Structural and Functional Peculiarities of Cytoplasmic Tropomyosin Isoforms, the Products of TPM1 and TPM4 Genes
Abstract
1. Introduction
2. Results
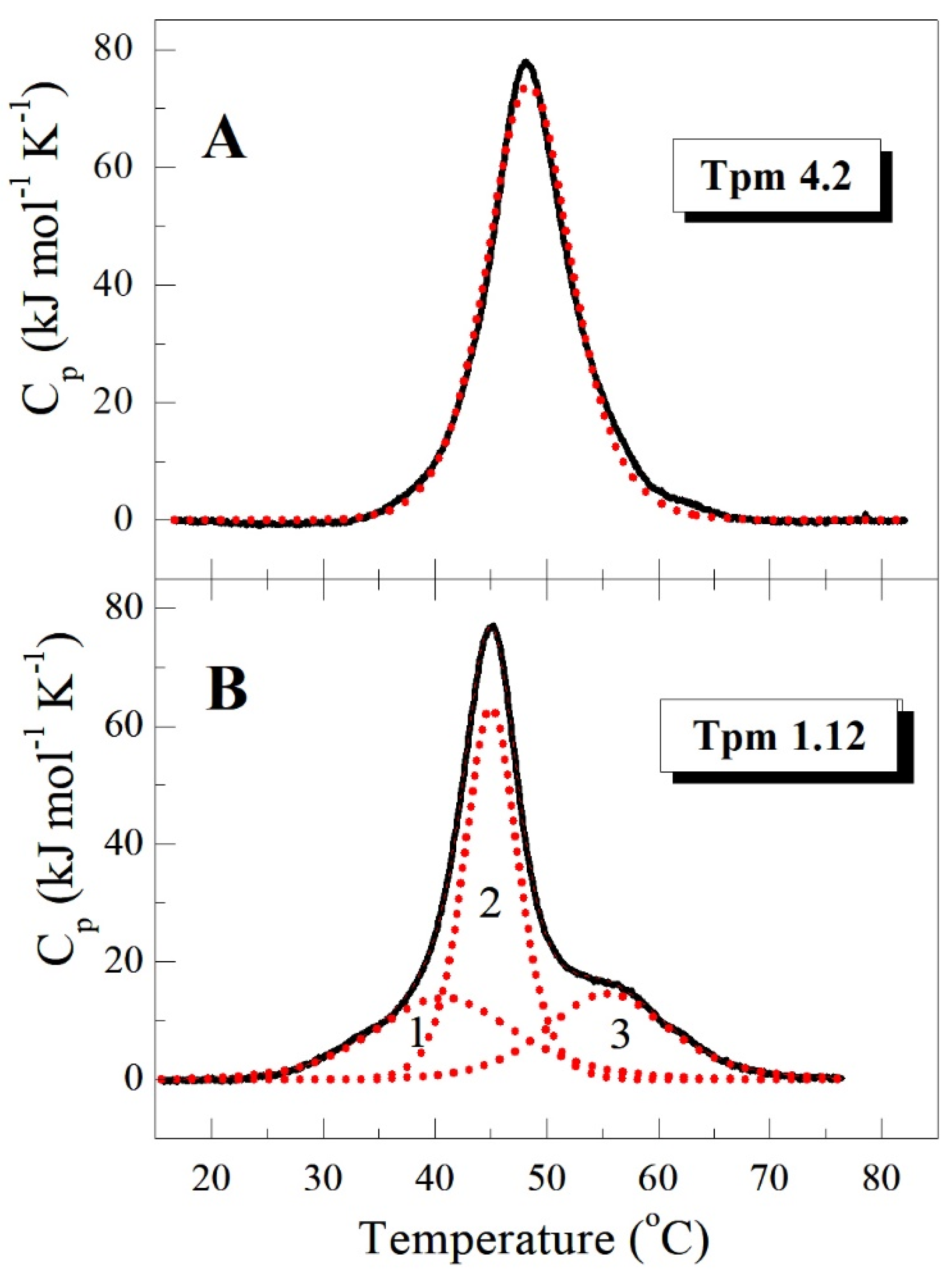
2.1. Thermal Stability of Cytoplasmic Tpm Isoforms (CD and DSC Studies)
2.2. Interaction between the N- and C-Ends of Tpm Isoforms
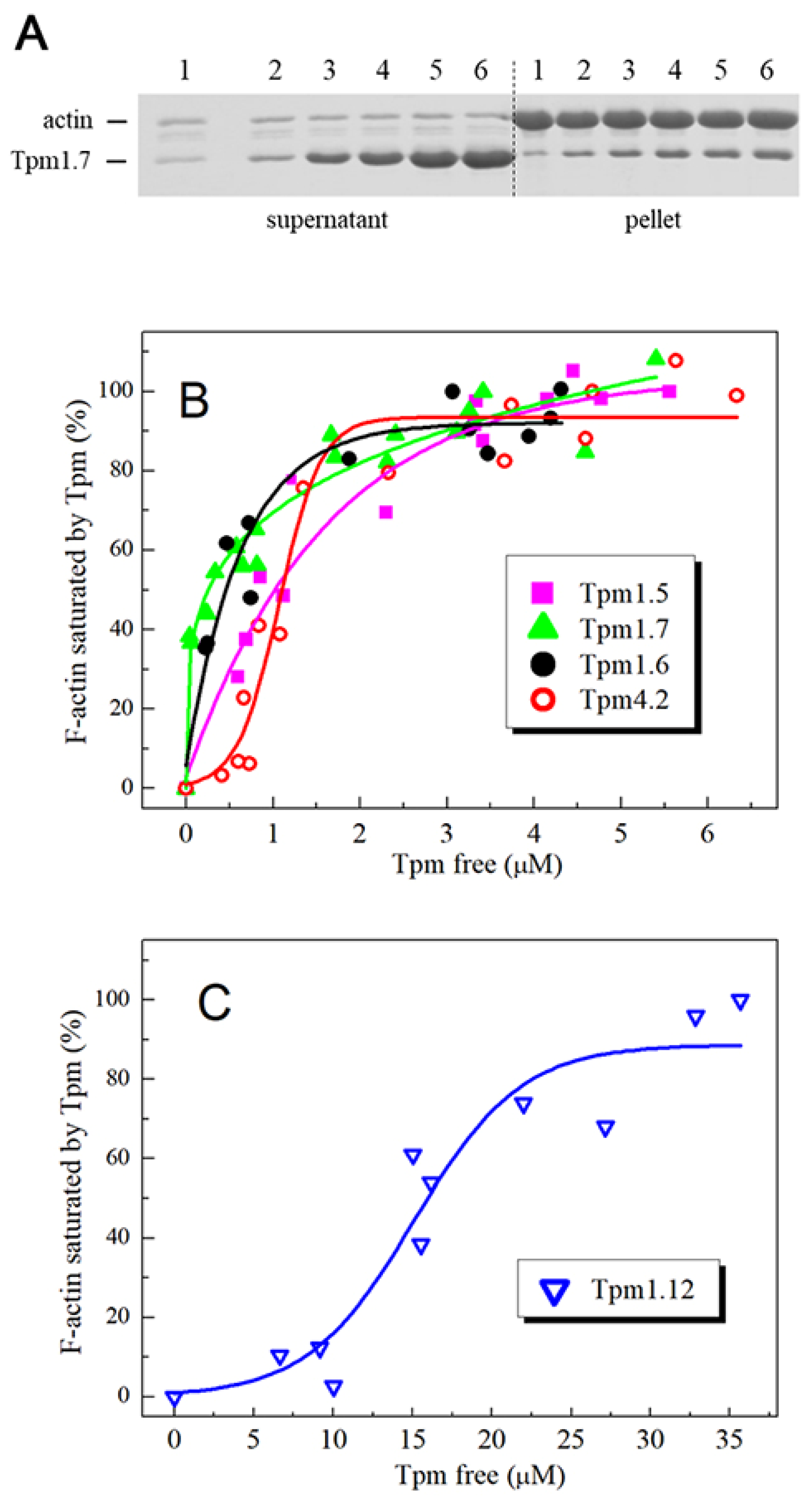
2.3. Affinity of Tpm Isoforms for F-actin
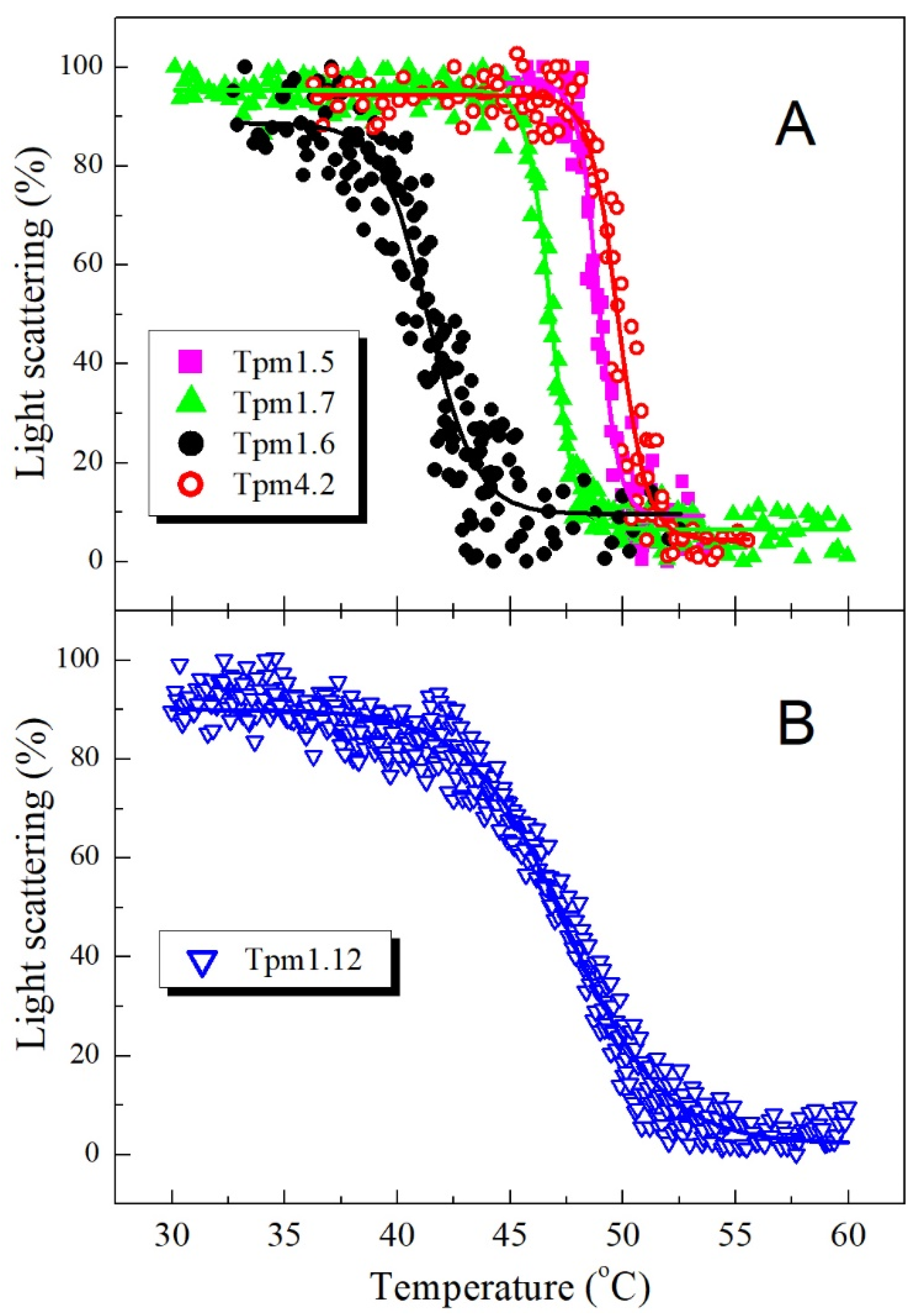
2.4. Thermally Induced Dissociation of the F-actin Complexes with Tpm Isoforms
3. Discussion
3.1. Properties of HMW Isoforms of Non-Muscle Tpm (Tpm1.5, Tpm1.6, and Tpm1.7)
3.2. Properties of LMW Isoforms Tpm1.12 and Tpm4.2
4. Materials and Methods
4.1. Protein Preparations
4.2. CD Measurements
4.3. Differential Scanning Calorimetry (DSC)
4.4. Viscosity Measurements
4.5. Cosedimentation of Tpm Species with F-actin
4.6. Temperature Dependences of Light Scattering
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gunning, P.; O’Neill, G.; Hardeman, E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 2008, 88, 1–35. [Google Scholar] [CrossRef]
- Perry, S. Vertebrate tropomyosin: Distribution, properties and function. J. Muscle Res. Cell Motil. 2001, 22, 5–49. [Google Scholar] [CrossRef]
- Nevzorov, A.I.; Levitsky, I.D. Tropomyosin: Double helix from the protein world. Biochemistry 2011, 76, 1507–1527. [Google Scholar] [CrossRef]
- Schevzov, G.; Vrhovski, B.; Bryce, N.S.; Elmir, S.; Qiu, M.R.; O’Neill, G.M.; Yang, N.; Verrills, N.M.; Kavallaris, M.; Gunning, P.W. Tissue-specific Tropomyosin Isoform Composition. J. Histochem. Cytochem. 2005, 53, 557–570. [Google Scholar] [CrossRef]
- Weinberger, R.P.; Henke, R.C.; Tolhurst, O.; Jeffrey, P.L.; Gunning, P. Induction of neuron-specific tropomyosin mRNAs by nerve growth factor is dependent on morphological differentiation. J. Cell Biol. 1993, 120, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Pelham, R.; Lin, J.; Wang, Y. A high molecular mass non-muscle tropomyosin isoform stimulates retrograde organelle transport. J. Cell Sci. 1996, 109, 981–989. [Google Scholar] [CrossRef]
- Caldwell, B.J.; Lucas, C.; Kee, A.J.; Gaus, K.; Gunning, P.W.; Hardeman, E.C.; Yap, A.S.; Gomez, G.A. Tropomyosin isoforms support actomyosin biogenesis to generate contractile tension at the epithelial zonula adherens. Cytoskeleton 2014, 71, 663–676. [Google Scholar] [CrossRef] [PubMed]
- McMichael, B.K.; Lee, B.S. Tropomyosin 4 regulates adhesion structures and resorptive capacity in osteoclasts. Exp. Cell Res. 2008, 314, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Craig, R.; Lehman, W. Crossbridge and tropomyosin positions observed in native, interacting thick and thin filaments. J. Mol. Biol. 2001, 311, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, W.H. Binding of Tropomyosin–Troponin to Actin Increases Filament Bending Stiffness. Biochem. Biophys. Res. Commun. 2000, 276, 1225–1228. [Google Scholar] [CrossRef]
- Nabiev, S.R.; Ovsyannikov, D.A.; Kopylova, G.V.; Shchepkin, D.V.; Matyushenko, A.M.; Koubassova, N.A.; Levitsky, D.I.; Tsaturyan, A.K.; Bershitsky, S.Y. Stabilizing the Central Part of Tropomyosin Increases the Bending Stiffness of the Thin Filament. Biophys. J. 2015, 109, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Weigt, C.; Schoepper, B.; Wegner, A. Tropomyosin-troponin complex stabilizes the pointed ends of actin filaments against polymerization and depolymerization. FEBS Lett. 1990, 260, 266–268. [Google Scholar] [CrossRef]
- Broschat, K.O. Tropomyosin prevents depolymerization of actin filaments from the pointed end. J. Biol. Chem. 1990, 265, 21323–21329. [Google Scholar] [CrossRef]
- Khaitlina, S.Y. Tropomyosin as a Regulator of Actin Dynamics. Int. Rev. Cell Mol. Biol. 2015, 318, 255–291. [Google Scholar] [CrossRef] [PubMed]
- Geeves, M.A.; Hitchcock-DeGregori, S.E.; Gunning, P.W. A systematic nomenclature for mammalian tropomyosin isoforms. J. Muscle Res. Cell Motil. 2015, 36, 147–153. [Google Scholar] [CrossRef]
- Lees-Miller, J.P.; Helfman, D.M. The molecular basis for tropomyosin isoform diversity. BioEssays 1991, 13, 429–437. [Google Scholar] [CrossRef]
- Brettle, M.; Patel, S.; Fath, T. Tropomyosins in the healthy and diseased nervous system. Brain Res. Bull. 2016, 126, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.T.; Kostyukova, A.S.; Fath, T. Actin regulation by tropomodulin and tropomyosin in neuronal morphogenesis and function. Mol. Cell. Neurosci. 2017, 84, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Schevzov, G.; Curthoys, N.M.; Gunning, P.W.; Fath, T. Functional Diversity of Actin Cytoskeleton in Neurons and its Regulation by Tropomyosin. Int. Rev. Cell Mol. Biol. 2012, 298, 33–94. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.A.; Cooke-Yarborough, C.M.; Chadwick, N.C.; Schevzov, G.; Arbuckle, S.M.; Gunning, P.; Weinberger, R.P. High-molecular-weight tropomyosins localize to the contractile rings of dividing CNS cells but are absent from malignant pe-diatric and adult CNS tumors. Glia 2003, 42, 25–35. [Google Scholar] [CrossRef]
- Parreno, J.; Amadeo, M.B.; Kwon, E.H.; Fowler, V.M. Tropomyosin 3.1 Association with Actin Stress Fibers is Required for Lens Epithelial to Mesenchymal Transition. Investig. Opthalmol. Vis. Sci. 2020, 61, 2. [Google Scholar] [CrossRef]
- Manstein, D.J.; Meiring, J.C.M.; Hardeman, E.C.; Gunning, P.W. Actin–tropomyosin distribution in non-muscle cells. J. Muscle Res. Cell Motil. 2020, 41, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Had, L.; Faivre-Sarrailh, C.; Legrand, C.; Mery, J.; Brugidou, J.; Rabie, A. Tropomyosin isoforms in rat neurons: The different developmental profiles and distributions of TM-4 and TMBr-3 are consistent with different functions. J. Cell Sci. 1994, 107, 2961–2973. [Google Scholar] [CrossRef] [PubMed]
- Gateva, G.; Kremneva, E.; Reindl, T.; Kotila, T.; Kogan, K.; Gressin, L.; Gunning, P.W.; Manstein, D.J.; Michelot, A.; Lappalainen, P. Tropomyosin Isoforms Specify Functionally Distinct Actin Filament Populations In Vitro. Curr. Biol. 2017, 27, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Janco, M.; Bonello, T.T.; Byun, A.; Coster, A.C.F.; Lebhar, H.; Dedova, I.; Gunning, P.W.; Böcking, T. The impact of tropomyosins on actin filament assembly is isoform specific. BioArchitecture 2016, 6, 61–75. [Google Scholar] [CrossRef]
- Kis-Bicskei, N.; Vig, A.; Nyitrai, M.; Bugyi, B.; Talian, G.C. Purification of tropomyosin Br-3 and 5NM1 and characterization of their interactions with actin. Cytoskeleton 2013, 70, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Moraczewska, J.; Nicholson-Flynn, K.; Hitchcock-DeGregori, S.E. The Ends of Tropomyosin Are Major Determinants of Actin Affinity and Myosin Subfragment 1-Induced Binding to F-Actin in the Open State†. Biochemistry 1999, 38, 15885–15892. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, V.V.; Marchenko, M.A.; Kleymenov, S.Y.; Datskevich, P.N.; Levitsky, D.I.; Matyushenko, A.M. Thermal unfolding of various human non-muscle isoforms of tropomyosin. Biochem. Biophys. Res. Commun. 2019, 514, 613–617. [Google Scholar] [CrossRef]
- Kremneva, E.; Nikolaeva, O.; Maytum, R.; Arutyunyan, A.M.; Kleimenov, S.Y.; Geeves, M.A.; Levitsky, D.I. Thermal unfolding of smooth muscle and non-muscle tropomyosin α-homodimers with alternatively spliced exons. FEBS J. 2006, 273, 588–600. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Shchepkin, D.V.; Kopylova, G.V.; Bershitsky, S.Y.; Levitsky, D.I. Unique functional properties of slow skeletal muscle tropomyosin. Biochimica 2020, 174, 1–8. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Artemova, N.V.; Shchepkin, D.V.; Kopylova, G.V.; Bershitsky, S.Y.; Tsaturyan, A.K.; Sluchanko, N.N.; Levitsky, D.I. Structural and functional effects of two stabilizing substitutions, D137L and G126R, in the middle part of α-tropomyosin molecule. FEBS J. 2014, 281, 2004–2016. [Google Scholar] [CrossRef] [PubMed]
- Matyushenko, A.M.; Shchepkin, D.V.; Kopylova, G.V.; Popruga, K.E.; Artemova, N.V.; Pivovarova, A.V.; Bershitsky, S.Y.; Levitsky, D.I. Structural and Functional Effects of Cardiomyopathy-Causing Mutations in the Troponin T-Binding Region of Cardiac Tropomyosin. Biochemistry 2016, 56, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Kremneva, E.; Boussouf, S.; Nikolaeva, O.; Maytum, R.; Geeves, M.A.; Levitsky, D.I. Effects of two familial hypertrophic cardiomyopathy mutations in α-tropomyosin, Asp175Asn and Glu180Gly, on the thermal unfolding of actin-bound tropo-myosin. Biophys. J. 2004, 87, 3922–3933. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J.; Kotlyanskaya, L.; Hitchcock-DeGregori, S.E. Structure of the N terminus of a nonmuscle α-tropomyosin in complex with the C terminus: Implications for actin binding. Biochemistry 2009, 48, 1272–1283. [Google Scholar] [CrossRef][Green Version]
- Frye, J.; Klenchin, V.A.; Rayment, I. Structure of the tropomyosin overlap complex from chicken smooth muscle: Insight into the diversity of N-terminal recognition. Biochemistry 2010, 49, 4908–4920. [Google Scholar] [CrossRef] [PubMed]
- Mamidi, R.; Muthuchamy, M.; Chandra, M. Instability in the central region of tropomyosin modulates the function of its overlapping ends. Biophys. J. 2013, 105, 2104–2113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hammell, R.L.; Hitchcock-DeGregori, S.E. Mapping the Functional Domains within the Carboxyl Terminus of α-Tropomyosin Encoded by the Alternatively Spliced Ninth Exon. J. Biol. Chem. 1996, 271, 4236–4242. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Artemova, N.V.; Shchepkin, D.V.; Kopylova, G.V.; Nabiev, S.R.; Nikitina, L.V.; Levitsky, D.I.; Bershitsky, S.Y. The interchain disulfide cross-linking of tropomyosin alters its regulatory properties and interaction with actin filament. Biochem. Biophys. Res. Commun. 2017, 482, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Lataro, R.; Ferro, J.; Reinach, F.D.C. Functional alpha-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J. Biol. Chem. 1994, 269, 10461–10466. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Koubassova, N.A.; Shchepkin, D.V.; Kopylova, G.V.; Nabiev, S.R.; Nikitina, L.V.; Bershitsky, S.Y.; Levitsky, D.I.; Tsaturyan, A.K. The effects of cardiomyopathy-associated mutations in the head-to-tail overlap junction of α-tropomyosin on its properties and interaction with actin. Int. J. Biol. Macromol. 2019, 125, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Spudich, J.A.; Watt, S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971, 246, 4866–4871. [Google Scholar] [CrossRef]
- Freire, E.; Biltonen, R.L. Statistical mechanical deconvolution of thermal transitions in macromolecules. I. Theory and application to homogeneous systems. Biopolymer 1978, 17, 463–479. [Google Scholar] [CrossRef]
- Nevzorov, I.A.; Nikolaeva, O.P.; Kainov, Y.A.; Redwood, C.S.; Levitsky, D.I. Conserved Noncanonical Residue Gly-126 Confers Instability to the Middle Part of the Tropomyosin Molecule. J. Biol. Chem. 2011, 286, 15766–15772. [Google Scholar] [CrossRef]
- Wegner, A. Equilibrium of the actin-tropomyosin interaction. J. Mol. Biol. 1979, 131, 839–853. [Google Scholar] [CrossRef]


| Tpm Isoform | Viscosity, mP∙s | |
|---|---|---|
| 1 mg/mL | 2 mg/mL | |
| HMW | ||
| Tpm 1.5 | 0.179 ± 0.005 | 0.653 ± 0.002 |
| Tpm 1.6 | 0.474 ± 0.005 | 1.339 ± 0.001 |
| Tpm 1.7 | 1.027 ± 0.009 | 3.575 ± 0.001 |
| LMW | ||
| Tpm 1.12 | 0.026 ± 0.004 | 0.066 ± 0.001 |
| Tpm 4.2 | 0.064 ± 0.004 | 0.193 ± 0.005 |
| Tpm Isoform | K50% (μM) | Tdiss (°C) |
|---|---|---|
| HMW | ||
| Tpm 1.5 | 1.11 ± 0.02 | 48.92 ± 0.04 |
| Tpm 1.6 | 0.45 ± 0.08 | 41.38 ± 0.11 |
| Tpm 1.7 | 0.40 ± 0.10 | 46.78 ± 0.02 |
| LMW | ||
| Tpm 1.12 | 14.9 ± 2.0 | 47.49 ± 0.06 * |
| Tpm 4.2 | 1.07 ± 0.07 | 49.79 ± 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchenko, M.; Nefedova, V.; Artemova, N.; Kleymenov, S.; Levitsky, D.; Matyushenko, A. Structural and Functional Peculiarities of Cytoplasmic Tropomyosin Isoforms, the Products of TPM1 and TPM4 Genes. Int. J. Mol. Sci. 2021, 22, 5141. https://doi.org/10.3390/ijms22105141
Marchenko M, Nefedova V, Artemova N, Kleymenov S, Levitsky D, Matyushenko A. Structural and Functional Peculiarities of Cytoplasmic Tropomyosin Isoforms, the Products of TPM1 and TPM4 Genes. International Journal of Molecular Sciences. 2021; 22(10):5141. https://doi.org/10.3390/ijms22105141
Chicago/Turabian StyleMarchenko, Marina, Victoria Nefedova, Natalia Artemova, Sergey Kleymenov, Dmitrii Levitsky, and Alexander Matyushenko. 2021. "Structural and Functional Peculiarities of Cytoplasmic Tropomyosin Isoforms, the Products of TPM1 and TPM4 Genes" International Journal of Molecular Sciences 22, no. 10: 5141. https://doi.org/10.3390/ijms22105141
APA StyleMarchenko, M., Nefedova, V., Artemova, N., Kleymenov, S., Levitsky, D., & Matyushenko, A. (2021). Structural and Functional Peculiarities of Cytoplasmic Tropomyosin Isoforms, the Products of TPM1 and TPM4 Genes. International Journal of Molecular Sciences, 22(10), 5141. https://doi.org/10.3390/ijms22105141





