Extracellular Vesicles-Encapsulated Yeast Prions and What They Can Tell Us about the Physical Nature of Propagons
Abstract
1. Introduction
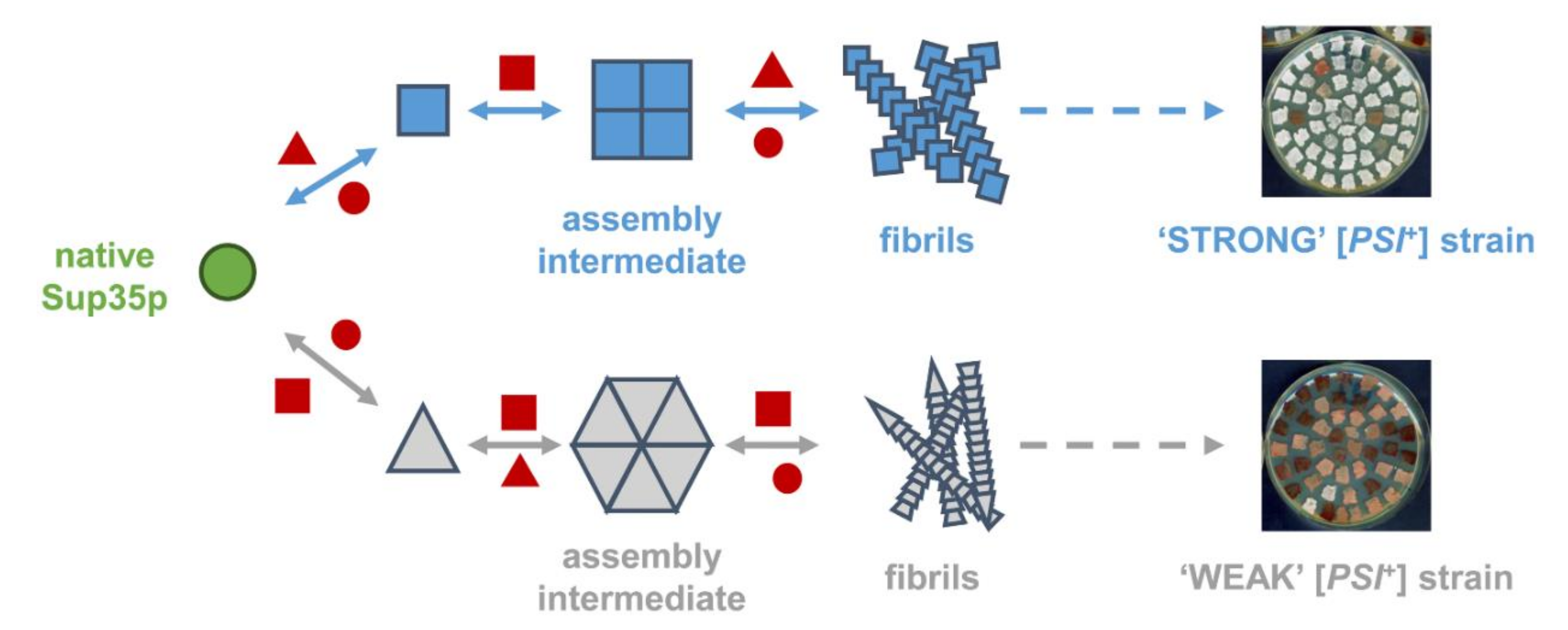
2. Questions Raised by Current [PSI+] Prion Propagation Models
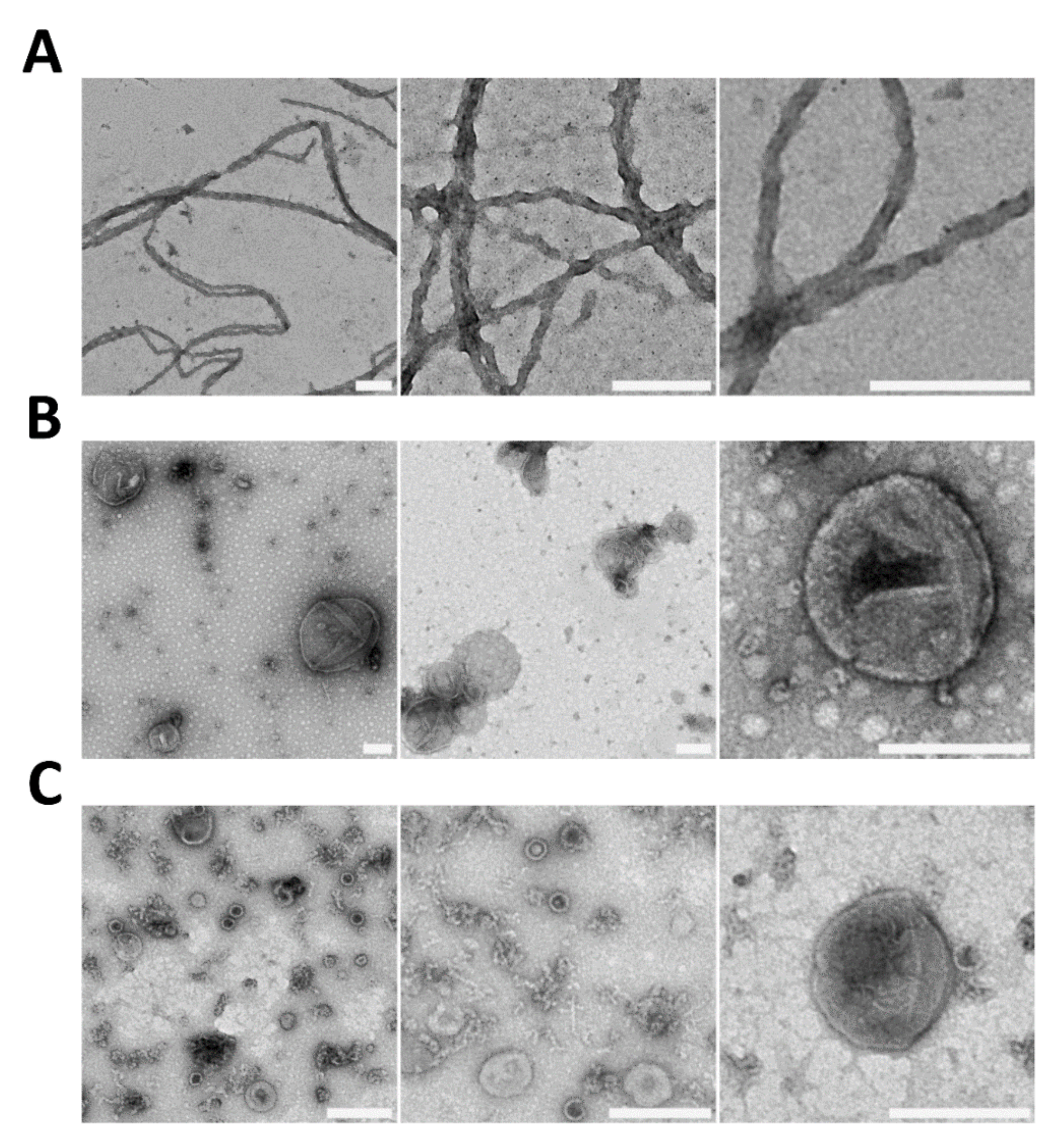
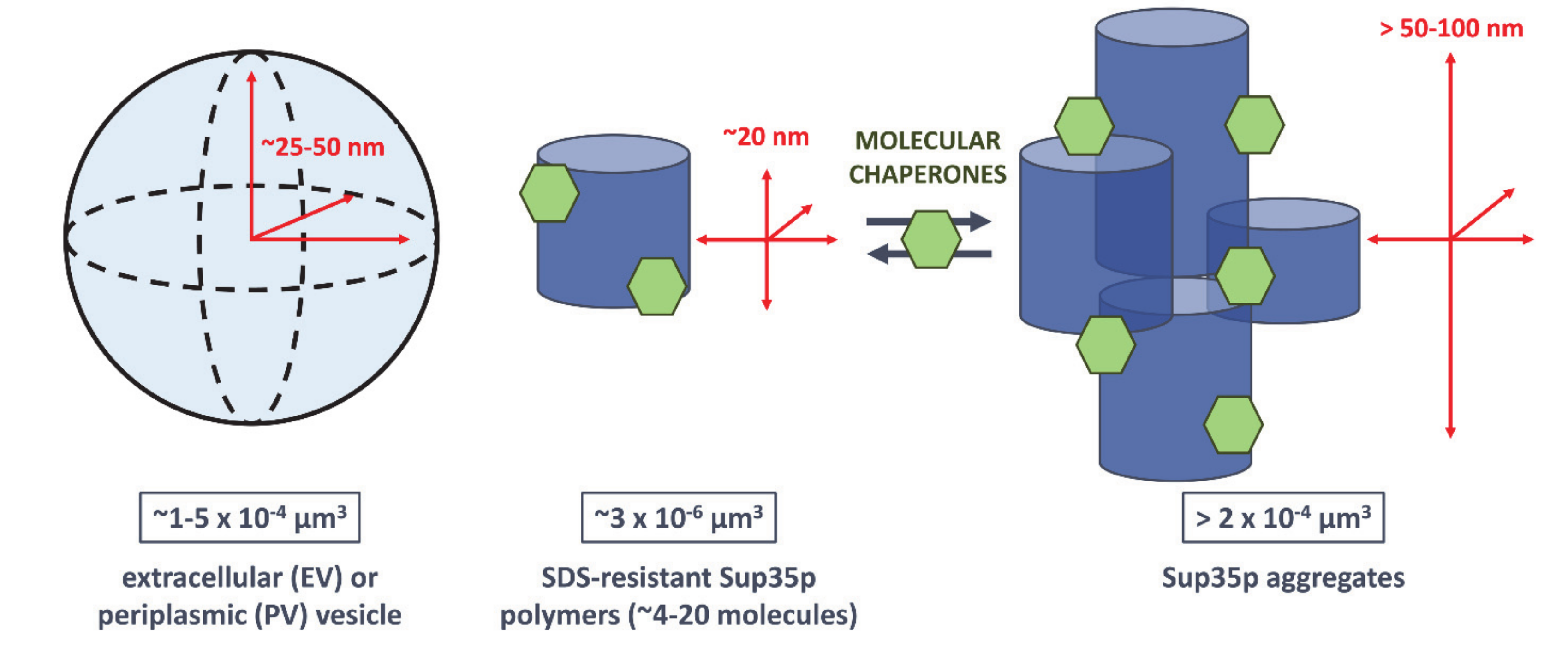
3. What We Can Learn about the Molecular Nature of Propagons Using EV
4. Vesicle-Encapsulated Propagons May Overcome Spatial Quality Control during Bud Formation
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EV | Extracellular vesicles |
| PV | Periplasmic vesicles |
| EM | Electron microscopy |
| VID | Vacuolar import and degradation |
| IVC | Intracellular vesicle clusters |
| IPOD | Insoluble protein deposit |
| JUNQ | Juxta nuclear quality control compartment |
References
- Aigle, M.; Lacroute, F. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. MGG Mol. Gen. Genet. 1975, 136, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Coustou, V.; Deleu, C.; Saupe, S.; Begueret, J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA 1997, 94, 9773–9778. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.S. PSI, a cytoplasmic suppressor of super-supressor in yeast. Heredity 1965, 20, 505–521. [Google Scholar] [CrossRef]
- Prusiner, S.B. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Kabani, M.; Melki, R. More than just trash bins? Potential roles for extracellular vesicles in the vertical and horizontal transmission of yeast prions. Curr. Genet. 2016, 62. [Google Scholar] [CrossRef] [PubMed]
- Kabani, M.; Melki, R. Yeast prions assembly and propagation: Contributions of the prion and non-prion moieties and the nature of assemblies. Prion 2011, 5, 277–284. [Google Scholar] [CrossRef]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef]
- Sindi, S.S.; Serio, T.R. Prion dynamics and the quest for the genetic determinant in protein-only inheritance. Curr. Opin. Microbiol. 2009, 12, 623–630. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Son, M.; Wu, S.; Niznikiewicz, M. How Do Yeast Cells Contend with Prions? Int. J. Mol. Sci. 2020, 21, 4742. [Google Scholar] [CrossRef]
- Glover, J.R.; Kowal, A.S.; Schirmer, E.C.; Patino, M.M.; Liu, J.J.; Lindquist, S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997, 89, 811–819. [Google Scholar] [CrossRef]
- Krzewska, J.; Melki, R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006, 25, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.K.; Liebman, S.W. “Prion-proof” for [PIN+]: Infection with In Vitro-made Amyloid Aggregates of Rnq1p-(132-405) Induces [PIN+]. J. Mol. Biol. 2007, 365, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Thual, C.; Komar, A.A.; Bousset, L.; Fernandez-Bellot, E.; Cullin, C.; Melki, R. Structural characterization of Saccharomyces cerevisiae prion-like protein Ure2. J. Biol. Chem. 1999, 274, 13666–13674. [Google Scholar] [CrossRef]
- Taylor, K.L.; Cheng, N.; Williams, R.W.; Steven, A.C.; Wickner, R.B. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 1999, 283, 1339–1343. [Google Scholar] [CrossRef]
- Bradley, M.E.; Edskes, H.K.; Hong, J.Y.; Wickner, R.B.; Liebman, S.W. Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. 4), 16392–16399. [Google Scholar] [CrossRef]
- Bradley, M.E.; Liebman, S.W. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics 2003, 165, 1675–1685. [Google Scholar]
- Derkatch, I.L.; Chernoff, Y.O.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar]
- Brachmann, A.; Baxa, U.; Wickner, R.B. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005, 24, 3082–3092. [Google Scholar] [CrossRef]
- Winkler, J.; Tyedmers, J.; Bukau, B.; Mogk, A. Chaperone networks in protein disaggregation and prion propagation. J. Struct. Biol. 2012, 179, 152–160. [Google Scholar] [CrossRef]
- Oamen, H.P.; Lau, Y.; Caudron, F. Prion-like proteins as epigenetic devices of stress adaptation. Exp. Cell Res. 2020, 396. [Google Scholar] [CrossRef] [PubMed]
- Tuite, M.F.; Cox, B.S. The [PSI+] prion of yeast: A problem of inheritance. Methods 2006, 39, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Patino, M.M.; Liu, J.J.; Glover, J.R.; Lindquist, S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 1996, 273, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN+]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Patel, B.K.; Gavin-Smyth, J.; Liebman, S.W. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 2009, 11, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Park, K.W.; Yu, H.; Fan, Q.; Li, L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 2008, 40, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef]
- Suzuki, G.; Shimazu, N.; Tanaka, M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 2012, 336, 355–359. [Google Scholar] [CrossRef]
- Chernova, T.A.; Kiktev, D.A.; Romanyuk, A.V.; Shanks, J.R.; Laur, O.; Ali, M.; Ghosh, A.; Kim, D.; Yang, Z.; Mang, M.; et al. Yeast Short-Lived Actin-Associated Protein Forms a Metastable Prion in Response to Thermal Stress. Cell Rep. 2017, 18, 751–761. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Bateman, D.; Kelly, A.C.; Gorkovskiy, A. The yeast prions [PSI+] and [URE3] are molecular degenerative diseases. Prion 2011, 5, 258–262. [Google Scholar] [CrossRef]
- Tuite, M.F. Yeast prions: Paramutation at the protein level? Semin. Cell Dev. Biol. 2015, 44, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Shewmaker, F.P.; Bateman, D.A.; Edskes, H.K.; Gorkovskiy, A.; Dayani, Y.; Bezsonov, E.E. Yeast prions: Structure, biology, and prion-handling systems. Microbiol. Mol. Biol. Rev. 2015, 79, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.M.; Jarosz, D.F. Rebels with a cause: Molecular features and physiological consequences of yeast prions. FEMS Yeast Res. 2014, 14, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, R.; Alberti, S.; Lindquist, S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010, 20, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Newby, G.A.; Lindquist, S. Blessings in disguise: Biological benefits of prion-like mechanisms. Trends Cell Biol. 2013, 23, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Shorter, J.; Lindquist, S. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 2005, 6, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.T.; Wickner, R.B. Heritable activity: A prion that propagates by covalent autoactivation. Genes Dev. 2003, 17, 2083–2087. [Google Scholar] [CrossRef]
- Rogoza, T.; Goginashvili, A.; Rodionova, S.; Ivanov, M.; Viktorovskaya, O.; Rubel, A.; Volkov, K.; Mironova, L. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc. Natl. Acad. Sci. USA 2010, 107, 10573–10577. [Google Scholar] [CrossRef]
- Jarosz, D.F.; Lancaster, A.K.; Brown, J.C.; Lindquist, S. An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell 2014, 158, 1072–1082. [Google Scholar] [CrossRef][Green Version]
- Chakravarty, A.K.; Smejkal, T.; Itakura, A.K.; Garcia, D.M.; Jarosz, D.F. A Non-amyloid Prion Particle that Activates a Heritable Gene Expression Program. Mol. Cell 2020, 77, 251–265. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.; Ness, F.; Tuite, M. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics 2003, 165, 23–33. [Google Scholar] [PubMed]
- Kabani, M. Hiding in plain sight: Vesicle-mediated export and transmission of prion-like proteins. Microb. Cell 2020, 7, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Melki, R.; Kabani, M. Growth phase-dependent changes in the size and infectivity of SDS-resistant Sup35p assemblies associated with the [PSI+] prion in yeast. Mol. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kabani, M.; Melki, R. Sup35p in its soluble and prion states is packaged inside extracellular vesicles. MBio 2015, 6. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Casadevall, A. A two-way road: Novel roles for fungal extracellular vesicles. Mol. Microbiol. 2018, 110, 11–15. [Google Scholar] [CrossRef]
- Kabani, M.; Pilard, M.; Melki, R. Glucose availability dictates the export of the soluble and prion forms of Sup35p via periplasmic or extracellular vesicles. Mol. Microbiol. 2020. [Google Scholar] [CrossRef]
- Huang, P.H.; Chiang, H.L. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J. Cell Biol. 1997, 136, 803–810. [Google Scholar] [CrossRef]
- Giardina, B.J.; Stein, K.; Chiang, H.-L. The endocytosis gene END3 is essential for the glucose-induced rapid decline of small vesicles in the extracellular fraction in Saccharomyces cerevisiae. J. Extracell. Vesicles 2014, 3, 1–11. [Google Scholar] [CrossRef]
- Giardina, B.J.; Stanley, B.A.; Chiang, H.L. Glucose induces rapid changes in the secretome of Saccharomyces cerevisiae. Proteome Sci. 2014, 12, 9. [Google Scholar] [CrossRef]
- Winters, C.M.; Hong-Brown, L.Q.; Chiang, H.L. Intracellular vesicle clusters are organelles that synthesize extracellular vesicle-associated cargo proteins in yeast. J. Biol. Chem. 2020, 295, 2650–2663. [Google Scholar] [CrossRef] [PubMed]
- Derdowski, A.; Sindi, S.S.; Klaips, C.L.; DiSalvo, S.; Serio, T.R. A Size Threshold Limits Prion Transmission and Establishes Phenotypic Diversity. Science 2010, 330, 680–683. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Melki, R.; Kabani, M. A prolonged chronological lifespan is an unexpected benefit of the [PSI+] prion in yeast. PLoS ONE 2017, 12, e0184905. [Google Scholar] [CrossRef] [PubMed]
- Byrne, L.J.; Cole, D.J.; Cox, B.S.; Ridout, M.S.; Morgan, B.J.T.; Tuite, M.F. The Number and Transmission of [PSI+] Prion Seeds (Propagons) in the Yeast Saccharomyces cerevisiae. PLoS ONE 2009, 4, e4670. [Google Scholar] [CrossRef] [PubMed]
- Speldewinde, S.H.; Grant, C.M. The frequency of yeast [PSI+] prion formation is increased during chronological ageing. Microb. Cell 2017, 4, 127–132. [Google Scholar] [CrossRef]
- Bagriantsev, S.N.; Gracheva, E.O.; Richmond, J.E.; Liebman, S.W. Variant-specific [PSI+] Infection Is Transmitted by Sup35 Polymers within [PSI+] Aggregates with Heterogeneous Protein Composition. Mol. Biol. Cell 2008, 19, 2433–2443. [Google Scholar] [CrossRef]
- Villali, J.; Dark, J.; Brechtel, T.M.; Pei, F.; Sindi, S.S.; Serio, T.R. Nucleation seed size determines amyloid clearance and establishes a barrier to prion appearance in yeast. Nat. Struct. Mol. Biol. 2020, 27, 540–549. [Google Scholar] [CrossRef]
- Tanaka, M.; Collins, S.R.; Toyama, B.H.; Weissman, J.S. The physical basis of how prion conformations determine strain phenotypes. Nature 2006, 442, 585–589. [Google Scholar] [CrossRef]
- Juanes, M.A.; Piatti, S. The final cut: Cell polarity meets cytokinesis at the bud neck in S. cerevisiae. Cell. Mol. Life Sci. 2016, 73, 3115–3136. [Google Scholar] [CrossRef]
- Bhavsar-Jog, Y.P.; Bi, E. Mechanics and regulation of cytokinesis in budding yeast. Semin. Cell Dev. Biol. 2017, 66, 107–118. [Google Scholar] [CrossRef]
- Sentandreu, R.; Northcote, D.H. The formation of buds in yeast. J. Gen. Microbiol. 1969, 55, 393–398. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Knoblach, B.; Rachubinski, R.A. Sharing the cell’s bounty—Organelle inheritance in yeast. J. Cell Sci. 2015, 128, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Higuchi-Sanabria, R.; Pernice, W.M.A.; Vevea, J.D.; Alessi Wolken, D.M.; Boldogh, I.R.; Pon, L.A. Role of asymmetric cell division in lifespan control in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 1133–1146. [Google Scholar] [CrossRef] [PubMed]
- Sontag, E.M.; Samant, R.S.; Frydman, J. Mechanisms and Functions of Spatial Protein Quality Control. Annu. Rev. Biochem. 2017, 86, 97–122. [Google Scholar] [CrossRef]
- Hill, S.M.; Hanzén, S.; Nyström, T. Restricted access: Spatial sequestration of damaged proteins during stress and aging. EMBO Rep. 2017, 18, 377–391. [Google Scholar] [CrossRef]
- Chernova, T.A.; Wilkinson, K.D.; Chernoff, Y.O. Prions, chaperones, and proteostasis in yeast. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef]
- Kalastavadi, T.; True, H.L. Analysis of the [RNQ+] prion reveals stability of amyloid fibers as the key determinant of yeast prion variant propagation. J. Biol. Chem. 2010, 285, 20748–20755. [Google Scholar] [CrossRef]
- Huang, V.J.; Stein, K.C.; True, H.L. Spontaneous Variants of the [RNQ+] Prion in Yeast Demonstrate the Extensive Conformational Diversity Possible with Prion Proteins. PLoS ONE 2013, 8, e79582. [Google Scholar] [CrossRef][Green Version]
- McGlinchey, R.P.; Kryndushkin, D.; Wickner, R.B. Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA 2011, 108, 5337–5341. [Google Scholar] [CrossRef]
- Schlumpberger, M.; Prusiner, S.B.; Herskowitz, I. Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol. 2001, 21, 7035–7046. [Google Scholar] [CrossRef]
- Krzewska, J.; Tanaka, M.; Burston, S.G.; Melki, R. Biochemical and functional analysis of the assembly of full-length Sup35p and its prion-forming domain. J. Biol. Chem. 2007, 282, 1679–1686. [Google Scholar] [CrossRef] [PubMed]
- Schutz, A.K.; Habenstein, B.; Luckgei, N.; Bousset, L.; Sourigues, Y.; Nielsen, A.B.; Melki, R.; Bockmann, A.; Meier, B.H. Solid-state NMR sequential assignments of the amyloid core of full-length Sup35p. Biomol. NMR Assign. 2014, 8, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Baxa, U.; Keller, P.W.; Cheng, N.; Wall, J.S.; Steven, A.C. In Sup35p filaments (the [PSI+] prion), the globular C-terminal domains are widely offset from the amyloid fibril backbone. Mol. Microbiol. 2011, 79, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Saibil, H.R.; Seybert, A.; Habermann, A.; Winkler, J.; Eltsov, M.; Perkovic, M.; Castano-Diez, D.; Scheffer, M.P.; Haselmann, U.; Chlanda, P.; et al. Heritable yeast prions have a highly organized three-dimensional architecture with interfiber structures. Proc. Natl. Acad. Sci. USA 2012, 109, 14906–14911. [Google Scholar] [CrossRef]
- Arslan, F.; Hong, J.Y.; Kanneganti, V.; Park, S.-K.; Liebman, S.W. Heterologous Aggregates Promote De Novo Prion Appearance via More than One Mechanism. PLoS Genet. 2015, 11, e1004814. [Google Scholar] [CrossRef]
- Mathur, V.; Taneja, V.; Sun, Y.; Liebman, S.W. Analyzing the Birth and Propagation of Two Distinct Prions, [PSI+] and [Het-s]y, in Yeast. Mol. Biol. Cell 2010, 21, 1449–1461. [Google Scholar] [CrossRef][Green Version]
- Sharma, J.; Wisniewski, B.T.; Paulson, E.; Obaoye, J.O.; Merrill, S.J.; Manogaran, A.L. De novo [PSI (+)] prion formation involves multiple pathways to form infectious oligomers. Sci. Rep. 2017, 7, 76. [Google Scholar] [CrossRef]
- Song, Y.; Wu, Y.X.; Jung, G.; Tutar, Y.; Eisenberg, E.; Greene, L.E.; Masison, D.C. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 2005, 4, 289–297. [Google Scholar] [CrossRef]
- Satpute-Krishnan, P.; Serio, T.R. Prion protein remodelling confers an immediate phenotypic switch. Nature 2005, 437, 262–265. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Zhou, P.; Chernoff, Y.O.; Liebman, S.W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997, 147, 507–519. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Uptain, S.M.; Outeiro, T.F.; Krishnan, R.; Lindquist, S.L.; Liebman, S.W. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 12934–12939. [Google Scholar] [CrossRef] [PubMed]
- Bui, Q.; Sherma, J.; Hines, J.K. Using high performance thin layer chromatography-densitometry to study the influence of the prion [RNQ+] and its determinant prion protein Rnq1 on yeast lipid profiles. Separations 2018, 5, 6. [Google Scholar] [CrossRef]
- Serio, T.R. [PIN + ]ing down the mechanism of prion appearance. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Chien, P.; Naber, N.; Cooke, R.; Weissman, J.S. Conformational variations in an infectious protein determine prion strain differences. Nature 2004, 428, 323–328. [Google Scholar] [CrossRef]
- King, C.Y.; Diaz-Avalos, R. Protein-only transmission of three yeast prion strains. Nature 2004, 428, 319–323. [Google Scholar] [CrossRef]
- Kabani, M.; Cosnier, B.; Bousset, L.; Rousset, J.P.; Melki, R.; Fabret, C. A mutation within the C-terminal domain of Sup35p that affects [PSI+] prion propagation. Mol. Microbiol. 2011, 81, 640–658. [Google Scholar] [CrossRef]
- Kabani, M.; Redeker, V.; Melki, R. A role for the proteasome in the turnover of sup35p and in [PSI+] prion propagation. Mol. Microbiol. 2014, 92, 507–528. [Google Scholar] [CrossRef]
- Bousset, L.; Luckgei, N.; Kabani, M.; Gardiennet, C.; Schütz, A.K.; Melki, R.; Meier, B.H.; Böckmann, A. Prion Amyloid Polymorphs—The Tag Might Change It All. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef]
- Tanaka, M.; Weissman, J.S. An Efficient Protein Transformation Protocol for Introducing Prions into Yeast. Methods Enzymol. 2006, 412, 185–200. [Google Scholar] [CrossRef]
- Sharma, D.; Martineau, C.N.; Le Dall, M.T.; Reidy, M.; Masison, D.C.; Kabani, M. Function of SSA subfamily of Hsp70 within and across species varies widely in complementing Saccharomyces cerevisiae cell growth and prion propagation. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]
- Pruyne, D.; Legesse-Miller, A.; Gao, L.; Dong, Y.; Bretscher, A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004, 20, 559–591. [Google Scholar] [CrossRef] [PubMed]
- Ganusova, E.E.; Ozolins, L.N.; Bhagat, S.; Newnam, G.P.; Wegrzyn, R.D.; Sherman, M.Y.; Chernoff, Y.O. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 2006, 26, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Manogaran, A.L.; Hong, J.Y.; Hufana, J.; Tyedmers, J.; Lindquist, S.; Liebman, S.W. Prion formation and polyglutamine aggregation are controlled by two classes of genes. PLoS Genet. 2011, 7, e1001386. [Google Scholar] [CrossRef] [PubMed]
- Speldewinde, S.H.; Doronina, V.A.; Tuite, M.F.; Grant, C.M. Disrupting the cortical actin cytoskeleton points to two distinct mechanisms of yeast [PSI+] prion formation. PLoS Genet. 2017, 13. [Google Scholar] [CrossRef]
- Dorweiler, J.E.; Oddo, M.J.; Lyke, D.R.; Reilly, J.A.; Wisniewski, B.T.; Davis, E.E.; Kuborn, A.M.; Merrill, S.J.; Manogaran, A.L. The actin cytoskeletal network plays a role in yeast prion transmission and contributes to prion stability. Mol. Microbiol. 2020. [Google Scholar] [CrossRef]
- Kumar, R.; Nawroth, P.P.; Tyedmers, J. Prion Aggregates Are Recruited to the Insoluble Protein Deposit (IPOD) via Myosin 2-Based Vesicular Transport. PLoS Genet. 2016, 12, e1006324. [Google Scholar] [CrossRef]
- Ferreira, P.C.; Ness, F.; Edwards, S.R.; Cox, B.S.; Tuite, M.F. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 2001, 40, 1357–1369. [Google Scholar] [CrossRef]
- Allen, K.D.; Wegrzyn, R.D.; Chernova, T.A.; Muller, S.; Newnam, G.P.; Winslett, P.A.; Wittich, K.B.; Wilkinson, K.D.; Chernoff, Y.O. Hsp70 chaperones as modulators of prion life cycle: Novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 2005, 169, 1227–1242. [Google Scholar] [CrossRef]
- Tipton, K.A.; Verges, K.J.; Weissman, J.S. In Vivo Monitoring of the Prion Replication Cycle Reveals a Critical Role for Sis1 in Delivering Substrates to Hsp104. Mol. Cell 2008, 32, 584–591. [Google Scholar] [CrossRef]
- Sondheimer, N.; Lopez, N.; Craig, E.A.; Lindquist, S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001, 20, 2435–2442. [Google Scholar] [CrossRef]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef]
- Zhou, C.; Slaughter, B.D.; Unruh, J.R.; Eldakak, A.; Rubinstein, B.; Li, R. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell 2011, 147, 1186–1196. [Google Scholar] [CrossRef]
- Tessarz, P.; Schwarz, M.; Mogk, A.; Bukau, B. The yeast AAA+ chaperone Hsp104 is part of a network that links the actin cytoskeleton with the inheritance of damaged proteins. Mol. Cell. Biol. 2009, 29, 3738–3745. [Google Scholar] [CrossRef] [PubMed]
- Erjavec, N.; Larsson, L.; Grantham, J.; Nyström, T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007, 21, 2410–2421. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Larsson, L.; Caballero, A.; Hao, X.; Öling, D.; Grantham, J.; Nyström, T. The Polarisome Is Required for Segregation and Retrograde Transport of Protein Aggregates. Cell 2010, 140, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Larsson, L.; Franssens, V.; Hao, X.; Hill, S.M.; Andersson, V.; Hoglund, D.; Song, J.; Yang, X.; Oling, D.; et al. Segregation of protein aggregates involves actin and the polarity machinery. Cell 2011, 147, 959–961. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hill, S.M.; Hao, X.; Grönvall, J.; Spikings-Nordby, S.; Widlund, P.O.; Amen, T.; Jörhov, A.; Josefson, R.; Kaganovich, D.; Liu, B.; et al. Asymmetric Inheritance of Aggregated Proteins and Age Reset in Yeast Are Regulated by Vac17-Dependent Vacuolar Functions. Cell Rep. 2016, 16, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Byrne, L.J.; Cox, B.S.; Cole, D.J.; Ridout, M.S.; Morgan, B.J.T.; Tuite, M.F. Cell division is essential for elimination of the yeast [PSI+] prion by guanidine hydrochloride. Proc. Natl. Acad. Sci. USA 2007, 104, 11688–11693. [Google Scholar] [CrossRef]
- Eaglestone, S.S.; Ruddock, L.W.; Cox, B.S.; Tuite, M.F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 240–244. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Alexandrov, I.M.; Ter-Avanesyan, M.D.; Kushnirov, V. V Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003, 278, 49636–49643. [Google Scholar] [CrossRef]
- Fevrier, B.; Vilette, D.; Archer, F.; Loew, D.; Faigle, W.; Vidal, M.; Laude, H.; Raposo, G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA 2004, 101, 9683–9688. [Google Scholar] [CrossRef] [PubMed]
- Dujardin, S.; Bégard, S.; Caillierez, R.; Lachaud, C.; Delattre, L.; Carrier, S.; Loyens, A.; Galas, M.C.; Bousset, L.; Melki, R.; et al. Ectosomes: A new mechanism for non-exosomal secretion of Tau protein. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Rajendran, L.; Bali, J.; Barr, M.M.; Court, F.A.; Kramer-Albers, E.-M.; Picou, F.; Raposo, G.; van der Vos, K.E.; van Niel, G.; Wang, J.; et al. Emerging Roles of Extracellular Vesicles in the Nervous System. J. Neurosci. 2014, 34, 15482–15489. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Schapira, A.H.; Gardiner, C.; Sargent, I.L.; Wood, M.J.A.; Cooper, J.M. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 2011, 42, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Minakaki, G.; Menges, S.; Kittel, A.; Emmanouilidou, E.; Schaeffner, I.; Barkovits, K.; Bergmann, A.; Rockenstein, E.; Adame, A.; Marxreiter, F.; et al. Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy 2018, 14, 98–119. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Macreadie, I. Protein homeostasis networks and the use of yeast to guide interventions in alzheimer’s disease. Int. J. Mol. Sci. 2020, 21, 8014. [Google Scholar] [CrossRef]
- Ishikawa, T. Saccharomyces cerevisiae in neuroscience: How unicellular organism helps to better understand prion protein? Neural Regen. Res. 2020, 16, 489–495. [Google Scholar] [CrossRef]
- Tuite, M.F. Yeast models of neurodegenerative diseases. In Progress in Molecular Biology and Translational Science; Elsevier B.V.: Amsterdam, The Netherlands, 2019; Volume 168, pp. 351–379. ISBN 978012817874. [Google Scholar]

| Prion | Protein | Protein Function | Prion Phenotypes | References |
|---|---|---|---|---|
| [PSI+] | Sup35p | translation terminator factor (eRF3) | reduced function results in increased nonsense suppression; impaired growth; growth advantage under stress conditions; increased chronological lifespan § | [3,22,23] |
| [URE3] | Ure2p | nitrogen regulation (poor nitrogen sources repression) | loss of function; slow growth | [1,5] |
| [PIN+] | Rnq1p | unknown | induction of [PSI+] or [URE3] formation | [24] |
| [OCT+] | Cyc8p | transcription repression | impaired mating and sporulation | [25] |
| [SWI+] | Swi1p | chromatin remodeling; transcription regulation | poor growth on non-fermentable carbon sources | [26] |
| [MOT3+] | Mot3p | transcription regulation | acquisition of multicellular growth forms (biofilms) | [27] |
| [MOD+] | Mod5p | tRNA isopentenyltransferase | fluconazole resistance; slow growth | [28] |
| [LSB+] | Lsb2p | actin nucleation inhibition | thermal stress-induced [PIN+] activity | [29] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kabani, M. Extracellular Vesicles-Encapsulated Yeast Prions and What They Can Tell Us about the Physical Nature of Propagons. Int. J. Mol. Sci. 2021, 22, 90. https://doi.org/10.3390/ijms22010090
Kabani M. Extracellular Vesicles-Encapsulated Yeast Prions and What They Can Tell Us about the Physical Nature of Propagons. International Journal of Molecular Sciences. 2021; 22(1):90. https://doi.org/10.3390/ijms22010090
Chicago/Turabian StyleKabani, Mehdi. 2021. "Extracellular Vesicles-Encapsulated Yeast Prions and What They Can Tell Us about the Physical Nature of Propagons" International Journal of Molecular Sciences 22, no. 1: 90. https://doi.org/10.3390/ijms22010090
APA StyleKabani, M. (2021). Extracellular Vesicles-Encapsulated Yeast Prions and What They Can Tell Us about the Physical Nature of Propagons. International Journal of Molecular Sciences, 22(1), 90. https://doi.org/10.3390/ijms22010090





