Cell–Cell Communication at the Embryo Implantation Site of Mouse Uterus Revealed by Single-Cell Analysis
Abstract
:1. Introduction
2. Results
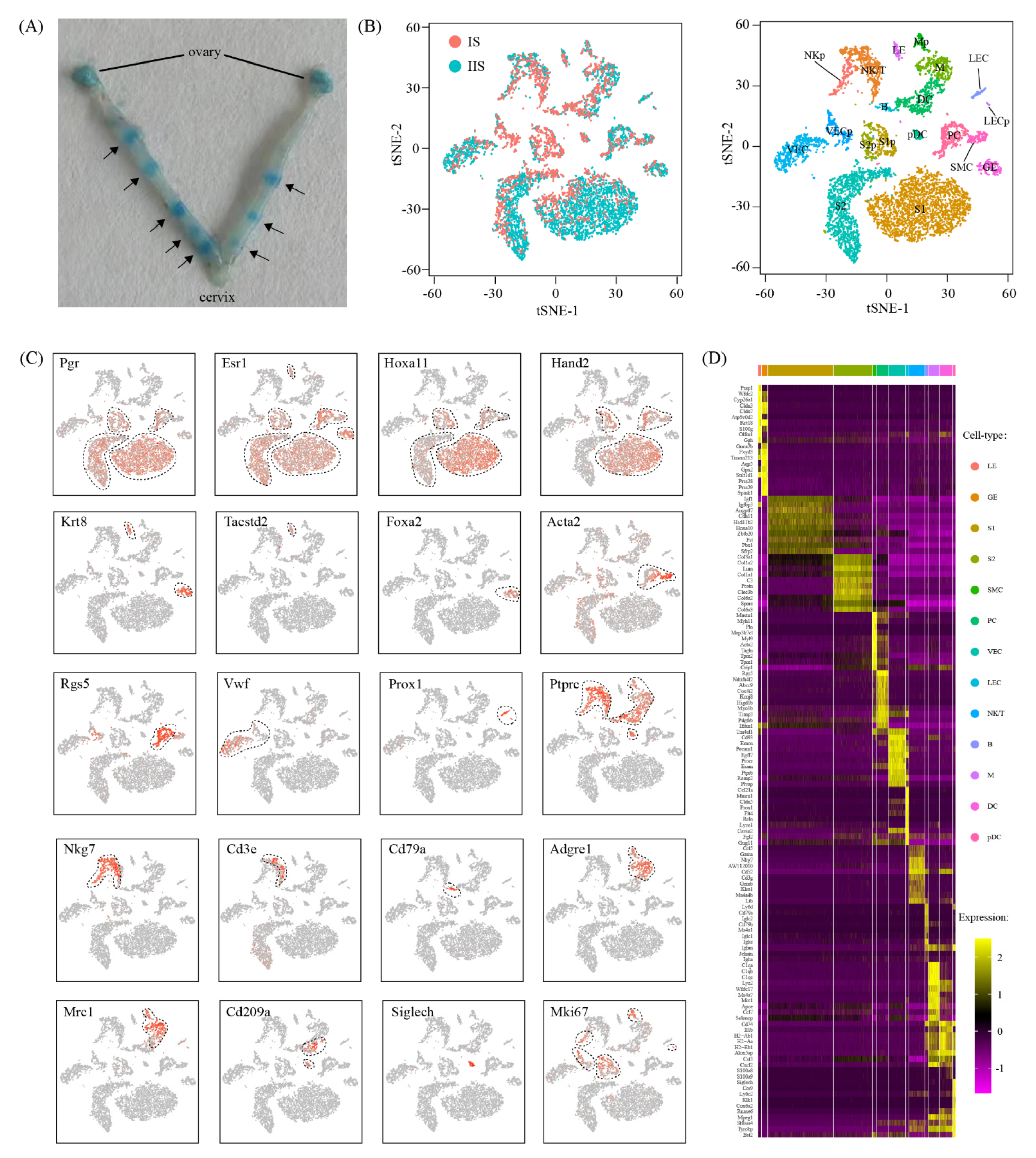
2.1. Single-Cell Analysis of Embryo Implantation Site in Mouse Uterus
2.2. Cell Type Proportion Changes at Implantation Site Compared to Inter-Implantation Site
2.3. Cell-Type-Specific Transcriptional Changes Associated with Embryo Implantation
2.4. Inferring Cell–Cell Communication between Blastocyst and Uterus at the Implantation Site
2.5. Inferring Uterine Epithelial-Stromal Crosstalk at the Implantation Site
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Single-Cell Dissociation of Mouse Uterus
4.3. Single-Cell RNA-Seq Library Preparation and Sequencing
4.4. Single-Cell RNA-Seq Data Analysis
4.5. Gene Ontology Analysis
4.6. Pathway Enrichment Analysis
4.7. Cell–Cell Communication Analysis
4.8. Pseudotime Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wilcox, L.S.; Peterson, H.B.; Haseltine, F.P.; Martin, M.C. Defining and interpreting pregnancy success rates for in vitro fertilization. Fertil. Steril. 1993, 60, 18–25. [Google Scholar] [CrossRef]
- Zinaman, M.J.; Clegg, E.D.; Brown, C.C.; O’Connor, J.; Selevan, S.G. Estimates of human fertility and pregnancy loss. Fertil. Steril. 1996, 65, 503–509. [Google Scholar] [CrossRef]
- Wang, H.; Dey, S.K. Roadmap to embryo implantation: Clues from mouse models. Nat. Rev. Genet. 2006, 7, 185–199. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, H.; Kong, S.; Wang, S.; Wang, H.; Wang, H.; Armant, D.R. Physiological and molecular determinants of embryo implantation. Mol. Asp. Med. 2013, 34, 939–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, J.; Sun, X.; Dey, S.K. Mechanisms of implantation: Strategies for successful pregnancy. Nat. Med. 2012, 18, 1754–1767. [Google Scholar] [CrossRef]
- Ma, X.H.; Hu, S.J.; Ni, H.; Zhao, Y.C.; Tian, Z.; Liu, J.L.; Ren, G.; Liang, X.H.; Yu, H.; Wan, P.; et al. Serial analysis of gene expression in mouse uterus at the implantation site. J. Biol. Chem. 2006, 281, 9351–9360. [Google Scholar] [CrossRef] [Green Version]
- Reese, J.; Das, S.K.; Paria, B.C.; Lim, H.; Song, H.; Matsumoto, H.; Knudtson, K.L.; DuBois, R.N.; Dey, S.K. Global gene expression analysis to identify molecular markers of uterine receptivity and embryo implantation. J. Biol. Chem. 2001, 276, 44137–44145. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.Q.; Zhao, M.; Huang, M.Y.; Liu, J.L. Comparative transcriptomic analysis of embryo implantation in mice and rats. Cell. Physiol. Biochem. 2018, 50, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ni, H.; Ma, X.H.; Hu, S.J.; Luan, L.M.; Ren, G.; Zhao, Y.C.; Li, S.J.; Diao, H.L.; Xu, X.; et al. Global analysis of differential luminal epithelial gene expression at mouse implantation sites. J. Mol. Endocrinol. 2006, 37, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.J.; Choi, D.H.; Lee, W.S.; Cha, K.Y.; Kim, S.N.; Lee, K.A. A molecular basis for embryo apposition at the luminal epithelium. Mol. Cell. Endocrinol. 2004, 219, 95–104. [Google Scholar] [CrossRef]
- Svensson, V.; Vento-Tormo, R.; Teichmann, S.A. Exponential scaling of single-cell rna-seq in the past decade. Nat. Protoc. 2018, 13, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Gendron, R.L.; Paradis, H.; Hsieh-Li, H.M.; Lee, D.W.; Potter, S.S.; Markoff, E. Abnormal uterine stromal and glandular function associated with maternal reproductive defects in hoxa-11 null mice. Biol. Reprod. 1997, 56, 1097–1105. [Google Scholar] [CrossRef]
- Reardon, S.N.; King, M.L.; MacLean, J.A., 2nd; Mann, J.L.; DeMayo, F.J.; Lydon, J.P.; Hayashi, K. Cdh1 is essential for endometrial differentiation, gland development, and adult function in the mouse uterus. Biol. Reprod. 2012, 86, 141. [Google Scholar] [CrossRef] [PubMed]
- Suryawanshi, H.; Morozov, P.; Straus, A.; Sahasrabudhe, N.; Max, K.E.A.; Garzia, A.; Kustagi, M.; Tuschl, T.; Williams, Z. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018, 4, eaau4788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mucenski, M.L.; Mahoney, R.; Adam, M.; Potter, A.S.; Potter, S.S. Single cell rna-seq study of wild type and hox9,10,11 mutant developing uterus. Sci. Rep. 2019, 9, 4557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalucka, J.; de Rooij, L.; Goveia, J.; Rohlenova, K.; Dumas, S.J.; Meta, E.; Conchinha, N.V.; Taverna, F.; Teuwen, L.A.; Veys, K.; et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 2020, 180, 764–779 e720. [Google Scholar] [CrossRef]
- Croy, B.A.; Chen, Z.; Hofmann, A.P.; Lord, E.M.; Sedlacek, A.L.; Gerber, S.A. Imaging of vascular development in early mouse decidua and its association with leukocytes and trophoblasts. Biol. Reprod. 2012, 87, 125. [Google Scholar] [CrossRef]
- Li, Q.; Kannan, A.; DeMayo, F.J.; Lydon, J.P.; Cooke, P.S.; Yamagishi, H.; Srivastava, D.; Bagchi, M.K.; Bagchi, I.C. The antiproliferative action of progesterone in uterine epithelium is mediated by hand2. Science 2011, 331, 912–916. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.J.; De Micheli, A.J.; Bidarimath, M.; Ellenson, L.H.; Cosgrove, B.D.; Flesken-Nikitin, A.; Nikitin, A.Y. Cells expressing pax8 are the main source of homeostatic regeneration of adult mouse endometrial epithelium and give rise to serous endometrial carcinoma. Dis. Model. Mech. 2020, 13. [Google Scholar] [CrossRef]
- Keenihan, S.N.; Robertson, S.A. Diversity in phenotype and steroid hormone dependence in dendritic cells and macrophages in the mouse uterus. Biol. Reprod. 2004, 70, 1562–1572. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Gaugler, B.; Mohty, M.; Malard, F. Plasmacytoid dendritic cell biology and its role in immune-mediated diseases. Clin. Transl. Immunol. 2020, 9, e1139. [Google Scholar] [CrossRef] [PubMed]
- Hilton, H.G.; Rubinstein, N.D.; Janki, P.; Ireland, A.T.; Bernstein, N.; Fong, N.L.; Wright, K.M.; Smith, M.; Finkle, D.; Martin-McNulty, B.; et al. Single-cell transcriptomics of the naked mole-rat reveals unexpected features of mammalian immunity. PLoS Biol. 2019, 17, e3000528. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yabuta, Y.; Okamoto, I.; Aramaki, S.; Yokobayashi, S.; Kurimoto, K.; Sekiguchi, K.; Nakagawa, M.; Yamamoto, T.; Saitou, M. Sc3-seq: A method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res. 2015, 43, e60. [Google Scholar] [CrossRef] [PubMed]
- Hantak, A.M.; Bagchi, I.C.; Bagchi, M.K. Role of uterine stromal-epithelial crosstalk in embryo implantation. Int. J. Dev. Biol. 2014, 58, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Dewar, A.; Kim, Y.S.; Dey, S.K.; Sun, X. Pregnancy success in mice requires appropriate cannabinoid receptor signaling for primary decidua formation. Elife 2020, 9, e61762. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Aikawa, S.; Deng, W.; Bartos, A.; Walz, G.; Grahammer, F.; Huber, T.B.; Sun, X.; Dey, S.K. Primary decidual zone formation requires scribble for pregnancy success in mice. Nat. Commun. 2019, 10, 5425. [Google Scholar] [CrossRef] [Green Version]
- Ye, X. Uterine luminal epithelium as the transient gateway for embryo implantation. Trends Endocrinol. Metab. 2020, 31, 165–180. [Google Scholar] [CrossRef]
- Zenclussen, A.C.; Hammerling, G.J. Cellular regulation of the uterine microenvironment that enables embryo implantation. Front. Immunol. 2015, 6, 321. [Google Scholar] [CrossRef] [Green Version]
- Kelleher, A.M.; DeMayo, F.J.; Spencer, T.E. Uterine glands: Developmental biology and functional roles in pregnancy. Endocr. Rev. 2019, 40, 1424–1445. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive integration of single-cell data. Cell 2019, 177, 1888–1902.e1821. [Google Scholar] [CrossRef]
- Liu, J.L.; Zuo, R.J.; Peng, Y.; Fu, Y.S. The impact of multiparity on uterine gene expression and decidualization in mice. Reprod. Sci. 2016, 23, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Law, M.; Shaw, D.R. Mouse genome informatics (MGI) is the international resource for information on the laboratory mouse. Methods Mol. Biol. 2018, 1757, 141–161. [Google Scholar] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. David bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. Cellphonedb: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.A.; Trapnell, C. Single-cell mrna quantification and differential analysis with census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; He, J.-P.; Liu, J.-L. Cell–Cell Communication at the Embryo Implantation Site of Mouse Uterus Revealed by Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 5177. https://doi.org/10.3390/ijms22105177
Yang Y, He J-P, Liu J-L. Cell–Cell Communication at the Embryo Implantation Site of Mouse Uterus Revealed by Single-Cell Analysis. International Journal of Molecular Sciences. 2021; 22(10):5177. https://doi.org/10.3390/ijms22105177
Chicago/Turabian StyleYang, Yi, Jia-Peng He, and Ji-Long Liu. 2021. "Cell–Cell Communication at the Embryo Implantation Site of Mouse Uterus Revealed by Single-Cell Analysis" International Journal of Molecular Sciences 22, no. 10: 5177. https://doi.org/10.3390/ijms22105177







