A Fructan Exohydrolase from Maize Degrades Both Inulin and Levan and Co-Exists with 1-Kestotriose in Maize
Abstract
:1. Introduction
2. Results
2.1. Maize Cell-Wall Invertase-Related Enzyme Zm-6&1-FEH2 with Both Inulin-Type and Levan-Type Fructan Exohydrolase Activities
2.2. Zm-6&1-FEH2 Localized to the Apoplast
2.3. Zm-6&1-FEH2 Expressed Differently during Plant Development and in Response to Drought Stress
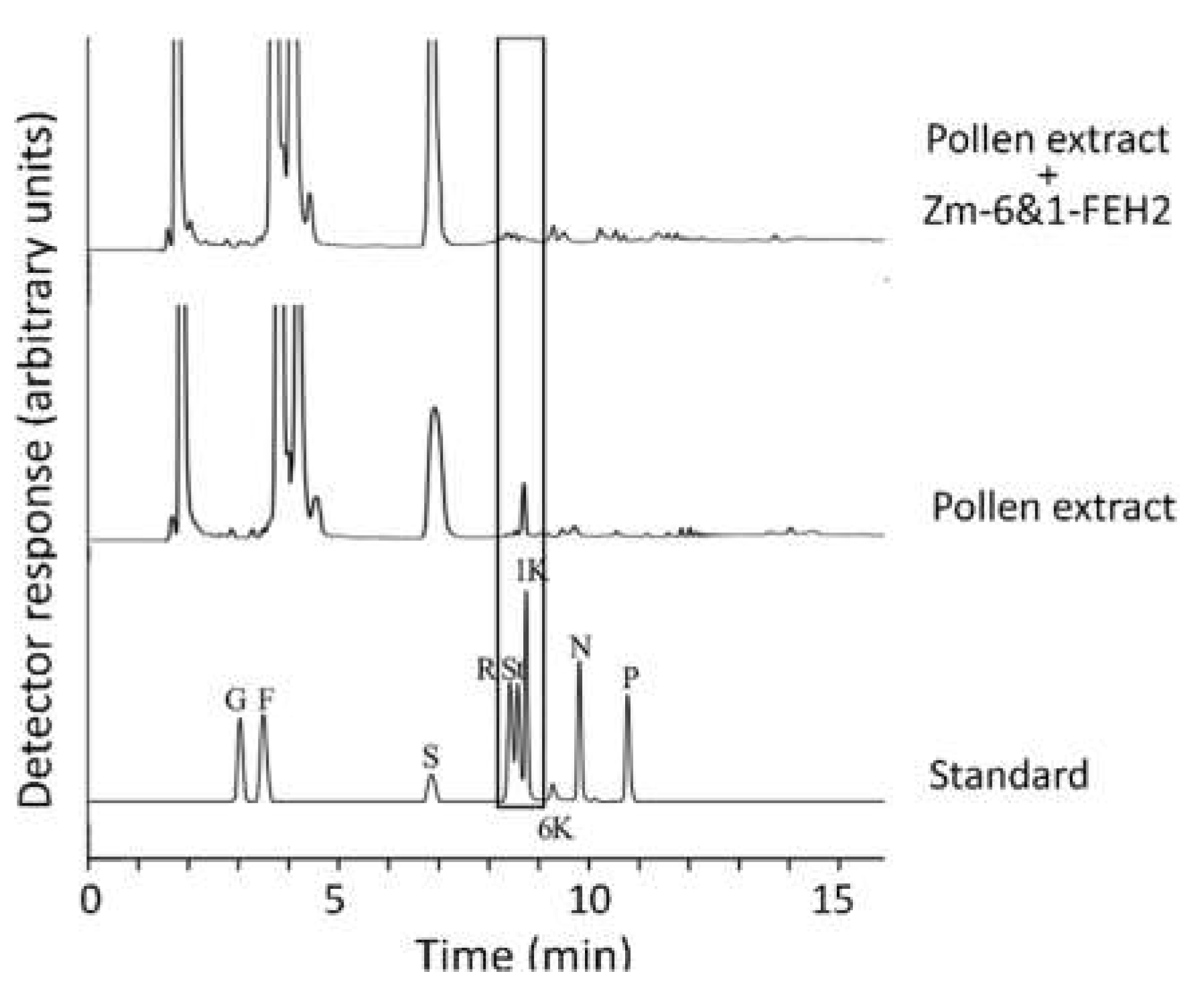
2.4. Zm-6&1-FEH2 Hydrolyze Oligofructan 1-Kestotriose from Maize
2.5. In Vitro Synthesis of Fructan Trisaccharides in Maize
3. Discussion
4. Materials and Methods
4.1. Plant Material and Cultivation
4.2. RNA Extraction, Cloning, Sequencing and Phylogeny
4.3. Gene Expression Analysis by qPCR
4.4. Expression of Recombinant FEH Protein in Pichia Pastoris
4.5. Plant Transformation and Protein Extraction
4.6. Determination of FEH Activity
4.7. Carbohydrate Extraction and Analysis
4.8. CLSM Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Sadeleer, E.D.; Vergauwen, R.; Struyf, T.; Le Roy, K.; Van den Ende, W. 1-FFT amino acids involved in high DP inulin accumulation in Viguiera discolor. Front. Plant Sci. 2015, 6, 616. [Google Scholar] [CrossRef] [Green Version]
- Lothier, J.; Van Laere, A.; Prud’homme, M.P.; Van den Ende, W.; Morvan-Bertrand, A. Cloning and characterization of a novel fructan 6-exohydrolase strongly inhibited by sucrose in Lolium perenne. Planta 2014, 240, 629–643. [Google Scholar] [CrossRef]
- Yang, S.; Sun, X.; Jiang, X.; Wang, L.; Tian, J.; Li, L.; Zhao, M.; Zhong, Q. Characterization of the Tibet plateau Jerusalem artichoke (Helianthus tuberosus L.) transcriptome by de novo assembly to discover genes associated with fructan synthesis and SSR analysis. Hereditas 2019, 156, 9. [Google Scholar] [CrossRef]
- Van den Ende, W. Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 2013, 4, 247. [Google Scholar]
- Chalmers, J.; Lidgett, A.; Cummings, N.; Cao, Y.; Forster, J.; Spangenberg, G. Molecular genetics of fructan metabolism in perennial ryegrass. Plant Biotechnol. J. 2005, 3, 459–474. [Google Scholar] [CrossRef]
- Livingston, D.P.; Hincha, D.K.; Heyer, A.G. Fructan and its relationship to abiotic stress tolerance in plants. Cell. Mol. Life Sci. 2009, 66, 2007–2023. [Google Scholar] [CrossRef] [Green Version]
- Bali, V.; Panesar, P.S.; Bera, M.B.; Panesar, R. Fructo-oligosaccharides: Production, Purification and Potential Applications. Crit. Rev. Food Sci. Nutr. 2015, 55, 1475–1490. [Google Scholar] [CrossRef]
- Playne, M.J.; Crittenden, R.G. Galacto-oligosaccharides and other products derived from lactose. In Advanced Dairy Chemistry; McSweeney, P., Fox, F.P., Eds.; Lactose, water, salts and minor constituents; Springer: New York, NY, USA, 2009; Volume 3, pp. 121–201. [Google Scholar]
- Hincha, D.K.; Livingston, D.P.; Premakumar, R.; Zuther, E.; Obel, N.; Cacela, C.; Heyer, A.G. Fructans from oat and rye: Composition and effects on membrane stability during drying. Biochim. Biophys. Acta Biomembr. 2007, 1768, 1611–1619. [Google Scholar] [CrossRef] [Green Version]
- Abeynayake, S.W.; Etzerodt, T.P.; Jonavičienė, K.; Byrne, S.; Asp, T.; Boelt, B. Fructan metabolism and changes in fructan composition during cold acclimation in perennial ryegrass. Front. Plant Sci. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tochio, T.; Kadota, Y.; Tanaka, T.; Koga, Y. 1-Kestose, the smallest fructooligosaccharide component, which efficiently stimulates Faecalibacterium prausnitzii as well as Bifidobacteria in humans. Foods 2018, 7, 140. [Google Scholar] [CrossRef] [Green Version]
- De Coninck, B.; Le Roy, K.; Francis, I.; Clerens, S.; Vergauwen, R.; Halliday, A.M.; Smith, S.M.; Van Laere, A.; Van Den Ende, W. Arabidopsis AtcwINV3 and 6 are not invertases but are fructan exohydrolases (FEHs) with different substrate specificities. Plant Cell Environ. 2005, 28, 432–443. [Google Scholar] [CrossRef]
- Hendry, G.A.F. Evolutionary origins and natural functions of fructans—A climatological, biogeographic and mechanistic appraisal. New Phytol. 1993, 123, 3–14. [Google Scholar] [CrossRef]
- Hou, J.; Huang, X.; Sun, W.; Du, C.; Wang, C.; Xie, Y.; Ma, Y.; Ma, D. Accumulation of water-soluble carbohydrates and gene expression in wheat stems correlates with drought resistance. J. Plant Physiol. 2018, 231, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Marx, S.P.; Nösberger, J.; Frehner, M. Seasonal variation of fructan-β-fructosidase (FEH) activity and characterization of a β-(2-1)-linkage specific FEH from tubers of Jerusalem artichoke (Helianthus tuberosus). New Phytol. 1997, 135, 267–277. [Google Scholar] [CrossRef]
- Van Den Ende, W.; De Coninck, B.; Van Laere, A. Plant fructan exohydrolases: A role in signaling and defense? Trends Plant Sci. 2004, 9, 523–528. [Google Scholar] [CrossRef]
- Van Laere, A.; Van den Ende, W. Inulin metabolism in dicots: Chicory as a model system. Plant Cell Environ. 2002, 25, 803–813. [Google Scholar] [CrossRef]
- Chatterton, N.J.; Harrison, P.A. Fructan oligomers in Poa ampla. New Phytol. 1997, 136, 3–10. [Google Scholar] [CrossRef]
- Wang, C.; Hua, D.; Yan, C. Structural characterization and antioxidant activities of a novel fructan from Achyranthes bidentata Blume, a famous medicinal plant in China. Ind. Crops Prod. 2015, 70, 427–434. [Google Scholar] [CrossRef]
- Van Den Ende, W.; De Coninck, B.; Clerens, S.; Vergauwen, R.; Van Laere, A. Unexpected presence of fructan 6-exohydrolases (6-FEHs) in non-fructan plants: Characterization, cloning, mass mapping and functional analysis of a novel “cell-wall invertase-like” specific 6-FEH from sugar beet (Beta vulgaris L.). Plant J. 2003, 36, 697–710. [Google Scholar] [CrossRef]
- Ueno, K.; Sonoda, T.; Yoshida, M.; Shiomi, N.; Onodera, S. Purification, characterization, and functional analysis of a novel 6G&1-FEH mainly hydrolyzing neokestose from asparagus. J. Exp. Bot. 2018, 69, 4295–4308. [Google Scholar] [PubMed] [Green Version]
- Kawakami, A.; Yoshida, M. Graminan breakdown by fructan exohydrolase induced in winter wheat inoculated with snow mold. J. Plant Physiol. 2012, 169, 294–302. [Google Scholar] [CrossRef]
- Meguro-Maoka, A.; Yoshida, M. Analysis of seasonal expression levels of wheat fructan exohydrolase (FEH) genes regulating fructan metabolism involved in wintering ability. J. Plant Physiol. 2016, 191, 54–62. [Google Scholar] [CrossRef]
- Le Roy, K.; Lammens, W.; Verhaest, M.; De Coninck, B.; Rabijns, A.; Van Laere, A.; Van Den Ende, W. Unraveling the difference between invertases and fructan exohydrolases: A single amino acid (Asp-239) substitution transforms arabidopsis cell wall invertase1 into a fructan 1-exohydrolase. Plant Physiol. 2007, 145, 616–625. [Google Scholar] [CrossRef]
- Van den Ende, W.; Michiels, A.; De Roover, J.; Verhaert, P.; Van Laere, A. Cloning and functional analysis of chicory root fructan 1-exohydrolase I (1-FEH I): A vacuolar enzyme derived from a cell-wall invertase ancestor? Mass fingerprint of the 1-FEH I enzyme. Plant J. 2000, 24, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Van Den Ende, W.; Clerens, S.; Vergauwen, R.; Van Riet, L.; Van Laere, A.; Yoshida, M.; Kawakami, A. Fructan 1-exohydrolases. β-(2,1)-Trimmers during graminan biosynthesis in stems of wheat? Purification, characterization, mass mapping, and cloning of two fructan 1-exohydrolase isoforms. Plant Physiol. 2003, 131, 621–631. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Luo, W.; Wu, S.; Long, Y.; Li, R.; Zheng, F.; Greiner, S.; Rausch, T.; Zhao, H. Apoplastic maize fructan exohydrolase Zm-6-FEH displays substrate specificity for levan and is induced by exposure to levan-producing bacteria. Int. J. Biol. Macromol. 2020, 163, 630–639. [Google Scholar] [CrossRef]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef] [PubMed]
- Ritsema, T.; Hernández, L.; Verhaar, A.; Altenbach, D.; Boller, T.; Wiemken, A.; Smeekens, S. Developing fructan-synthesizing capability in a plant invertase via mutations in the sucrose-binding box. Plant J. 2006, 48, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Van Den Ende, W.; Schroeven, L.; Clerens, S.; Geuten, K.; Cheng, S.; Bennett, J. The rice genome encodes two vacuolar invertases with fructan exohydrolase activity but lacks the related fructan biosynthesis genes of the Pooideae. New Phytol. 2007, 173, 50–62. [Google Scholar] [CrossRef]
- Bach Knudsen, K.E. Carbohydrate and lignin contents of plant materials used in animal feeding. Anim. Feed Sci. Technol. 1997, 67, 319–338. [Google Scholar] [CrossRef]
- Muir, J.G.; Shepherd, S.J.; Rosella, O.; Rose, M.; Barrett, J.S.; Gibson, P.R. Fructan and free fructose content of common Australian vegetables and fruit. J. Agric. Food Chem. 2007, 55, 6619–6627. [Google Scholar] [CrossRef]
- Kim, J.Y.; Mahé, A.; Guy, S.; Brangeon, J.; Roche, O.; Chourey, P.S.; Prioul, J.L. Characterization of two members of the maize gene family, Incw3 and Incw4, encoding cell-wall invertases. Gene 2000, 245, 89–102. [Google Scholar] [CrossRef]
- Van Den Ende, W.; Lammens, W.; Van Laere, A.; Schroeven, L.; Le Roy, K. Donor and acceptor substrate selectivity among plant glycoside hydrolase family 32 enzymes. FEBS J. 2009, 276, 5788–5798. [Google Scholar] [CrossRef]
- Verhaest, M.; Lammens, W.; Le Roy, K.; De Ranter, C.J.; Van Laere, A.; Rabijns, A.; Van Den Ende, W. Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-Kestose as substrate vs sucrose as inhibitor. New Phytol. 2007, 174, 90–100. [Google Scholar] [CrossRef]
- Bizzarri, M.; Delledonne, M.; Ferrarini, A.; Tononi, P.; Zago, E.; Vittori, D.; Damiani, F.; Paolocci, F. Whole-Transcriptome Analysis Unveils the Synchronized Activities of Genes for Fructans in Developing Tubers of the Jerusalem Artichoke. Front. Plant Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Rigui, A.P.; Carvalho, V.; Wendt dos Santos, A.L.; Morvan-Bertrand, A.; Prud’homme, M.P.; Machado de Carvalho, M.A.; Gaspar, M. Fructan and antioxidant metabolisms in plants of Lolium perenne under drought are modulated by exogenous nitric oxide. Plant Physiol. Biochem. 2019, 145, 205–215. [Google Scholar] [CrossRef]
- Trouverie, J.; Chateau-Joubert, S.; Thévenot, C.; Jacquemot, M.P.; Prioul, J.L. Regulation of vacuolar invertase by abscisic acid or glucose in leaves and roots from maize plantlets. Planta 2004, 219, 894–905. [Google Scholar] [CrossRef]
- Livingston, D.P.; Henson, C.A. Apoplastic sugars, fructans, fructan exohydrolase, and invertase in winter oat: Responses to second-phase cold hardening. Plant Physiol. 1998, 116, 403–408. [Google Scholar] [CrossRef] [Green Version]
- Blanch, M.; Sanchez-Ballesta, M.T.; Escribano, M.I.; Merodio, C. Fructo-oligosaccharides in table grapes and response to storage. Food Chem. 2011, 129, 724–730. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Guo, M.; Zhao, W.; Chen, K.; Zhang, P. Burdock fructooligosaccharide induces stomatal closure in Pisum sativum. Carbohydr. Polym. 2013, 97, 731–735. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, P.; et al. ClustalW and ClustalX version 2. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Zhao, H.; Su, T.; Bausewein, A.; Greiner, S.; Harms, K.; Rausch, T. Chicory R2R3-MYB transcription factors CiMYB5 and CiMYB3 regulate fructan 1-exohydrolase expression in response to abiotic stress and hormonal cues. J. Exp. Bot. 2017, 68, 4323–4338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, T.; Han, M.; Min, J.; Zhou, H.; Zhang, Q.; Zhao, J.; Fang, Y. Functional Characterization of Invertase Inhibitors PtC/VIF1 and 2 Revealed Their Involvements in the Defense Response to Fungal Pathogen in Populus trichocarpa. Front. Plant Sci. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Greiner, S.; Scheffzek, K.; Rausch, T.; Wang, G. A 6&1-FEH encodes an enzyme for fructan degradation and interact with invertase inhibitor protein in maize (Zea mays L.). Int. J. Mol. Sci. 2019, 20, 1–17. [Google Scholar]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Greiner, S.; Ma, C.; Zhong, J.; Huang, X.; Rausch, T.; Zhao, H. A Fructan Exohydrolase from Maize Degrades Both Inulin and Levan and Co-Exists with 1-Kestotriose in Maize. Int. J. Mol. Sci. 2021, 22, 5149. https://doi.org/10.3390/ijms22105149
Wu S, Greiner S, Ma C, Zhong J, Huang X, Rausch T, Zhao H. A Fructan Exohydrolase from Maize Degrades Both Inulin and Levan and Co-Exists with 1-Kestotriose in Maize. International Journal of Molecular Sciences. 2021; 22(10):5149. https://doi.org/10.3390/ijms22105149
Chicago/Turabian StyleWu, Silin, Steffen Greiner, Chongjian Ma, Jiaxin Zhong, Xiaojia Huang, Thomas Rausch, and Hongbo Zhao. 2021. "A Fructan Exohydrolase from Maize Degrades Both Inulin and Levan and Co-Exists with 1-Kestotriose in Maize" International Journal of Molecular Sciences 22, no. 10: 5149. https://doi.org/10.3390/ijms22105149
APA StyleWu, S., Greiner, S., Ma, C., Zhong, J., Huang, X., Rausch, T., & Zhao, H. (2021). A Fructan Exohydrolase from Maize Degrades Both Inulin and Levan and Co-Exists with 1-Kestotriose in Maize. International Journal of Molecular Sciences, 22(10), 5149. https://doi.org/10.3390/ijms22105149




