ABA Metabolism and Homeostasis in Seed Dormancy and Germination
Abstract
:1. Introduction
2. An Overview of the ABA Metabolism Pathway
2.1. Carotenoid Interconversion and Cleavage in Plastids
2.2. Cytosolic Steps from Xanthoxin to ABA
2.3. ABA Conjugation and Hydroxylation
3. ABA Metabolism and Seed Dormancy Induction
3.1. Control of ABA Levels by Carotenoid Cleavage and ABA Hydroxylation
3.2. Influence of Maternal Environmental Conditions on Dormancy and Germination
3.3. Regulatory Factors of ABA Metabolism in Developing Seeds
4. ABA Metabolism during Dormancy Release and Germination
4.1. Importance of ABA Hydroxylation in the Control of Germination
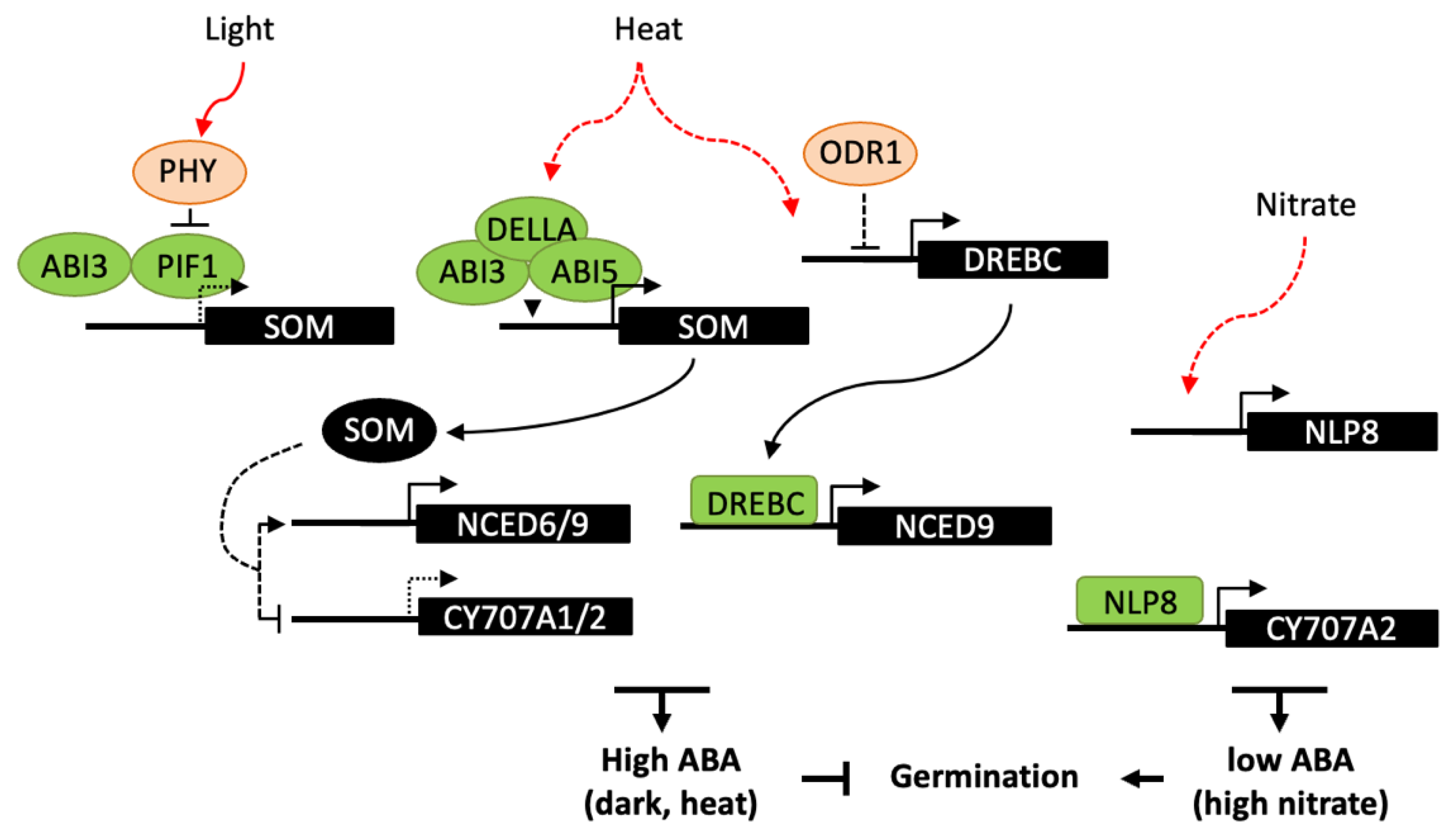
4.2. Influence of Light, Temperature and Nitrate on Germination
4.3. Regulatory Factors of ABA Metabolism in Imbibed Seeds
5. Natural Variations of Genes Affecting ABA Metabolism in Seeds
5.1. Relationship between ABA Levels and Germination in Natural Lines
5.2. Natural Variations of ABA Metabolism Genes Associated with Germination-Related Phenotypes
| Gene | Species | Types of Genetic Variation | Involvement in ABA Metabolism and Phenotype | Ref. |
|---|---|---|---|---|
| ZEP | Arabidopsis | Intergenic SNPs/QTL region | Affecting mRNA levels of ZEP and ratio of violaxanthin to antheraxanthin | [156] |
| ZEP1 | Maize | SNPs in coding region | Associated with carotenoid composition | [157] |
| Intergenic SNPs | Associated with kernel color | [158] | ||
| QTL regions | Colocating with QTLs for kernel desiccation and ABA content | [160] | ||
| ZEP | Sorghum | SNPs inside the gene | Associated with zeaxanthin levels | [159] |
| NCED4 | Lettuce | SNPs in promoter | Affecting mRNA levels of NCED4, ABA levels and thermoinhibition | [126] |
| NCED3 | Rice | Low similarity of regulatory region SNPs and Indels in coding region | Higher expression at mRNA level in variety seeds with higher ABA level and stronger dormancy | [163] |
| NCED2 | Nonsynonymous SNPs | Affecting ABA levels in leaves and tolerance to drought but not clearly associated with pericarp color | [166] [167] | |
| NCED3 | Arabidopsis | QTL region/Nonsynonymous substitutions | Affecting ABA accumulation in seedling under low water potential stress | [164] |
| QTL region | Colocating with QTLs for germination speed and ability on media containing salt or ABA | [165] | ||
| ABA2 AO3 | Wheat | SNPs in coding region | Associated with yellow pigment content and yellow index of grain | [168] |
| AO3 | No clear polymorphisms in promoter | Highly expressed at the mRNA level in variety seeds with lower carotenoid content | [169] | |
| ABA3 | Soybean | Intergenic SNPs | Associated with seed germination under salt stress | [170] |
| CYP707A5 | Rice | SNPs and Indels in regulatory and coding region | Lower expression at mRNA level in variety seeds with higher ABA level and stronger dormancy | [163] |
| ABA8′OH-2 | Wheat | QTL region/Nonsynonymous substitutions | Colocating with QTL for seed dormancy | [171] |
5.3. Perspectives for Understanding Natural Variations That Underlie ABA Metabolism in Seeds
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Penfield, S.; MacGregor, D.R. Effects of environmental variation during seed production on seed dormancy and germination. J. Exp. Bot. 2017, 68, 819–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burghardt, L.T.; Edwards, B.R.; Donohue, K. Multiple paths to similar germination behavior in Arabidopsis thaliana. New Phytol. 2016, 209, 1301–1312. [Google Scholar] [CrossRef] [Green Version]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Yan, D.; Duermeyer, L.; Leoveanu, C.; Nambara, E. The functions of endosperm during seed germination. Plant Cell Physiol. 2014, 55, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Carrera-Castano, G.; Calleja-Cabrera, J.; Pernas, M.; Gomez, L.; Onate-Sanchez, L. An updated overview on the regulation of seed germination. Plants 2020, 9, 703. [Google Scholar] [CrossRef] [PubMed]
- Chahtane, H.; Kim, W.; Lopez-Molina, L. Primary seed dormancy: A temporally multilayered riddle waiting to be unlocked. J. Exp. Bot. 2017, 68, 857–869. [Google Scholar] [CrossRef]
- Tuan, P.A.; Kumar, R.; Rehal, P.K.; Toora, P.K.; Ayele, B.T. Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 2018, 9, 668. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Hou, X. Regulation of ABA and GA in metabolism and signaling. Front. Plant Sci. 2018, 9, 251. [Google Scholar] [CrossRef] [Green Version]
- Cadman, C.S.C.; Toorop, P.E.; Hilhorst, H.W.M.; Finch-Savage, W.E. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006, 46, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Douterelo-Soler, I.; Clay, H.; Finch-Savage, W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 20236–20241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [Green Version]
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, I.J.R.; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 2132. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; Rodríguez-Gacio, M.C.; Matilla, A.J. Delay of germination-1 (DOG1): A key to understanding seed dormancy. Plants 2020, 9, 480. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Okamoto, M.; Tatematsu, K.; Yano, R.; Seo, M.; Kamiya, Y. Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 2010, 20, 55–67. [Google Scholar] [CrossRef]
- Matilla, A.J.; Carrillo-Barral, N.; Rodriguez-Gacio, M.d.C. An update on the role of NCED and CYP707A ABA metabolism genes in seed dormancy induction and the response to after-ripening and nitrate. J. Plant Growth Regul. 2015, 34, 274–293. [Google Scholar] [CrossRef]
- Seo, M.; Marion-Poll, A. Abscisic acid metabolism and transport. In Abscisic Acid in Plants; Seo, M., Marion-Poll, A., Eds.; Advances in Botanical Research; Academic Press: London, UK, 2019; Volume 92, pp. 1–49. [Google Scholar]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zha, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A.D. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003, 131, 1591–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koornneef, M.; Jorna, M.L.; Brinkhorst-van der Swan, D.L.C.; Karssen, C.M. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 1982, 61, 385–393. [Google Scholar] [CrossRef]
- Rock, C.D.; Zeevaart, J.A.D. The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 1991, 88, 7496–7499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marin, E.; Nussaume, L.; Quesada, A.; Gonneau, M.; Sotta, B.; Hugueney, P.; Frey, A.; Marion-Poll, A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996, 15, 2331–2342. [Google Scholar] [CrossRef]
- Buch, K.; Stransky, H.; Hager, A. FAD is a further essential cofactor of the NAD(P)H and O2-dependent zeaxanthin-epoxidase. FEBS Lett. 1995, 376, 45–48. [Google Scholar] [CrossRef] [Green Version]
- Bouvier, F.; d’Harlingue, A.; Hugueney, P.; Marin, E.; Marion-Poll, A.; Camara, B. Xanthophyll biosynthesis. Cloning, expression, functional reconstitution, and regulation of b-cyclohexenyl carotenoid epoxidase from pepper (Capsicum annuum). J. Biol. Chem. 1996, 271, 28861–28867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bugos, R.C.; Hieber, A.D.; Yamamoto, H.Y. Xanthophyll cycle enzymes are members of the lipocalin family, the first identified from plants. J. Biol. Chem. 1998, 273, 15321–15324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- North, H.M.; Frey, A.; Boutin, J.P.; Sotta, B.; Marion-Poll, A. Analysis of xanthophyll cycle gene expression during the adaptation of Arabidopsis to excess light and drought stress: Changes in RNA steady state levels do not contribute to short-term responses. Plant Sci. 2005, 169, 115–124. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; McCarty, D.R.; Welch, W.; Zeevaart, J.A.D. Substrate specificity and kinetics for VP14, a carotenoid cleavage dioxy- genase in the ABA biosynthetic pathway. Biochim. Biophys. Acta 2003, 1619, 9–14. [Google Scholar] [CrossRef]
- North, H.; De Almeida, A.; Boutin, J.P.; Frey, A.; To, A.; Botran, L.; Sotta, B.; Marion-Poll, A. The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 2007, 50, 810–824. [Google Scholar] [CrossRef]
- Neuman, H.; Galpaz, N.; Cunningham, F.X.; Zamir, D.; Hirschberg, J. The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis. Plant J. 2014, 78, 80–93. [Google Scholar] [CrossRef]
- Perreau, F.; Frey, A.; Effroy-Cuzzi, D.; Savane, P.; Berger, A.; Gissot, L.; Marion-Poll, A. ABA4 has an essential function in both cis-violaxanthin and cis-neoxanthin synthesis. Plant Physiol. 2020, 184, 1303–1316. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A.D. Characterization of a novel carotenoid cleavage dioxygenase from plants. J. Biol. Chem. 2001, 276, 25208–25211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D.; Limon, M.C.; Meléndez-Martínez, A.J.; Olmedilla-Alonso, B.; Palou, A.; et al. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Progr. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef] [Green Version]
- Tan, B.C.; Cline, K.; McCarty, D.R. Localization and targeting of the VP14 epoxy-carotenoid dioxygenase to chloroplast membranes. Plant J. 2001, 27, 373–382. [Google Scholar] [CrossRef] [Green Version]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef] [Green Version]
- Messing, S.A.J.; Gabelli, S.B.; Echeverria, I.; Vogel, J.T.; Guan, J.C.; Tan, B.C.; Klee, H.J.; McCarty, D.R.; Amzel, L.M. Structural insights into maize Viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 2010, 22, 2970–2980. [Google Scholar] [CrossRef] [Green Version]
- Bouchnak, I.; Brugière, S.; Moyet, L.; Le Gall, S.; Salvi, D.; Kuntz, M.; Tardif, M.; Rolland, N. Unraveling hidden components of the chloroplast envelope proteome: Opportunities and limits of better MS sensitivity. Mol. Cell Proteom. 2019, 18, 1285–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leon-Kloosterziel, K.M.; Alvarez Gil, M.; Ruijs, G.J.; Jacobsen, S.E.; Olszewski, N.E.; Schwartz, S.H.; Zeevaart, J.A.D.; Koornneef, M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996, 10, 655–661. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Leon-Kloosterziel, K.M.; Koornneef, M.; Zeevaart, J.A.D. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997, 114, 161–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Guzman, M.; Apostolova, N.; Belles, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodriguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, W.H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. Unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef]
- Rook, F.; Corke, F.; Card, R.; Munz, G.; Smith, C.; Bevan, M.W. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001, 26, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Merlot, S.; Mustilli, A.C.; Genty, B.; North, H.; Lefebvre, V.; Sotta, B.; Vavasseur, A.; Giraudat, J. Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J. 2002, 30, 601–609. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.; Peeters, A.J.; Koiwai, H.; Oritani, T.; Marion-Poll, A.; Zeevaart, J.A.D.; Koornneef, M.; Kamiya, Y.; Koshiba, T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 2000, 97, 12908–12913. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.; Aoki, H.; Koiwai, H.; Kamiya, Y.; Nambara, E.; Koshiba, T. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 2004, 45, 1694–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Guzman, M.; Abia, D.; Salinas, J.; Serrano, R.; Rodriguez, P.L. Two new alleles of the abscisic aldehyde oxidase 3 gene reveal its role in abscisic acid biosynthesis in seeds. Plant Physiol. 2004, 135, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Bittner, F.; Oreb, M.; Mendel, R.R. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 40381–40384. [Google Scholar] [CrossRef] [Green Version]
- Xiong, L.M.; Ishitani, M.; Lee, H.; Zhu, J.K. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 2001, 13, 2063–2083. [Google Scholar] [PubMed] [Green Version]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′–hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [Green Version]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As encode (+)-abscisic acid 8’-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Cutler, A.J.; Ambrose, S.J.; Galka, M.M.; Nelson, K.M.; Squires, T.M.; Loewen, M.K.; Jadhav, A.S.; Ross, A.R.S.; Taylor, D.C.; et al. A new abscisic acid catabolic pathway. Plant Physiol. 2004, 134, 361–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamoto, M.; Kushiro, T.; Jikumaru, Y.; Abrams, S.R.; Kamiya, Y.; Seki, M.; Nambara, E. ABA 9′-hydroxylation is catalysed by CYP707A in Arabidopsis. Phytochemistry 2011, 72, 717–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, J.K.; Ye, M.; Li, B.; Noel, J.P. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 2016, 166, 881–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, E.K.; Doucet, C.J.; Hou, B.; Jackson, R.G.; Abrams, S.R.; Bowles, D.J. Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron Asymmetry 2005, 16, 143–147. [Google Scholar] [CrossRef]
- Priest, D.M.; Ambrose, S.J.; Vaistij, F.E.; Elias, L.; Higgins, G.S.; Ross, A.R.; Abrams, S.R.; Bowles, D.J. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006, 46, 492–502. [Google Scholar] [CrossRef]
- Dong, T.; Xu, Z.Y.; Park, Y.; Kim, D.H.; Lee, Y.; Hwang, I. Abscisic acid uridine diphosphate glucosyltransferases play a crucial role in abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2014, 165, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Yan, J.P.; Li, D.K.; Luo, Q.; Yan, Q.; Liu, Z.B.; Ye, L.M.; Wang, J.M.; Li, X.F.; Yang, Y. UDP-Glucosyltransferase71C5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.T.; Liu, F.F.; Xiao, D.W.; Jiang, X.Y.; Li, P.; Zhao, S.M.; Hou, B.K.; Li, Y.J. The Arabidopsis UDP-glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol. 2020, 102, 389–401. [Google Scholar] [CrossRef]
- Lee, K.H.; Piao, H.L.; Kim, H.Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.J.; Hwang, I. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A.; et al. A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 2012, 24, 2184–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karssen, C.M.; Brinkhorstvanderswan, D.L.C.; Breekland, A.E.; Koornneef, M. Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 1983, 157, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.; Godin, B.; Bonnet, M.; Sotta, B.; Marion-Poll, A. Maternal synthesis of abscisic acid controls seed development and yield in Nicotiana plumbaginifolia. Planta 2004, 218, 958–964. [Google Scholar] [CrossRef]
- Kanno, Y.; Jikumaru, Y.; Hanada, A.; Nambara, E.; Abrams, S.R.; Kamiya, Y.; Seo, M. Comprehensive hormone profiling in developing Arabidopsis seeds: Examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010, 51, 1988–2001. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.; Kanno, Y.; Frey, A.; North, H.M.; Marion-Poll, A. Dissection of Arabidopsis NCED9 promoter regulatory regions reveals a role for ABA synthesized in embryos in the regulation of GA-dependent seed germination. Plant Sci. 2016, 246, 91–97. [Google Scholar] [CrossRef]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef]
- Chauffour, F.; Bailly, M.; Perreau, F.; Cueff, G.; Suzuki, H.; Collet, B.; Frey, A.; Clément, G.; Soubigou-Taconnat, L.; Balliau, T.; et al. Multi-omics analysis reveals sequential roles for ABA during Arabidopsis seed maturation. Plant Physiol. 2019, 180, 1198–2018. [Google Scholar] [CrossRef] [PubMed]
- Chono, M.; Honda, I.; Shinoda, S.; Kushiro, T.; Kamiya, Y.; Nambara, E.; Kawakami, N.; Kaneko, S.; Watanabe, Y. Field studies on the regulation of abscisic acid content and germinability during grain development of barley: Molecular and chemical analysis of pre-harvest sprouting. J. Exp. Bot. 2006, 57, 2421–2434. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.D.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–122240. [Google Scholar] [CrossRef] [Green Version]
- Chono, M.; Matsunaka, H.; Seki, M.; Fujita, M.; Kiribuchi-Otobe, C.; Oda, S.; Kojima, H.; Kobayashi, D.; Kawakami, N. Isolation of a wheat (Triticum aestivum L.) mutant in ABA 8′-hydroxylase gene: Effect of reduced ABA catabolism on germination inhibition under field condition. Breed. Sci. 2013, 63, 104–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kendall, S.L.; Hellwege, A.; Marriot, P.; Whalley, C.; Graham, I.A.; Penfield, S. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 2011, 23, 2568–2580. [Google Scholar] [CrossRef] [Green Version]
- Tuan, P.A.; Nguyen, T.N.; Jordan, M.C.; Ayele, B.T. A shift in abscisic acid/gibberellin balance underlies retention of dormancy induced by seed development temperature. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Brunel-Muguet, S.; D’Hooghe, P.; Bataillé, M.-P.; Larré, C.; Kim, T.-H.; Trouverie, J.; Avice, J.-C.; Etienne, P.; Dürr, C. Heat stress during seed filling interferes with sulfur restriction on grain composition and seed germination in oilseed rape (Brassica napus L.). Front. Plant Sci. 2015, 6, 213. [Google Scholar] [CrossRef] [Green Version]
- Suriyasak, C.; Oyama, Y.; Ishida, T.; Mashiguchi, K.; Yamaguchi, S.; Hamaoka, N.; Iwaya-Inoue, M.; Ishibashi, Y. Mechanism of delayed seed germination caused by high temperature during grain filling in rice (Oryza sativa L.). Sci. Rep. 2020, 10, 17378. [Google Scholar] [CrossRef]
- Alboresi, A.; Gestin, C.; Leydecker, M.T.; Bedu, M.; Meyer, C.; Truong, H.N. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 2005, 28, 500–512. [Google Scholar] [CrossRef]
- He, H.; de Souza Vidigal, D.; Snoek, L.B.; Schnabel, S.; Nijveen, H.; Hilhorst, H.; Bentsink, L. Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J. Exp. Bot. 2014, 65, 6603–6615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duermeyer, L.; Khodapanahi, E.; Yan, D.; Krapp, A.; Rothstein, S.J.; Nambara, E. Regulation of seed dormancy and germination by nitrate. Seed Sci. Res. 2018, 28, 150–157. [Google Scholar] [CrossRef]
- Matakiadis, T.; Alboresi, A.; Jikumaru, Y.; Tatematsu, K.; Pichon, O.; Renou, J.P.; Kamiya, Y.; Nambara, E.; Truong, H.N. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2009, 149, 949–960. [Google Scholar] [CrossRef] [Green Version]
- Yan, A.; Chen, Z. The control of seed dormancy and germination by temperature, light and nitrate. Bot. Rev. 2020, 86, 39–75. [Google Scholar] [CrossRef]
- He, H.; Willems, L.; Batushansky, A.; Fait, A.; Hanson, J.; Nijveen, H.; Hilhorst, H.W.M.; Bentsink, L. Effects of parental temperature and nitrate on seed performance are reflected by partly overlapping genetic and metabolic pathways. Plant Cell Physiol. 2016, 57, 473–487. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Zhou, W.; Yin, H.; Luo, X.; Chen, W.; Liu, X.; Wang, X.; Meng, Y.; Feng, L.; Qin, Y.; et al. Shading of the mother plant during seed development promotes subsequent seed germination in soybean. J. Exp. Bot. 2020, 71, 2072–2084. [Google Scholar] [CrossRef]
- Carbonero, P.; Iglesias-Fernandez, R.; Vicente-Carbajosa, J. The AFL subfamily of B3 transcription factors: Evolution and function in angiosperms seeds. J. Exp. Bot. 2017, 68, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Devic, M.; Roscoe, T.; Bouyer, D.; Zhou, D.X.; Boulard, C.; Baud, S.; Dubreucq, B. Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 2018, 31, 291–307. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Tsuchiya, Y.; Lumba, S.; Okamoto, M.; McCourt, P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the action of the hormones gibberellins and abscisic acid. Dev. Cell 2004, 7, 373–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koornneef, M.; Reuling, G.; Karssen, C.M. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 1984, 61, 377–383. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, H.; Ding, L.; Soppe, W.J.J.; Xiang, Y. REVERSAL OF RDO5 1, a homolog of rice Seed dormancy4, interacts with bHLH57 and controls ABA biosynthesis and seed dormancy in Arabidopsis. Plant Cell 2020, 32, 1933–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Y.; Nakabayashi, K.; Ding, J.; He, F.; Bentsink, L.; Soppe, W.J.J. Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 2014, 26, 4362–4375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, K.; Takeuchi, Y.; Ebana, K.; Miyao, A.; Hirochika, H.; Hara, N.; Ishiyama, K.; Kobayashi, M.; Ban, Y.; Hattori, T.; et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 2010, 107, 5792–5797. [Google Scholar] [CrossRef] [Green Version]
- Je, J.; Chen, H.; Song, C.; Lim, C.O. Arabidopsis DREB2C modulates ABA biosynthesis during germination. Biochem. Biophys. Res. Commun. 2014, 452, 91–98. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, X.D.; Zhang, H.; Wang, P.; Li, Y.; Wang, S.; Tang, S.; Liu, C.; et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016, 85, 348–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrasekaran, U.; Luo, X.; Zhou, W.; Shu, K. Multifaceted signaling networks mediated by Abscisic Acid Insensitive 4. Plant Commun. 2020, 1, 100040. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Lee, K.; Seo, P.J. The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 2015, 87, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Seo, P.J. Coordination of seed dormancy and germination processes by MYB96. Plant Signal. Behav. 2015, 10, e1056423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Li, W.; Fang, C.; Xu, F.; Liu, Y.; Wang, Z.; Yang, R.; Zhang, M.; Liu, S.; Lu, S.; et al. Parallel selection on a dormancy gene during domestication of crops from multiple families. Nature Genet. 2018, 50, 1435–1441. [Google Scholar] [CrossRef]
- Chen, H.; Tong, J.; Fu, W.; Liang, Z.; Ruan, J.; Yu, Y.; Song, X.; Yuan, L.; Xiao, L.; Liu, J.; et al. The H3K27me3 demethylase RELATIVE OF EARLY FLOWERING6 suppresses seed dormancy by inducing abscisic acid catabolism. Plant Physiol. 2020, 184, 1969–1978. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, S.; Liu, X.; Wu, K. Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front. Plant Sci. 2015, 6, 159. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Koornneef, M.; Soppe, W.J.J. The absence of histone h2b monoubiquitination in the arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 2007, 19, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Cao, H.; Sun, Y.; Li, X.; Chen, F.; Carles, A.; Li, Y.; Ding, M.; Zhang, C.; Deng, X.; et al. Arabidopsis paired amphipathic helix proteins SLN1 and SLN2 redundantly regulate primary seed dormancy via abscisic acid–ethylene antagonism mediated by histone deacetylation. Plant Cell 2013, 25, 149–166. [Google Scholar] [CrossRef] [Green Version]
- Yao, M.; Chen, W.; Kong, J.; Zhang, X.; Shi, N.; Zhong, S.; Ma, P.; Gallusci, P.; Jackson, S.; Liu, Y.; et al. METHYLTRANSFERASE1 and ripening modulate vivipary during tomato fruit development. Plant Physiol. 2020, 183, 1883–1897. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Rajjou, L.; North, H.M. Lost in Translation: Physiological roles of stored mRNAs in seed germination. Plants 2020, 9, 347. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, J.V.; Pearce, D.W.; Poole, A.T.; Pharis, R.P.; Mander, L.N. Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant. 2002, 115, 428–441. [Google Scholar] [CrossRef]
- Ali-Rachedi, S.; Bouinot, D.; Wagner, M.H.; Bonnet, M.; Sotta, B.; Grappin, P.; Jullien, M. Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: Studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 2004, 219, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Jacobsen, J.V.; Ross, J.J.; Helliwell, C.A.; Poole, A.T.; Scofield, G.; Reid, J.B.; Gubler, F. Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 2006, 45, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Lechat, M.M.; Pouvreau, J.B.; Péron, T.; Gauthier, M.; Montiel, G.; Véronési, C.; Todoroki, Y.; Le Bizec, B.; Monteau, F.; Macherel, D.; et al. PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J. Exp. Bot. 2012, 63, 5311–5322. [Google Scholar] [CrossRef] [PubMed]
- Brun, G.; Thoiron, S.; Braem, L.; Pouvreau, J.B.; Montiel, G.; Lechat, M.-M.; Simier, P.; Gevaert, K.; Goormachtig, S.; Delavault, P. CYP707As are effectors of karrikin and strigolactone signalling pathways in Arabidopsis thaliana and parasitic plants. Plant Cell Environ. 2019, 42, 2612–2626. [Google Scholar] [CrossRef]
- Debeaujon, I.; Koornneef, M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 2000, 122, 415–424. [Google Scholar] [CrossRef] [Green Version]
- Barrero, J.M.; Jacobsen, J.V.; Talbot, M.J.; White, R.G.; Swain, S.M.; Garvin, D.F.; Gubler, F. Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytol. 2012, 193, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Basbouss-Serhal, I.; Pateyron, S.; Cochet, F.; Leymarie, J.; Bailly, C. 5′ to 3′ mRNA decay contributes to the regulation of Arabidopsis seed germination by dormancy. Plant Physiol. 2017, 173, 1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bethke, P.C.; Libourel, I.G.L.; Aoyama, N.; Chung, Y.Y.; Still, D.W.; Jones, R.L. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 2007, 143, 1173–1188. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.P.; Piskurewicz, U.; Tureckova, V.; Strnad, M.; Lopez-Molina, L. A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. USA 2010, 107, 19108–19113. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Yim, S.; Choi, H.; Kim, A.; Lee, K.P.; Lopez-Molina, L.; Martinoia, E.; Lee, Y. Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 2015, 6, 8113. [Google Scholar] [CrossRef] [Green Version]
- Topham, A.T.; Taylor, R.E.; Yan, D.; Nambara, E.; Johnston, I.G.; Bassel, G.W. Temperature variability is integrated by a spatially embedded decision-making center to break dormancy in Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 2017, 114, 6629–6634. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Liu, S.; Lin, R. The role of light in regulating seed dormancy and germination. J. Integr. Plant Biol. 2020, 62, 1310–1326. [Google Scholar] [CrossRef]
- Seo, M.; Hanada, A.; Kuwahara, A.; Endo, A.; Okamoto, M.; Yamauchi, Y.; North, H.; Marion-Poll, A.; Sun, T.P.; Koshiba, T.; et al. Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006, 48, 354–366. [Google Scholar] [CrossRef]
- Sawada, Y.; Aoki, M.; Nakaminami, K.; Mitsuhashi, W.; Tatematsu, K.; Kushiro, T.; Koshiba, T.; Kamiya, Y.; Inoue, Y.; Nambara, E.; et al. Phytochrome- and gibberellin-mediated regulation of abscisic acid metabolism during germination of photoblastic lettuce seeds. Plant Physiol. 2008, 146, 1386–1396. [Google Scholar] [CrossRef] [Green Version]
- Gubler, F.; Hughes, T.; Waterhouse, P.; Jacobsen, J. Regulation of dormancy in barley by blue light and after-ripening: Effects on abscisic acid and gibberellin metabolism. Plant Physiol. 2008, 147, 886–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, H.H.; Sechet, J.; Bailly, C.; Leymarie, J.; Corbineau, F. Inhibition of germination of dormant barley (Hordeum vulgare L.) grains by blue light as related to oxygen and hormonal regulation. Plant Cell Environ. 2014, 37, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.; Imamura, A.; Watanabe, A.; Nakabayashi, K.; Okamoto, M.; Jikumaru, Y.; Hanada, A.; Aso, Y.; Ishiyama, K.; Tamura, N.; et al. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 2008, 146, 1368–1385. [Google Scholar] [CrossRef] [Green Version]
- Huo, H.; Dahal, P.; Kunusoth, K.; McCallum, C.M.; Bradford, K.J. Expression of 9-cis-EPOXYCAROTENOID DIOXYGENASE4 is essential for thermoinhibition of lettuce seed germination but not for seed development or stress tolerance. Plant Cell 2013, 25, 884–900. [Google Scholar] [CrossRef] [Green Version]
- Izydorczyk, C.; Nguyen, T.N.; Jo, S.H.; Son, S.H.; Tuan, P.A.; Ayele, B.T. Spatiotemporal modulation of abscisic acid and gibberellin metabolism and signalling mediates the effects of suboptimal and supraoptimal temperatures on seed germination in wheat (Triticum aestivum L.). Plant Cell Environ. 2018, 41, 1022–1037. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; Matilla, A.J.; Rodriguez-Gacio, M.C.; Iglesias-Fernandez, R. Nitrate affects sensu-stricto germination of after-ripened Sisymbrium officinale seeds by modifying expression of SoNCED5, SoCYP707A2 and SoGA3ox2 genes. Plant Sci. 2014, 217–218, 99–108. [Google Scholar] [CrossRef]
- Lee, K.; Lee, H.G.; Yoon, S.; Kim, H.U.; Seo, P.J. The Arabidopsis MYB96 transcription factor is a positive regulator of ABSCISIC ACID-INSENSITIVE4 in the control of seed germination. Plant Physiol. 2015, 168, 677–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.M.; Rothstein, S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef] [Green Version]
- De Wit, M.; Galvão, V.C.; Fankhauser, C. Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 2008, 20, 1260–1277. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Lee, N.; Kim, W.; Lim, S.; Cho, G. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 2011, 23, 1404–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.P.; Piskurewicz, U.; Tureckova, V.; Carat, S.; Chappuis, R.; Strnad, M.; Fankhauser, C.; Lopez-Molina, L. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 2012, 26, 1984–1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barros-Galvao, T.; Dave, A.; Gilday, A.D.; Harvey, D.; Vaistij, F.E.; Graham, I.A. ABA INSENSITIVE4 promotes rather than represses PHYA-dependent seed germination in Arabidopsis thaliana. New Phytol. 2020, 226, 953–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, L.D.; Xu, X.; Gevaert, K.; De Smet, I. Developmental plasticity at high temperature. Plant Physiol. 2019, 181, 399–411. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.; Park, J.; Lee, N.; Jeong, J.; Toh, S.; Watanabe, A.; Kim, J.; Kang, H.; Kim, D.H.; Kawakami, N.; et al. ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 2013, 25, 4863–4878. [Google Scholar] [CrossRef] [Green Version]
- Buijs, G. A perspective on secondary seed dormancy in Arabidopsis thaliana. Plants 2020, 9, 749. [Google Scholar] [CrossRef]
- Martel, C.; Blair, L.K.; Donohue, K. PHYD prevents secondary dormancy establishment of seeds exposed to high temperature and is associated with lower PIL5 accumulation. J. Exp. Bot. 2018, 69, 3157–3169. [Google Scholar] [CrossRef]
- Chiu, R.S.; Nahal, H.; Provart, N.J.; Gazzarrini, S. The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biol. 2012, 12, 15. [Google Scholar] [CrossRef] [Green Version]
- Yan, D.; Easwaran, V.; Chau, V.; Okamoto, M.; Ierullo, M.; Kimura, M.; Endo, A.; Yano, R.; Pasha, A.; Gong, Y.; et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 2016, 7, 13179. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef]
- Ogura, T.; Busch, W. From phenotypes to causal sequences: Using genome wide association studies to dissect the sequence basis for variation of plant development. Curr. Opin. Plant Biol. 2015, 23, 98–108. [Google Scholar] [CrossRef]
- Bazakos, C.; Hanemian, M.; Trontin, C.; Jiménez-Gómez, J.M.; Loudet, O. New strategies and tools in quantitative genetics: How to go from the phenotype to the genotype. Annu. Rev. Plant Biol. 2017, 68, 435–455. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Sinha, P.; Singh, V.K.; Kumar, A.; Zhang, Q.; Bennetzen, J.L. 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 2020, 56, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Mott, R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- The 1001 Genomes Consortium. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 2016, 166, 481–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso-Blanco, C.; Koornneef, M. Naturally occurring variation in Arabidopsis: An underexploited resource for plant genetics. Trends Plant Sci. 2000, 5, 22–29. [Google Scholar] [CrossRef]
- Weigel, D. Natural variation in Arabidopsis: From molecular genetics to ecological genomics. Plant Physiol. 2012, 158, 2–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preston, J.; Tatematsu, K.; Kanno, Y.; Hobo, T.; Kimura, M.; Jikumaru, Y.; Yano, R.; Kamiya, Y.; Nambara, E. Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: A comparative study on dormant and non-dormant accessions. Plant Cell Physiol. 2009, 50, 1786–1800. [Google Scholar] [CrossRef] [Green Version]
- Kashiwakura, Y.; Kobayashi, D.; Jikumaru, Y.; Takebayashi, Y.; Nambara, E.; Seo, M.; Kamiya, Y.; Kushiro, T.; Kawakami, N. Highly sprouting-tolerant wheat grain exhibits extreme dormancy and cold imbibition-resistant accumulation of abscisic acid. Plant Cell Physiol. 2016, 57, 715–732. [Google Scholar] [CrossRef] [Green Version]
- Argyris, J.; Dahal, P.; Hayashi, E.; Still, D.W.; Bradford, K.J. Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiol. 2008, 148, 926–947. [Google Scholar] [CrossRef] [Green Version]
- Howitt, C.A.; Pogson, B.J. Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ. 2006, 29, 435–445. [Google Scholar] [CrossRef]
- Fang, J.; Chai, C.; Qian, Q.; Li, C.; Tang, J.; Sun, L.; Huang, Z.; Guo, X.; Sun, C.; Liu, M.; et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 2008, 54, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Stanley, L.; Yuan, Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Jorge, S.; Mehrshahi, P.; Magallanes-Lundback, M.; Lipka, A.E.; Angelovici, R.; Gore, M.A.; DellaPenna, D. ZEAXANTHIN EPOXIDASE activity potentiates carotenoid degradation in maturing seed. Plant Physiol. 2016, 171, 1837–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, B.F.; Lipka, A.E.; Magallanes-Lundback, M.; Tiede, T.; Diepenbrock, C.H.; Kandianis, C.B.; Kim, E.; Cepela, J.; Mateos-Hernandez, M.; Buell, C.R.; et al. foundation for provitamin a biofortification of maize: Genome-wide association and genomic prediction models of carotenoid levels. Genetics 2014, 198, 1699–1716. [Google Scholar] [CrossRef] [Green Version]
- Owens, B.F.; Mathew, D.; Diepenbrock, C.H.; Tiede, T.; Wu, D.; Mateos-Hernandez, M.; Gore, M.A.; Rocheford, T. Genome-wide association study and pathway-level analysis of kernel color in maize. G3 2019, 9, 1945–1955. [Google Scholar] [CrossRef] [Green Version]
- Cruet-Burgos, C.; Cox, S.; loerger, B.P.; Perumal, R.; Hu, Z.; Herald, T.J.; Bean, S.R.; Rhodes, D.H. Advancing provitamin a biofortification in sorghum: Genome-wide association studies of grain carotenoids in global germplasm. Plant Genome 2020, 13, e20013. [Google Scholar] [CrossRef] [Green Version]
- Capelle, V.; Remoué, C.; Moreau, L.; Reyss, A.; Mahé, A.; Massonneau, A.; Falque, M.; Charcosset, A.; Thévenot, C.; Rogowsky, P.; et al. QTLs and candidate genes for desiccation and abscisic acid content in maize kernels. BMC Plant Biol. 2010, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argyris, J.; Truco, M.J.; Ochoa, O.; McHale, L.; Dahal, P.; Van Deynze, A.; Michelmore, R.W.; Bradford, K.J. A gene encoding an abscisic acid biosynthetic enzyme (LsNCED4) collocates with the high temperature germination locus Htg6.1 in lettuce (Lactuca sp.). Theor. Appl. Genet. 2011, 122, 95–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, H.; Wu, G.; Sun, J.; Hao, L.; Ge, X.; Yu, J.; Wang, W. Sequence variation and expression analysis of seed dormancy- and germination-associated ABA- and GA-related genes in rice cultivars. Front. Plant Sci. 2011, 2, 17. [Google Scholar] [CrossRef] [Green Version]
- Kalladan, R.; Lasky, J.R.; Sharma, S.; Kumar, M.N.; Juenger, T.E.; Des Marais, D.L.; Verslues, P.E. Natural variation in 9-cis-epoxycartenoid dioxygenase 3 and ABA accumulation. Plant Physiol. 2019, 179, 1620–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clerkx, E.J.; El-Lithy, M.E.; Vierling, E.; Ruys, G.J.; Blankestijn-De Vries, H.; Groot, S.P.; Vreugdenhil, D.; Koornneef, M. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 2004, 135, 432–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amer Hamzah, M.; Mohd Kasim, N.A.; Shamsuddin, A.; Mustafa, N.; Mohamad Rusli, N.I.; Teh, C.Y.; Ho, C.L. Nucleotide variations of 9-cis-epoxycarotenoid dioxygenase 2 (NCED2) and pericarp coloration genes (Rc and Rd) from upland rice varieties. 3 Biotech 2020, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Zhang, S.; Dong, Y.; He, W.; Zhang, J.; Deng, X.; Zhang, Y.; Li, X.; Li, B.; Huang, W.; et al. Analysis of elite variety tag SNPs reveals an important allele in upland rice. Nat. Commun. 2013, 4, 2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colasuonno, P.; Lozito, M.L.; Marcotuli, I.; Nigro, D.; Giancaspro, A.; Mangini, G.; De Vita, P.; Mastrangelo, A.M.; Pecchioni, N.; Houston, K.; et al. The carotenoid biosynthetic and catabolic genes in wheat and their association with yellow pigments. BMC Genom. 2017, 18, 122. [Google Scholar] [CrossRef] [Green Version]
- Colasuonno, P.; Marcotuli, I.; Lozito, M.L.; Simeone, R.; Blanco, A.; Gadaleta, A. Characterization of aldehyde oxidase (AO) genes involved in the accumulation of carotenoid pigments in wheat grain. Front. Plant Sci. 2017, 8, 863. [Google Scholar] [CrossRef] [Green Version]
- Kan, G.; Zhang, W.; Yang, W.; Ma, D.; Zhang, D.; Hao, D.; Hu, Z.; Yu, D. Association mapping of soybean seed germination under salt stress. Mol. Genet. Genom. 2015, 290, 2147–2162. [Google Scholar] [CrossRef]
- Nakamura, S.; Chono, M.; Abe, F.; Miura, H. Mapping a diploid wheat abscisic acid 8′-hydroxylase homologue in the seed dormancy QTL region on chromosome 5Am. Euphytica 2010, 171, 111–120. [Google Scholar] [CrossRef]
- Kalladan, R.; Lasky, J.R.; Chang, T.Z.; Sharma, S.; Juenger, T.E.; Verslues, P.E. Natural variation identifies genes affecting drought-induced abscisic acid accumulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 11536–11541. [Google Scholar] [CrossRef] [Green Version]
- Farh, K.K.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.; Shishkin, A.A.; et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef]
- Liu, S.; Li, C.; Wang, H.; Wang, S.; Yang, S.; Liu, X.; Yan, J.; Li, B.; Beatty, M.; Zastrow-Hayes, G.; et al. Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol. 2020, 21, 163. [Google Scholar] [CrossRef]
- De Villemereuil, P.; Gaggiotti, O.E.; Mouterde, M.; Till-Bottraud, I. Common garden experiments in the genomic era: New perspectives and opportunities. Heredity 2016, 116, 249–254. [Google Scholar] [CrossRef] [Green Version]
- Ferrero-Serrano, Á.; Assmann, S.M. Phenotypic and genome-wide association with the local environment of Arabidopsis. Nat. Ecol. Evol. 2019, 3, 274–285. [Google Scholar] [CrossRef]
- Lee, G.A.; Jeon, Y.A.; Lee, H.S.; Hyun, D.Y.; Lee, J.R.; Lee, M.C.; Lee, S.Y.; Ma, K.H.; Koh, H.J. Variation in pre-harvest sprouting resistance, seed germination and changes in abscisic acid levels during grain development in diverse rice genetic resources. Plant Genet. Resour. 2016, 16, 18–27. [Google Scholar] [CrossRef]
- Ali, A.; Cao, J.; Jiang, H.; Chang, C.; Zhang, H.P.; Sheikh, S.W.; Shah, L.; Ma, C. Unraveling molecular and genetic studies of wheat (Triticum aestivum L.) Resistance against factors causing pre-harvest sprouting. Agronomy 2019, 9, 117. [Google Scholar] [CrossRef] [Green Version]
- Tai, L.; Wang, H.J.; Xu, X.J.; Sun, W.H.; Ju, L.; Liu, W.T.; Li, W.Q.; Sun, J.; Chen, K.M. Cereal pre-harvest sprouting: A global agricultural disaster regulated by complex genetic and biochemical mechanisms. J. Exp. Bot. 2021, in press. [Google Scholar]
- Dejonghe, W.; Cutler, S.R. Abscisic acid as a gateway for the crops of tomorrow. In Abscisic Acid in Plants; Seo, M., Marion-Poll, A., Eds.; Advances in Botanical Research; Academic Press: London, UK, 2019; Volume 92, pp. 341–370. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sano, N.; Marion-Poll, A. ABA Metabolism and Homeostasis in Seed Dormancy and Germination. Int. J. Mol. Sci. 2021, 22, 5069. https://doi.org/10.3390/ijms22105069
Sano N, Marion-Poll A. ABA Metabolism and Homeostasis in Seed Dormancy and Germination. International Journal of Molecular Sciences. 2021; 22(10):5069. https://doi.org/10.3390/ijms22105069
Chicago/Turabian StyleSano, Naoto, and Annie Marion-Poll. 2021. "ABA Metabolism and Homeostasis in Seed Dormancy and Germination" International Journal of Molecular Sciences 22, no. 10: 5069. https://doi.org/10.3390/ijms22105069
APA StyleSano, N., & Marion-Poll, A. (2021). ABA Metabolism and Homeostasis in Seed Dormancy and Germination. International Journal of Molecular Sciences, 22(10), 5069. https://doi.org/10.3390/ijms22105069






