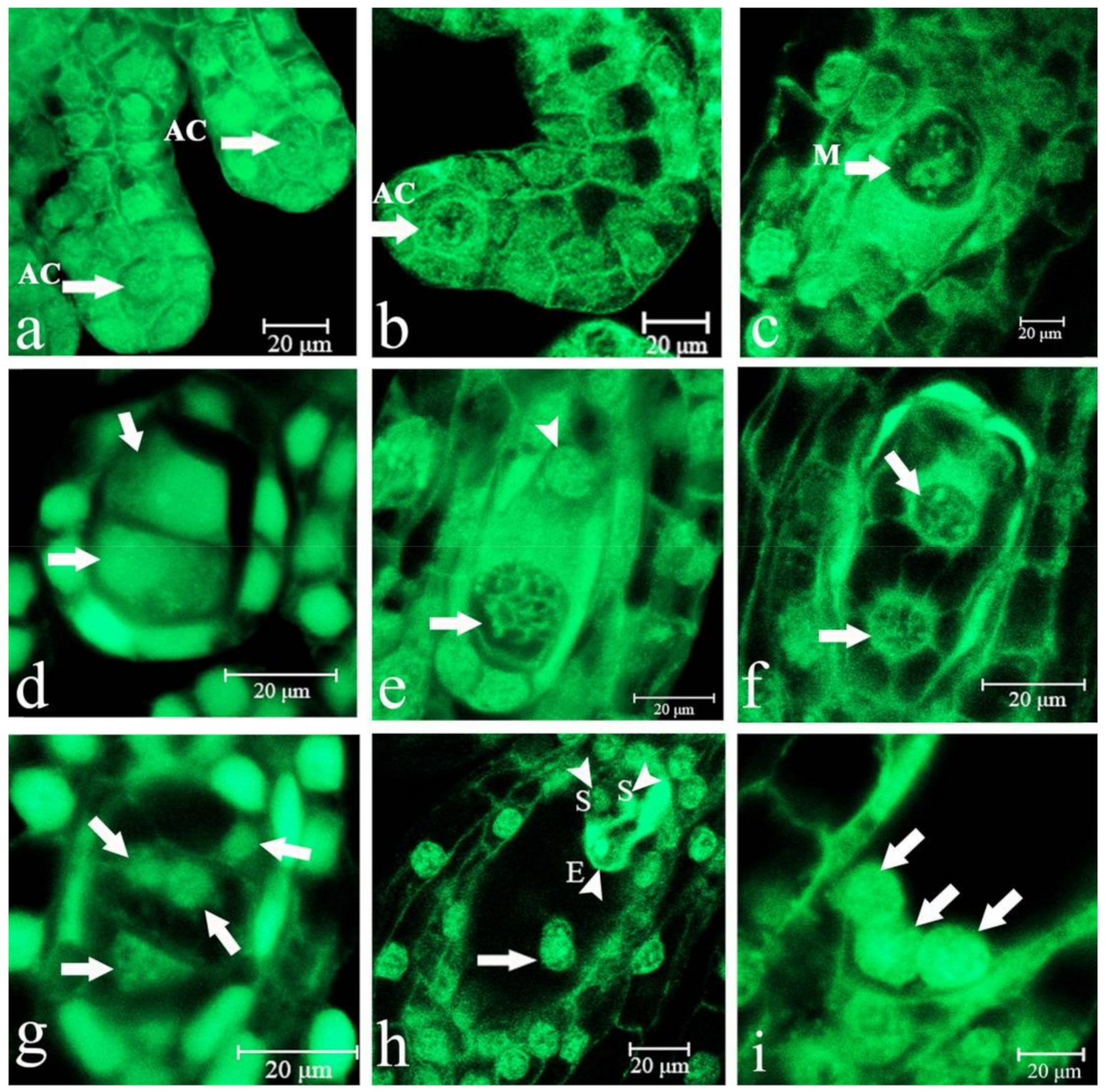
2.1. Embryo Sac Development, Megasporogenesis, and Female Gametophyte Formation
2.2. Fertilization
2.3. Embryonic Development
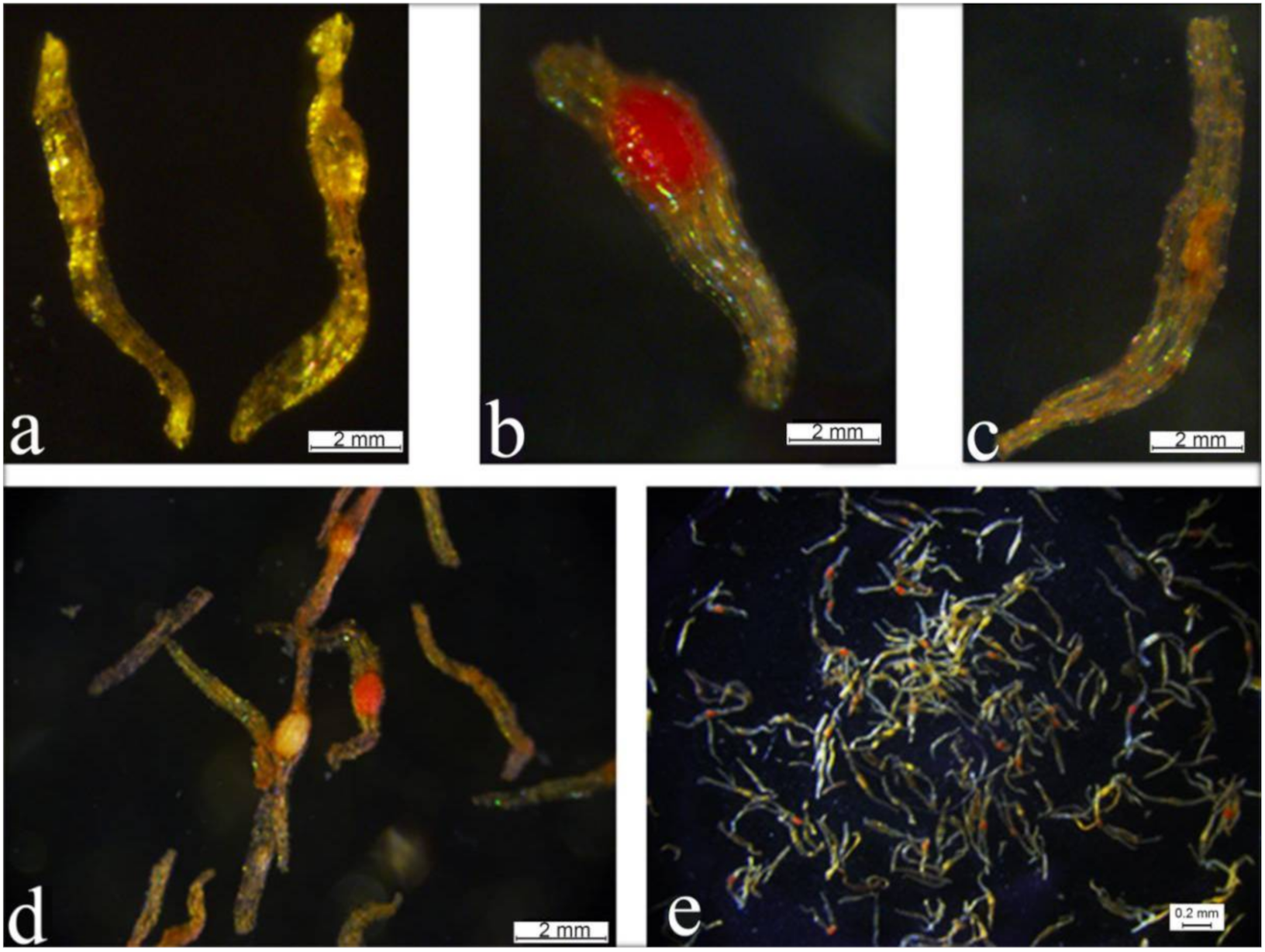
2.4. Morphological Characteristics and Seed Viability Tests
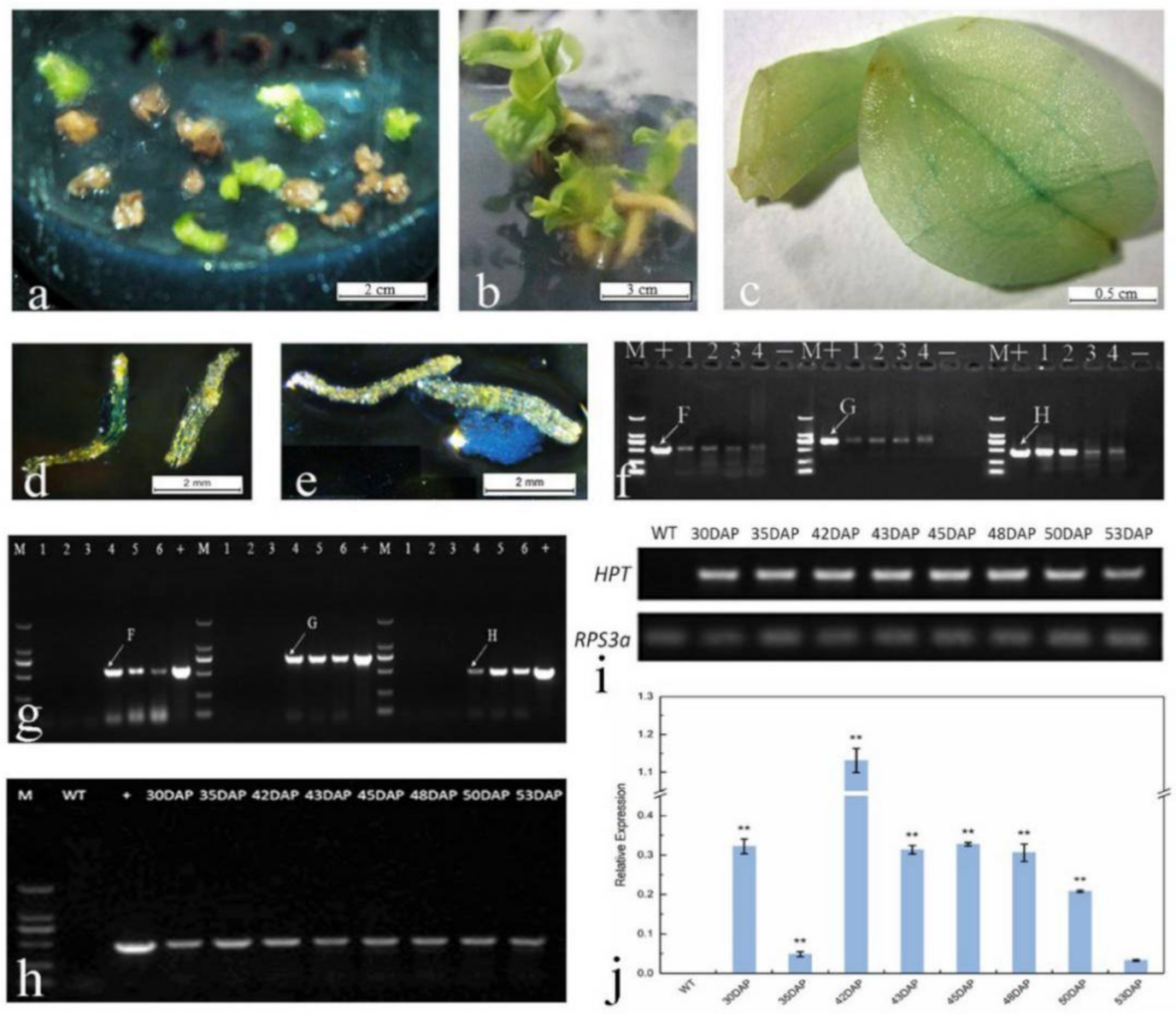
2.5. In Planta Transformation by Agrobacterium-Mediated Ovary-Injection
2.6. TTC and GUS Staining and Preliminary Detection of Transgenic Seeds
2.7. In Vitro Germination
2.8. Selection Pressure and Hygromycin Screening
2.9. GUS Staining and Molecular Detection of Hyg-Resistant Plants
3. Discussion
3.1. Ovules and Embryo Development
3.2. Agrobacterium-Mediated Ovary-Injection
4. Materials and Methods
4.1. Plant Material and Agrobacterium Strain
4.2. Histological and Histochemical Studies
4.3. Injection of Ovaries with Agrobacterium
4.4. Aseptic Sowing
4.5. Seed Viability Test
4.6. GUS Assay for Seeds and Transgenic Seedlings
4.7. Selection Pressure and Resistance Screening of Protocorms
4.8. PCR Detection of Seeds and Resistant Seedlings
4.9. Semi-Quantitative PCR and Quantitative Real-Time PCR Analysis
4.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Z.J.; Chen, S.C.; Chen, L.J.; Lei, S.P. The Genus Paphiopedilum in China; Science Press: Beijing, China, 2009; pp. 13–349. [Google Scholar]
- Zeng, S.J.; Wu, K.L.; Teixeira da Silva, J.A.; Zhang, J.X.; Chen, Z.L.; Xia, N.H.; Duan, J. Asymboiotic seed germination, seedling development and reintroduction of Paphiopedilum wardii Sumerh., an endangered terrestrial orchid. Sci. Hortic. 2012, 138, 198–209. [Google Scholar] [CrossRef]
- Cribb, P. The Genus Paphiopedilum, 2nd ed.; National History Publications: Borneo, Malaysia, 1998; pp. 48–396. [Google Scholar]
- Wu, Z.Y.; Raven, P.H.; Hong, D.Y. Flora of China; Science Press: Beijing, China; Missouri Botanical Garden Press: St., Louis, MI, USA, 2009; Volume 25, pp. 33–44. [Google Scholar]
- World Checklist of Selected Plant Families. Available online: http://apps.kew.org/wcsp/qsearch.do (accessed on 22 December 2020).
- The IUCN Red List of Threatened Species. Version 2020-2. Available online: http://www.iucnredlist.org/search (accessed on 22 December 2020).
- Zeng, S.J.; Huang, W.C.; Wu, K.L.; Zhang, J.X.; Teixeira da Silva, J.A.; Duan, J. In vitro propagation of Paphiopedilum orchids. Crit. Rev. Biotechnol. 2016, 36, 521–534. [Google Scholar]
- Zeng, S.J.; Tian, R.X.; Chen, Z.L.; Wu, K.L.; Duan, J. Research progress on cross breeding of Paphiopedilum. J. Trop. Subtrop. Bot. 2010, 18, 459–468. [Google Scholar]
- Huang, L.C. A procedure for asexual multiplication of Paphiopedilum in vitro. Am. Orchid Soc. Bull. 1988, 57, 274–278. [Google Scholar]
- Liao, Y.J.; Tsai, Y.C.; Sun, Y.W.; Lin, R.S.; Wu, F.S. In vitro shoot induction and plant regeneration from flower buds in Paphiopedilum orchids. In Vitro Cell. Dev. Biol. Plant 2011, 47, 702–709. [Google Scholar]
- Fu, Y.Y.; Jiang, N.; Wu, K.L.; Zhang, J.X.; Teixeira da Silva, J.A.; Duan, J.; Liu, H.T.; Zeng, S.J. Stimulatory effects of sodium hypochlorite and ultrasonic treatments on tetrazolium staining and seed germination in vitro of Paphiopedilum SCBG Red Jewel. Seed Sci. Technol. 2016, 44, 77–90. [Google Scholar]
- Zhang, X.S.; O’Neill, S.D. Ovary and gametophyte development are coordinately regulated by auxin and ethylene following pollination. Plant Cell 1993, 5, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.L.; Guo, J.Y. Female gametophyte development and embryogenesis in Cymbidium sinense (Andr.) Willd. J. Trop. Subtrop. Bot. 1995, 3, 54–58. [Google Scholar]
- Tang, Y.J.; Ye, X.L.; Chen, Z.L. The studies of the female gametophyte development and the embryogenesis in Doritis pulcherrima Lindl. J. Trop. Subtrop. Bot. 1998, 6, 289–292. [Google Scholar]
- Li, D.M.; Wu, C.H.; Ye, X.L.; Liang, C.Y. Ultrastructural observation on embryo sac development in Phaius tankervilliae (Aiton) Bl. Plant Sci. J. 2012, 30, 188–198. [Google Scholar] [CrossRef]
- Liu, F.; Tian, M.; Wang, C.X.; Gong, M.J.; Li, Q.J. Observation on fruit growth dynamics and embryo development process of Cymbidium japonicum. J. Plant Res. Dev. 2012, 21, 28–35. [Google Scholar]
- Zhang, Y.; Zhang, Q.X.; Zhao, S.W.; Ling, C.Y. Embryo and integument development of the endangered species Cypripedium macrathos Sw. Acta Hortic. Sin. 2010, 37, 72–76. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Wu, K.L.; Zhang, J.X.; Deng, R.F.; Duan, J.; Teixeira da Silva, J.A.; Huang, W.C.; Zeng, S.J. Embryo development in association with asymbiotic seed germination in vitro of Paphiopedilum armeniacum S. C. Chen et F. Y. Liu. Sci. Rep. 2015, 5, 16356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.H.; Huang, G.W. Application of laser scanning confocal microscope in botany. Acta Laser Biol. Sin. 2005, 14, 76–79. [Google Scholar]
- Li, J.X.; Dai, X.M.; Zhang, J.J.; Zhang, Y.S. Using a whole stain-clearing technique to observe embryo sac of cotton. Exp. Technol. Manag. 2014, 31, 60–62. [Google Scholar]
- Dai, X.M.; Huang, Q.C.; Li, G.P.; Hu, X.M.; Qin, G.Y. Observation on the process of double fertilization in rice using laser scanning confocal microscopy. J. Henan Agric. Sci. 2003, 6, 25–30. [Google Scholar]
- Zhang, H.H.; Feng, J.H.; Lu, Y.G.; Yang, B.Y.; Liu, X.D. Observation on formation and development of autotetraploid rice embryo sac using laser scanning confocal microscope. J. Chin. Electron Microsc. Soc. 2003, 22, 380–384. [Google Scholar]
- Zhu, D.Z.; Wang, J.W.; Zong, X.J.; Wei, H.R.; Liu, Q.Z. Observation of female gametophyte development of sweet cheery with confocal scanning microscope. Int. J. Fruit Sci. 2014, 31, 36–40. [Google Scholar]
- Johnson, T.R.; Kane, M.E. Asymbiotic germination of ornamental Vanda: In vitro germination and development of three hybrids. Plant Cell Tissue Organ Cult. 2007, 91, 251–261. [Google Scholar] [CrossRef]
- Hossain, M.M.; Kant, R.; Van, P.T.; Winarto, B.; Liu, J.J.; Zeng, S.J.; Teixeira da Silva, J.A. The application of biotechnology to orchids. Crit. Rev. Plant Sci. 2013, 32, 69–139. [Google Scholar] [CrossRef]
- Teixeira da Silva, J.A. Orchids: Advances in tissue culture, genetics, phytochemistry and transgenic biotechnology. Floricult. Ornam. Biotechnol. 2013, 7, 1–52. [Google Scholar]
- Zhou, G.Y.; Weng, J.; Zeng, Y.; Huang, J.; Qian, S.; Liu, G. Introduction of exogenous DNA into cotton embryos. Meth. Enzymol. 1983, 101, 433–481. [Google Scholar]
- Yang, A.F.; Su, L.J.; An, L.J. Ovary-drip transformation: A simple method for directly generating vector- and marker-free transgenic maize (Zea mays L.) with a linear GFP cassette transformation. Planta 2009, 229, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.F.; Dong, H.; Zeng, S.J.; Chen, Z.L.; Wu, K.L.; Zhang, J.X.; Duan, J. In planta transformation of Dendrobium nobile by ovary-injection of Agrobacterium. J. South China Agric. Univ. 2013, 34, 378–382. [Google Scholar]
- Wang, J.; Zeng, S.J.; Chen, Z.L.; Wu, K.L.; Zhang, J.X.; Duan, J. Genetic transformation of Doritis pulcherima (Orchidaceae) via ovary-injection. Chin. J. Trop. Crops 2013, 34, 1498–1501. [Google Scholar]
- Teixeira da Silva, J.A.; Kerbauy, G.B.; Zeng, S.J.; Chen, Z.L.; Duan, J. In vitro flowering of orchids. Crit. Rev. Biotechnol. 2014, 34, 56–76. [Google Scholar] [CrossRef]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation tagging of the floral inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.; Zhao, F.Y.; Wu, C.X. Pleiotropic effects of FT and its orthologues on plant development. Acta Bot. Boreal. Occid. Sin. 2011, 31, 2558–2564. [Google Scholar]
- Lee, Y.I.; Lee, N.; Yeung, E.C.; Chung, M.C. Embryo development of Cypripedium formosanum in relation to seed germination in vitro. J. Am. Soc. Hortic. Sci. 2005, 130, 747–753. [Google Scholar] [CrossRef] [Green Version]
- Zeng, S.J.; Chen, Z.L.; Wu, K.L.; Zhang, J.X.; Bai, C.K.; Teixeira da Silva, J.A.; Duan, J. Asymbiotic seed germination, induction of calli and protocorm-like bodies, and in vitro seedling development of the rare and endangered Nothodoritis zhejiangensis Chinese orchid. HortScience 2011, 46, 460–465. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.F. The time interval from pollination to fertilization. Biol. Bull. 1990, 10, 8. [Google Scholar]
- Peng, Z.Z.; Zhao, Y.; Zeng, Q.; Huang, L.H.; Chen, H.D.; Li, Y.G.; Zhang, X.W. Inplanta transformation of Gossypium hirsutum by ovary-injection of Agrobacterium. J. Cotton Sci. 2011, 23, 311–316. [Google Scholar]
- Yam, T.W.; Yeung, E.C.; Ye, X.L.; Zee, S.Y.; Arditti, J. Orchid embryos. In Orchid Biology: Reviews and Perspectives, 8th ed.; Kull, T., Arditti, J., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; pp. 287–385. [Google Scholar]
- Liu, J.P. The embryonic development of angiosperms. Biol. Bull. 1984, 5, 4–6. [Google Scholar]
- Ren, L.; Wang, F.H. Embryological studies of Paphiopedilum godefroy Srein. Acta Bot. Sin. 1987, 29, 14–21. [Google Scholar]
- Chin, D.P.; Mishiba, K.; Mii, M. Agrobacterium-mediated transformation of protocorm-like bodies in Cymbidium. Plant Cell Rep. 2007, 26, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.L.; Lin, K.H.; Sanjaya; Liao, L.J.; Chen, W.H.; Chan, M.T. Gene stacking in Phalaenopsis orchid enhances dual tolerance to pathogen attack. Transgenic Res. 2005, 14, 279–288. [Google Scholar] [CrossRef]
- Liau, C.H.; You, S.J.; Prasad, V.; Hsiao, H.H.; Lu, J.C.; Yang, N.S.; Chan, M.T. Agrobacterium tumefaciens-mediated transformation of an Oncidium orchid. Plant Cell Rep. 2003, 21, 993–998. [Google Scholar] [CrossRef]
- Mishiba, K.; Chin, D.P.; Mii, M. Agrobacterium-mediated transformation of Phalaenopsis by targeting protocorms at an early stage after germination. Plant Cell Rep. 2005, 24, 297–303. [Google Scholar] [CrossRef]
- Sjahril, R.; Mii, M. High-efficiency Agrobacterium-mediated transformation of Phalaenopsis using meropenem, a novel antibiotic to eliminate Agrobacterium. J. Hortic. Sci. Biotech. 2006, 81, 458–464. [Google Scholar] [CrossRef]
- Nan, G.L.; Kuehnle, A.R. Factors affecting gene delivery by particle bombardment of Dendrobium orchids. Biol. Plant. 1995, 31, 131–136. [Google Scholar]
- Yang, J.; Lee, H.J.; Shin, D.H.; Oh, S.K.; Seon, J.H.; Paek, K.Y.; Han, K.H. Genetic transformation of Cymbidium orchid by particle bombardment. Plant Cell Rep. 1999, 18, 978–984. [Google Scholar] [CrossRef]
- Knapp, J.E.; Kausch, A.P.; Chandlee, J.M. Transformation of three genera of orchid using the bar gene as a selectable marker. Plant Cell Rep. 2000, 19, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Men, S.; Ming, X.; Wang, Y.; Liu, R.; Wei, C.; Li, Y. Genetic transformation of two species of orchid by biolistic bombardment. Plant Cell Rep. 2003, 21, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Tee, C.S.; Marziah, M.; Tan, C.S.; Abdullah, M.P. Evaluation of different promoters driving the GFP report gene and selected target tissues for particle bombardment of Dendrobium Sonia 17. Plant Cell Rep. 2003, 21, 452–458. [Google Scholar] [CrossRef]
- Li, S.H.; Kuoh, C.S.; Chen, Y.H.; Chen, H.H.; Chen, W.H. Osmotic sucrose enhancement of single-cell embryogenesis and transformation efficiency in Oncidium. Plant Cell Tissue Organ Cult. 2005, 81, 183–192. [Google Scholar] [CrossRef]
- Teixeira da Silva, J.A.; Chin, D.P.; Van, P.T.; Mii, M. Transgenic orchids. Sci. Hortic. 2011, 130, 673–680. [Google Scholar] [CrossRef]
- Zhou, G.Y.; Weng, J.; Gong, Z.Z.; Zhen, Y.S.; Yang, W.X.; Shen, W.F.; Wang, Z.F.; Tao, Q.Z.; Huang, J.G.; Qian, S.Y.; et al. Molecular breeding of agriculture: A technique for introducing exogenous DNA into plants after self-pollination. Sci. Agric. Sin. 1988, 21, 1–6. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Bent, A.F. Arabidopsisin planta transformation. uses, mechanisms, and prospects for transformation of other species. Plant Physiol. 2000, 124, 1540–1547. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Li, G.; Li, M.G.; Wang, J.H. Transgenic soybean with low phytate content constructed by Agrobacterium transformation and pollen-tube pathway. Euphytica 2010, 177, 375–382. [Google Scholar] [CrossRef]
- Teixeira da Silva, J.A.; Tsavkelova, E.; Zeng, S.J.; Ng, T.B.; Dobránszki, J.; Parthibhan, S.; Cardoso, J.C.; Rao, M.V. Symbiotic in vitro seed propagation of Dendrobium: Fungal and bacterial partners and their influence on plant growth and development. Planta 2015, 242, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Teixeira da Silva, J.A.; Tsavkelova, E.; Ng, T.B.; Dobránszki, J.; Parthibhan, S.; Cardoso, J.C.; Rao, M.V.; Zeng, S.J. Asymbiotic in vitro seed propagation of Dendrobium. Plant Cell Rep. 2015, 34, 1685–1706. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.L.; Duan, J.; Zeng, S.J.; Liang, C.Y.; Ye, X.L. Agrobacterium-mediated transformation of Dendrobium orchid by targeting protocorms. Acta Sci. Nat. Univ. Sunyatseni 2007, 46, 86–90. [Google Scholar]
- Huang, W.T.; Fang, Z.M.; Zeng, S.J.; Zhang, J.X.; Wu, K.L.; Chen, Z.L.; Teixeira da Silva, J.A.; Duan, J. Molecular cloning and functional analysis of three FLOWERING LOCUS T (FT) homologous genes from Chinese Cymbidium. Int. J. Mol. Sci. 2012, 13, 11385–11398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, G.Q.; Walworth, A.; Zhao, D.Y.; Jiang, N.; Hancock, J.F. The Vaccinium corymbosum Flowering Locus T-like gene (VcFT): A flowering activator reverses photoperiodic and chilling requirements in blueberry. Plant Cell Rep. 2013, 32, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Smart, M.; Roden, L.C. Initiation of flowering in Protea compacta × Protea neriifolia hybrid ‘Carnival’ coincides with expression of the Flowering Locus T homologue. Plant Mol. Biol. Rep. 2014, 32, 372–381. [Google Scholar] [CrossRef]
- Yu, G.R.; Yin, J.; Ren, J.P.; Guo, X.Y. Transferring Anti-TrxS gene into wheat by methods of pollen tube pathway and ovary injection. J. Henan Agric. Sci. 2004, 6, 6–10. [Google Scholar]
- Gao, X.R.; Wang, G.K.; Su, Q.; Wang, Y.; An, L.J. Phytase expression in transgenic soybeans: Stable transformation with a vector-less construct. Biotechnol. Lett. 2007, 29, 1781–1787. [Google Scholar] [CrossRef]
- Chen, Y. Study on techniques of introduction of exogenous DNA into plants via pollen tube pathway. Northern Hortic. 2010, 13, 226–228. [Google Scholar]
- Wang, Y.; Wu, Y.; Wang, Y.; Li, Z.B.; Wang, G.X. The five kinds of DNA transformation techniques. Rainfed Crops 2010, 30, 186–189. [Google Scholar]
- Wang, G.L.; Fang, H.J.; Zhu, Y.M. Plant Genetic Engineering; Science Press: Beijing, China, 2009; pp. 287–288. [Google Scholar]
- Li, W.J.; Zhang, J.P.; Chen, F.; Sun, W.J. Transformation of herbicide resistant gene (bar) into flax by pollen tube pathway. Chin. Agric. Sci. Bull. 2013, 29, 96–100. [Google Scholar]
- Wang, M.; Zhang, B.H.; Wang, Q.L. Transgenic Cotton Methods and Protocols; Humana Press: Totowa, NJ, USA, 2013; pp. 71–77. [Google Scholar]
- Luo, H.L.; Luo, K.C.; Luo, L.P.; Li, E.X.; Guan, B.C.; Xiong, D.J.; Sun, B.T.; Peng, K.; Yang, B.Y. Evaluation of candidate reference genes for gene expression studies in Cymbidium kanran. Sci. Hortic. 2014, 167, 43–48. [Google Scholar] [CrossRef]
- NCBI Primer-BLAST. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome (accessed on 22 December 2020).



| Days after Pollination (DAP) | Developmental Stage |
|---|---|
| 6, 10, 20 | Formation of an archesporial cell |
| 20, 25 | Formation of megasporocyte |
| 25, 30 | Formation of functional megaspore |
| 35, 38, 40 | Two-nucleate embryo sac |
| 42, 44 | Four-nucleate embryo sac |
| 45, 50 | Eight-nucleate embryo sac |
| 50, 54 | Mature embryo sac, fertilization |
| 54, 60 | First division of the zygote |
| 70, 76, 80, 85, 90 | Pre-embryo with 2-8 cells, suspensor with 1–2 cells and two-nucleate endosperm |
| 95, 100, 105, 109, 114 | Pre-embryo with 10-40 cells, suspensor with 2–4 cells and nucleate endosperm |
| 120, 125, 130, 140 | Embryo with more than 100 cells, suspensor and nucleate endosperm degenerated, massive starch and lipid globule found in the embryonal cells |
| 150 | Seed mature |
| 160 | Seed fully mature |
| 180 | Capsules begin to open |
| Injection Time (DAP) | TTC Staining (%) * |
|---|---|
| Control (no injection) | 60.61 ± 0.05 a |
| 35 | 12.11 ± 4.41 ef |
| 40 | 14.63 ± 5.33 def |
| 44 | 18.63 ± 2.61 cde |
| 46 | 24.85 ± 3.70 cde |
| 50 | 43.01 ± 3.60 b |
| 53 | 32.98 ± 2.47 bc |
| 55 | 22.31 ± 0.84 cdef |
| 57 | 19.96 ± 6.79 cd |
| 60 | 14.04 ± 1.24 def |
| 65 | 11.91 ± 7.51 def |
| 70 | 10.43 ± 4.06 f |
| Injection Time (DAP) | Seed Germination (%) * |
|---|---|
| Control (no injection) | 36.30 ± 2.13 a |
| 35 | 7.26 ± 2.50 bc |
| 40 | 7.53 ± 1.65 bc |
| 44 | 8.78 ± 1.15 bc |
| 46 | 10.32 ± 1.26 bc |
| 50 | 15.21 ± 2.91 bc |
| 53 | 9.31 ± 0.43 bc |
| 55 | 5.46 ± 1.23 c |
| 57 | 4.71 ± 0.10 c |
| 60 | 4.61 ± 0.91 c |
| 65 | 3.66 ± 0.93 c |
| 70 | 2.84 ± 1.08 c |
| Injection Time (DAP) | Number of Hygromycin-Resistant Seedlings | Number of Germinated Seeds | Percentage of Resistant Seedlings (Transformation Frequency (%)) | Percentage of Hygromycin-Resistant Seedlings from all Seeds Sown * (Transformation Frequency (%)) |
|---|---|---|---|---|
| No injection | 0 | 530 | 0 g | 0 e |
| 30 | 2 | 443 | 0.45 f | 0.13 cd |
| 35 | 7 | 371 | 1.89 c | 0.47 ab |
| 42 | 5 | 231 | 2.16 b | 0.33 bc |
| 43 | 1 | 235 | 1.07 d | 0.07 de |
| 45 | 3 | 334 | 0.90 e | 0.20 cd |
| 48 | 4 | 163 | 2.45 a | 0.27 c |
| 50 | 9 | 354 | 2.54 a | 0.60 a |
| 53 | 7 | 282 | 2.48 a | 0.47 ab |
| 55 | 0 | 242 | 0 g | 0 e |
| 57 | 0 | 230 | 0 g | 0 e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, B.-X.; Zhang, L.; Zheng, F.; Wu, K.-L.; Li, L.; Zhang, X.-H.; Ma, G.-H.; Teixeira da Silva, J.A.; Fang, L.; Zeng, S.-J. Ovule Development and in Planta Transformation of Paphiopedilum Maudiae by Agrobacterium-Mediated Ovary-Injection. Int. J. Mol. Sci. 2021, 22, 84. https://doi.org/10.3390/ijms22010084
Luo B-X, Zhang L, Zheng F, Wu K-L, Li L, Zhang X-H, Ma G-H, Teixeira da Silva JA, Fang L, Zeng S-J. Ovule Development and in Planta Transformation of Paphiopedilum Maudiae by Agrobacterium-Mediated Ovary-Injection. International Journal of Molecular Sciences. 2021; 22(1):84. https://doi.org/10.3390/ijms22010084
Chicago/Turabian StyleLuo, Bai-Xue, Li Zhang, Feng Zheng, Kun-Lin Wu, Lin Li, Xin-Hua Zhang, Guo-Hua Ma, Jaime A. Teixeira da Silva, Lin Fang, and Song-Jun Zeng. 2021. "Ovule Development and in Planta Transformation of Paphiopedilum Maudiae by Agrobacterium-Mediated Ovary-Injection" International Journal of Molecular Sciences 22, no. 1: 84. https://doi.org/10.3390/ijms22010084
APA StyleLuo, B. -X., Zhang, L., Zheng, F., Wu, K. -L., Li, L., Zhang, X. -H., Ma, G. -H., Teixeira da Silva, J. A., Fang, L., & Zeng, S. -J. (2021). Ovule Development and in Planta Transformation of Paphiopedilum Maudiae by Agrobacterium-Mediated Ovary-Injection. International Journal of Molecular Sciences, 22(1), 84. https://doi.org/10.3390/ijms22010084





