Genome-Wide Identification and Analysis of the APETALA2 (AP2) Transcription Factor in Dendrobium officinale
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of AP2 Gene Family in D. officinale
2.2. Prediction of Protein–Protein Interaction Network of AP2 Proteins
2.3. Analysis of Cis-Regulatory Elements in the Promoters of DoAP2 Genes
2.4. Expression Analysis of DoAP2 Genes at Different Developmental Stages
2.5. Expression Analysis of DoAP2 Genes in Response to Abiotic Stresses
2.6. Subcellular Localization of Selected DoAP2 Proteins
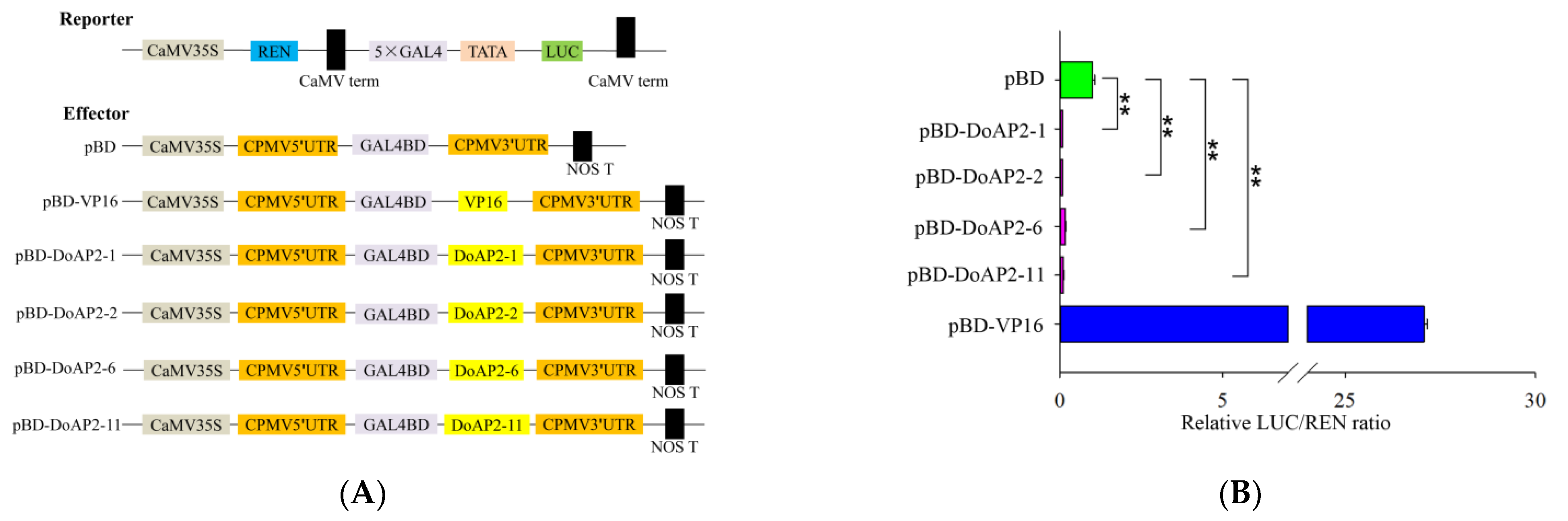
2.7. Four DoAP2 Proteins Displayed Transcriptional Repression in Tobacco Leaves
3. Discussion
3.1. Bioinformatics Analysis of DoAP2 TFs
3.2. The AP2 TF Play Important Roles in Plant Regeneration and Flower Development
3.3. DoAP2 Genes May Play a Role in Abiotic Stress Response
3.4. The DoAP2 Proteins Are Localized in the Nucleus and Display Transcription Activity
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Identification of DoAP2 Genes from the D. officinale Genome
4.3. Bioinformatics Analysis of DoAP2 Proteins
4.4. RNA Extraction, cDNA Synthesis and qRT-PCR
4.5. Subcellular Localization Analysis
4.6. Dual-Luciferase Reporter (DLR) Assay
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 1/2MS | half-strength Murashige and Skoog (1962) medium |
| AD | transcriptional activation domain |
| AP2 | APETALA2 |
| AP2/ERF | APETALA2/Ethylene Response Factor |
| BD | DNA-binding domain |
| CDD | conserved domain database |
| Co | column |
| DBD | DNA binding domain |
| EREB | ethylene response element binding factor |
| FB1 | small flower bud |
| FB2 | medium flower bud |
| FBF | fully bloomed flower |
| HMM | Hidden Markov mode |
| Li | lip |
| LUC | firefly luciferase |
| MEGA | Molecular Evolutionary Genetics Analysis |
| MS | multiple shoots |
| PEG | polyethylene glycol |
| PLB | protocorm-like body |
| Pe | petal |
| qRT-PCR | quantificational real-time polymerase chain reaction |
| REN | renilla luciferase |
| Se | sepal |
| TF | transcription factor |
| YFP | yellow fluorescent protein |
References
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Weirauch, M.T. The human transcription factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.; Gerentes, D.; Smyth, P. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Campagne, M.M.V.L. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef]
- Graaff, E. A new role for the Arabidopsis AP2 transcription factor, LEAFY PETIOLE, in gibberellin-induced germination is revealed by the misexpression of a homologous gene, SOB2/DRN-LIKE. Plant Cell 2006, 18, 29–39. [Google Scholar] [CrossRef][Green Version]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Koichiro, A.; Tokunori, H.; Kanna, S.I.; Miyako, U.T.; Hidemi, K.; Makoto, M. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol. 2014, 897–912. [Google Scholar] [CrossRef]
- Chuck, G.; Meeley, R.B.; Hake, S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 1998, 12, 1145. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Florez, S.L.; Erwin, R.L.; Maximova, S.N.; Guiltinan, M.J.; Curtis, W.R. Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC Plant Biol. 2015, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef]
- Chandler, J.W. Class VIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci. 2017, 23, 151–162. [Google Scholar] [CrossRef]
- Shigyo, M.; Hasebe, M.; Ito, M. Molecular evolution of the AP2 subfamily. Gene 2006, 366, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Ohto, M.A.; Fischer, R.L.; Goldberg, R.B.; Nakamura, K.; Harada, J.J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3123–3128. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef]
- Zhuang, J.; Cai, B.; Peng, R.-H.; Zhu, B.; Jin, X.-F.; Xue, Y.; Gao, F.; Fu, X.-Y.; Tian, Y.-S.; Zhao, W.; et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Shigyo, M.; Ito, M. Analysis of gymnosperm two-AP2-domain-containing genes. Dev. Genes Evol. 2004, 214, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Willemsen, V.; Boutilier, K.; Heidstra, R. AINTEGUMENTA-LIKE proteins: Hubs in a plethora of networks. Trends Plant Sci. 2014, 19, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Soltis, P.S.; Wall, K.; Soltis, D.E. Phylogeny and domain Evolution in the APETALA2-like gene family. Mol. Biol. Evol. 2006, 23, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Komaki, M.K.; Okada, K.; Nishino, E.; Shimura, Y. Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development 1988, 104, 1536–1546. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell Online 1989, 1, 37–52. [Google Scholar] [CrossRef]
- Kunst, L.; Klenz, J.E.; Haughn, M. AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1989, 1, 1195–1208. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef]
- Krizek, B.A. AINTEGUMENTA-LIKE genes have partly overlapping functions with AINTEGUMENTA but make distinct contributions to Arabidopsis thaliana flower development. J. Exp. Bot. 2015, 66, 4537–4549. [Google Scholar] [CrossRef] [PubMed]
- Klucher, K.M.; Chow, H.; Fischer, R. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell Online 1996, 8, 137–153. [Google Scholar] [CrossRef][Green Version]
- Dash, M.; Malladi, A. The AINTEGUMENTA genes, MdANT1 and MdANT2, are associated with the regulation of cell production during fruit growth in apple (Malus × domestica Borkh.). BMC Plant Biol. 2012, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chialva, C.; Eichler, E.; Grissi, C.; Mu?Oz, C.; Gomez-Talquenca, S.; Martínez-Zapater, J.; Lijavetzky, D. Expression of grapevine AINTEGUMENTA-like genes is associated with variation in ovary and berry size. Plant Mol. Biol. 2016, 91, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Ma, X.; Zhao, S.; Tang, Y.; Liu, F.; Gu, P.; Fu, Y.; Zhu, Z.; Cai, H.; Sun, C. The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size. Plant Cell 2019, 31, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.K.; Li, X.; Xu, Q.; Chen, J.Y.; Yin, X.R.; Ferguson, I.B.; Chen, K.S. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 2016, 13, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Chuck, G.; Meeley, R.; Hake, S. Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 2008, 135, 3013–3019. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Boer, B.G.d.; Montagu, M.V.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.J.; Choi, Y.D. Methyl jasmonate as a vital substance in plants. Trends Genet. 2003, 19, 409–413. [Google Scholar] [CrossRef]
- Krizek, B.A. AINTEGUMENTA and AINTEGUMENTA-like6 act redundantly to regulate arabidopsis floral growth and patterning. Plant Physiol. 2009, 150, 1916–1929. [Google Scholar] [CrossRef]
- Rudall, P.J.; Bateman, R.M. Roles of synorganisation, zygomorphy and heterotopy in floral evolution: The gynostemium and labellum of orchids and other lilioid monocots. Biol. Rev. 2010, 77, 403–441. [Google Scholar] [CrossRef] [PubMed]
- Hawku, M.D.; Goher, F.; Islam, M.A.; Guo, J.; He, F.; Bai, X.; Yuan, P.; Kang, Z.; Guo, J. TaAP2-15, An AP2/ERF transcription factor, is positively involved in wheat resistance to Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2021, 22, 2080. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Ahmeda, S.; Rashid, M.A.R.; Zafar, S.A.; Azhar, M.T.; Waqas, M.; Uzair, M.; Rana, I.A.; Azeem, F.; Chung, G.; Ali, Z.; et al. Genome-wide investigation and expression analysis of APETALA-2 transcription factor subfamily reveals its evolution, expansion and regulatory role in abiotic stress responses in Indica rice (Oryza sativa L. ssp. indica). Genomics 2020, 113, 1029–1043. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, R.; Xu, D.; Bi, H.; Xia, Z.; Peng, H. Genome-wide identification and analysis of the AP2 transcription factor gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 1286. [Google Scholar] [CrossRef] [PubMed]
- Dipp-Álvarez, M.; Cruz-Ramírez, A. A phylogenetic study of the ANT family points to a preANT gene as the ancestor of basal and euANT transcription factors in land plants. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Zhao, Y.; Cheng, S.; Song, Y.; Huang, Y.; Zhou, S.; Liu, X.; Zhou, D.X. The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 2015, 27, 2469–2483. [Google Scholar] [CrossRef]
- Diaz-Martin, J.; Almoguera, C.N.; Prieto-Dapena, P.; Espinosa, J.M.; Jordano, J. Functional interaction between two transcription factors involved in the developmental regulation of a small heat stress protein gene promoter. Plant Physiol. 2005, 139, 1483–1494. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Son, G.H.; Wan, J.; Kim, H.J.; Xuan Canh, N.; Chung, W.S.; Hong, J.C.; Stacey, G. Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe Interact. 2012, 25, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Moffat, C.S.; Ingle, R.A.; Wathugala, D.L.; Saunders, N.J.; Knight, H.; Knight, M.R. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against botrytis cinerea in Arabidopsis. PLoS ONE 2012, 7, e35995. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar] [CrossRef]
- Shukla, R.K.; Raha, S.; Tripathi, V.; Chattopadhyay, D. Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol. 2006, 142, 113–123. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Chen, S.; Jiang, J.; Teng, N.; Chen, Y.; Chen, F. The AP2-like gene NsAP2 from water lily is involved in floral organogenesis and plant height. J. Plant Physiol. 2012, 169, 992–998. [Google Scholar] [CrossRef]
- Shoji, T.; Mishima, M.; Hashimoto, T. Divergent DNA-binding specificities of a group of ETHYLENE RESPONSE FACTOR transcription factors involved in plant defense. Plant Physiol. 2013, 162, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qin, L.; Liu, W.; Zhang, D.; Wang, Y. A novel ethylene-responsive factor from Tamarix hispida, ThERF1, is a GCC-box- and DRE-motif binding protein that negatively modulates abiotic stress tolerance in Arabidopsis. Physiol. Plant. 2014, 152, 84–97. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. Regulation of Apetala2/Ethylene response factors in plants. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Che, P.; Lall, S.; Nettleton, D.; Howell, S.H. Gene expression programs during shoot, root and callus development in Arabidopsis tissue culture. Plant Biol. 2006, 2006, 282–283. [Google Scholar] [CrossRef]
- Du, X.; Fang, T.; Liu, Y.; Huang, L.; Zang, M.; Wang, G.; Liu, Y.; Fu, J. Transcriptome profiling predicts new genes to promote maize callus formation and transformation. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Piyatrakul, P.; Putranto, R.-A.; Martin, F.; Rio, M.; Dessailly, F.; Leclercq, J.; Dufayard, J.-F.; Lardet, L.; Montoro, P. Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis. BMC Plant Biol. 2012, 12, 1–20. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, D.M.; Lee, J.H.; Kim, J.; Bang, J.W.; Kim, W.T.; Pai, H.S. Functional characterization of NtCEF1, an AP2/EREBP-type transcriptional activator highly expressed in tobacco callus. Planta 2005, 222, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Harashima, H.; Ikeuchi, M.; Rymen, B.; Ohnuma, M.; Komaki, S.; Morohashi, K.; Kurata, T.; Nakata, M.; Ohme-Takagi, M.; et al. WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 2017, 29, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.-B.; Shang, G.-D.; Pan, Y.; Xu, Z.-G.; Zhou, C.-M.; Mao, Y.-B.; Bao, N.; Sun, L.; Xu, T.; Wang, J.-W. AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 2020, 32, 226–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhao, L.; Yang, X.; Li, M.; Sun, J.; Wang, K.; Li, Y.; Zheng, Y.; Yao, Y.; Li, W. GmRAV1 regulates regeneration of roots and adventitious buds by the cytokinin signaling pathway in Arabidopsis and soybean. Physiol. Plant. 2019, 165, 814–829. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Hsu, S.; Yeung, E.C. Orchid protocorm-like bodies are somatic embryos. Am. J. Bot. 2013, 100, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Teixeira da Silva, J.A.; Cardoso, J.C.; Dobránszki, J.; Zeng, S. Dendrobium micropropagation: A review. Plant Cell Rep. 2015, 34, 671–704. [Google Scholar] [CrossRef]
- Ripoll, J.J.; Roeder, A.; Ditta, G.S.; Yanofsky, M.F. A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 2011, 138, 5167–5176. [Google Scholar] [CrossRef]
- Samad, A.; Muhammad, S.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.; Zamri, Z.; Ismanizan, I. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Si, C.; Teixeira da Silva, J.A.; Li, M.; Duan, J. Genome-wide identification and classification of MIKC-type MADS-box genes in Streptophyte lineages and expression analyses to reveal their role in seed germination of orchid. BMC Plant Biol. 2019, 19, 1–15. [Google Scholar] [CrossRef]
- Yu, H.; Goh, C.J. Molecular genetics of reproductive biology in orchids. Plant Physiol. 2001, 127, 1390–1393. [Google Scholar] [CrossRef]
- Cao, Y.; Song, F.; Goodman, R.M.; Zheng, Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J. Plant Physiol. 2006, 163, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, D.; Ishida, S.; Fukazawa, J.; Takahashi, Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 2001, 13, 2483–2497. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Phukan, U.J.; Tripathi, V.; Singh, D.K.; Luqman, S.; Shukla, R.K. PsAP2 an AP2/ERF family transcription factor from Papaver somniferum enhances abiotic and biotic stress tolerance in transgenic tobacco. Plant Mol. Biol. 2015, 89, 173–186. [Google Scholar] [CrossRef]
- Chen, J.-Q.; Dong, Y.; Wang, Y.-J.; Liu, Q.; Zhang, J.-S.; Chen, S.-Y. An AP2/EREBP-type transcription-factor gene from rice is cold-inducible and encodes a nuclear-localized protein. Theor. Appl. Genet. 2003, 107, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Duan, X.; Ding, X.; Chen, C.; Zhu, D.; Yin, K.; Cao, L.; Song, X.; Zhu, P.; Li, Q.; et al. A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol. Biol. 2017, 94, 509–530. [Google Scholar] [CrossRef]
- Thiel, G.; Lietz, M.; Hohl, M. How mammalian transcriptional repressors work. Eur. J. Biochem. 2004, 271, 2855–2862. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006, 11, 109–112. [Google Scholar] [CrossRef]
- Yaish, M.W.; El-kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.-M.; Rothstein, S.J. The APETALA-2-Like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef] [PubMed]
- Pan, I.C.; Li, C.-W.; Su, R.-C.; Cheng, C.-P.; Lin, C.-S.; Chan, M.-T. Ectopic expression of an EAR motif deletion mutant of SlERF3 enhances tolerance to salt stress and Ralstonia solanacearum in tomato. Planta 2010, 232, 1075–1086. [Google Scholar] [CrossRef]
- Drews, G.N.; Bowman, J.L.; Meyerowitz, E.M. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 1991, 65, 991–1002. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Omidyar, P.K.; Gee, Z.; Okamuro, J.K. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3117–3122. [Google Scholar] [CrossRef]
- Zhou, Y.; Lu, D.; Li, C.; Luo, J.; Zhu, B.-F.; Zhu, J.; Shangguan, Y.; Wang, Z.; Sang, T.; Zhou, B.; et al. Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 2012, 24, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.S.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- He, C.; Zeng, S.; Teixeira da Silva, J.A.; Yu, Z.; Tan, J.; Duan, J. Molecular cloning and functional analysis of the phosphomannomutase (PMM) gene from Dendrobium officinale and evidence for the involvement of an abiotic stress response during germination. Protoplasma 2017, 254, 1693–1704. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method - a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhang, J.; Liu, X.; Zeng, S.; Wu, K.; Yu, Z.; Wang, X.; Teixeira da Silva, J.A.; Lin, Z.; Duan, J. Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol. 2015, 88, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Citovsky, V.; Lee, L.-Y.; Vyas, S.; Glick, E.; Chen, M.-H.; Vainstein, A.; Gafni, Y.; Gelvin, S.B.; Tzfira, T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 2006, 362, 1120–1131. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-C.; Kuang, J.-F.; Chen, J.-Y.; Liu, X.-C.; Xiao, Y.-Y.; Fu, C.-C.; Wang, J.-N.; Wu, K.-Q.; Lu, W.-J. Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and Represses the expression of MaACO1 and Expansins during fruit ripening. Plant Physiol. 2016, 171, 1070–1084. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, D.; Teixeira da Silva, J.A.; Zhang, M.; Yu, Z.; Si, C.; Zhao, C.; Dai, G.; He, C.; Duan, J. Genome-Wide Identification and Analysis of the APETALA2 (AP2) Transcription Factor in Dendrobium officinale. Int. J. Mol. Sci. 2021, 22, 5221. https://doi.org/10.3390/ijms22105221
Zeng D, Teixeira da Silva JA, Zhang M, Yu Z, Si C, Zhao C, Dai G, He C, Duan J. Genome-Wide Identification and Analysis of the APETALA2 (AP2) Transcription Factor in Dendrobium officinale. International Journal of Molecular Sciences. 2021; 22(10):5221. https://doi.org/10.3390/ijms22105221
Chicago/Turabian StyleZeng, Danqi, Jaime A. Teixeira da Silva, Mingze Zhang, Zhenming Yu, Can Si, Conghui Zhao, Guangyi Dai, Chunmei He, and Jun Duan. 2021. "Genome-Wide Identification and Analysis of the APETALA2 (AP2) Transcription Factor in Dendrobium officinale" International Journal of Molecular Sciences 22, no. 10: 5221. https://doi.org/10.3390/ijms22105221
APA StyleZeng, D., Teixeira da Silva, J. A., Zhang, M., Yu, Z., Si, C., Zhao, C., Dai, G., He, C., & Duan, J. (2021). Genome-Wide Identification and Analysis of the APETALA2 (AP2) Transcription Factor in Dendrobium officinale. International Journal of Molecular Sciences, 22(10), 5221. https://doi.org/10.3390/ijms22105221





