Viral Interactions with Adaptor-Protein Complexes: A Ubiquitous Trait among Viral Species
Abstract
:1. Introduction
2. Main Body
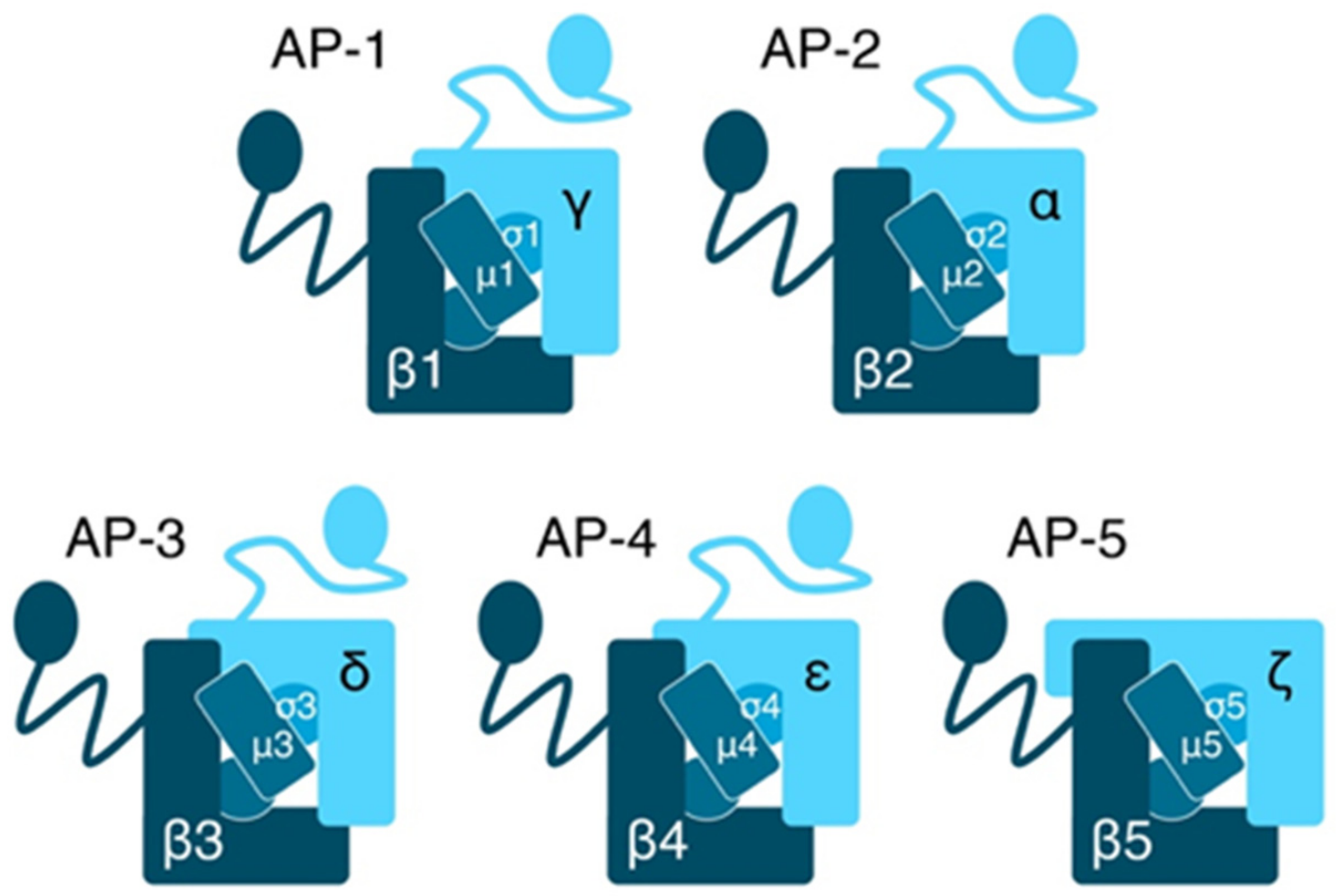
2.1. Composition and Function of AP Complexes
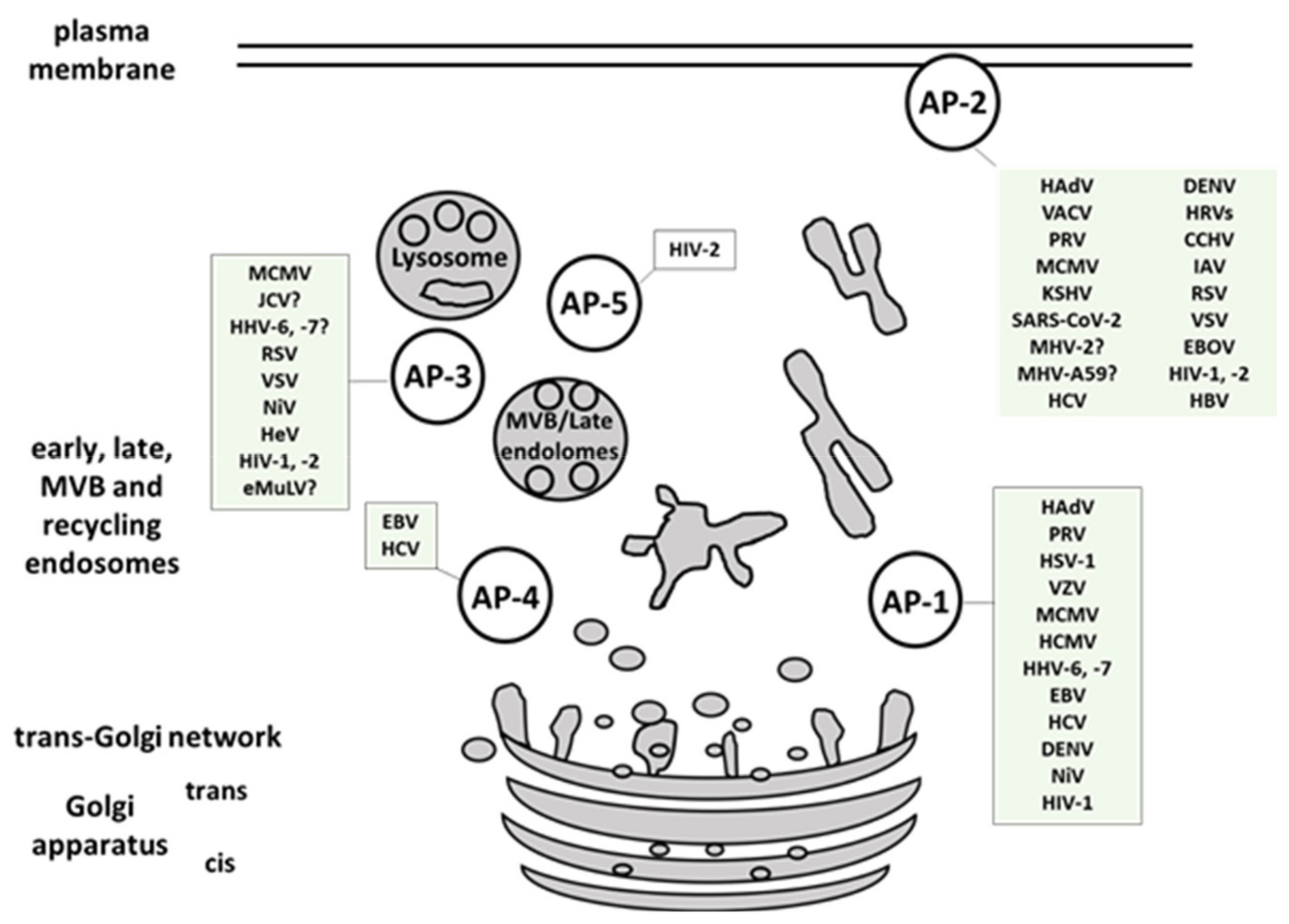
2.2. Viral Interaction with AP Complexes
2.2.1. Adenoviruses
2.2.2. Herpesviruses
Alphaherpesviruses
Betaherpesviruses
Gammaherpesviruses
2.2.3. Human JC Polyomavirus (JCV)
2.2.4. Vaccinia Virus (VACV)
2.2.5. Coronaviridae
2.2.6. Flaviviridae
Dengue Virus (DENV)
Hepatitis C Virus (HCV)
2.2.7. Human Rhinoviruses (HRVs)
2.2.8. Zaire Ebolavirus (Common Ebola Virus (EBOV))
2.2.9. Crimean–Congo Hemorrhagic Fever Virus (CCHFV)
2.2.10. Influenza A Virus (IAV)
2.2.11. Paramyxoviridae
2.2.12. Human Respiratory Syncytial Virus (RSV)
2.2.13. Indiana Vesiculovirus (Former Vesicular Stomatitis Indiana Virus (VSIV or VSV))
2.2.14. Retroviridae
Ecotropic Murine Leukemia Virus (eMuLV)
Human Immunodeficiency Virus Type 1 (HIV-1)
Human Immunodeficiency Virus Type 2 (HIV-2)
2.2.15. Hepatitis B Virus (HBV)
3. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| AMPA | α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| ACA | N-(p-amylcinnamoyl) anthranilic acid |
| AAK-1 | adaptor-associated protein kinase 1 |
| AP | adaptor-protein-complex |
| ADP | adenosine 5′-diphosphate |
| Arf | ADP-ribosylation factor |
| ACE2 | angiotensin converting enzyme 2 |
| AIDS | acquired immunodeficiency syndrome |
| ATG9A | autophagy-related protein 9A |
| BSL | biosafety level |
| BST2 | bone Marrow Stromal Cell Antigen 2 |
| BIKE | bone morphogenic protein 2-inducible kinase |
| CAT-1 | cationic amino acid transporter |
| CALM | clathrin-assembly lymphoid myeloid leukemia protein |
| CME | clathrin-mediated endocytosis |
| CCHFV | Crimean-Congo hemorrhagic fever virus |
| GAK | cyclin G-associated kinase |
| DENV | Dengue virus |
| EBOV | Ebola virus |
| eMuLV | Ecotropic murine leukemia virus |
| EGFR | epidermal growth factor receptor |
| Eps15 | epidermal growth factor receptor substrate 15 |
| EBV | Epstein-Barr virus |
| EXOC6 | exocyst complex component 6 |
| FFAR2 | free fatty acid receptor 2 |
| FDA | Food and Drug Administration |
| GPI | glycosylphosphatidylinositol |
| GAP | GTPase-activating proteins |
| GTP | guanosine-5’-triphosphate |
| HA | hemagglutinin |
| HeV | Hendra virus |
| HBV | hepatitis B virus |
| HCV | hepatitis C virus |
| HSV | herpes simplex virus |
| HAdV | human Adenoviruses |
| HHV | human betaherpesvirus |
| HCMV | human cytomegalovirus |
| Huh-7 cells | human hepatocellular carcinoma cell line |
| HIV | human immunodeficiency virus |
| HRVs | human rhinoviruses |
| VSV | Indiana vesiculovirus |
| IAV | Influenza A Virus |
| ICAM-1 | intercellular adhesion molecule-1 |
| JCV | JC polyomavirus |
| KSHV | Kaposi Sarcoma-associated Herpesvirus |
| LDLR | low-density lipoprotein receptor |
| LAMP1 | lysosomal-associated membrane protein 1 |
| MHC | major histocompatibility complex |
| MPR | mannose-6-phosphate receptors |
| mRNA | messenger ribonucleic acid |
| MERS | Middle East respiratory syndrome-related virus |
| MCMV | mouse cytomegalovirus |
| MHV | Mouse hepatitis virus |
| NKT cells | natural killer T cells |
| NiV | Nipah virus |
| PIP | phosphatidyl-inositol phosphates |
| PRV | pseudorabies virus |
| PHHs | primary human hepatocytes |
| PREPL | polyl endopeptidase-like |
| Rab5 | Ras-related protein Rab-5A |
| RBD | receptor-binding domain |
| RIDα | receptor internalization and degradation |
| RSV | respiratory syncytial virus |
| SARS-CoV | Severe acute respiratory syndrome coronavirus |
| SLAM | signalling lymphocyte activation molecule |
| SIV | Simian immunodeficiency virus |
| siRNA | small interfering RNA |
| NTCP | sodium taurocholate co-transporting polypeptide |
| SPG | spastic paraplegia protein |
| TGN | trans-Golgi network |
| TNF | tumor necrosis factor |
| TRAIL-R | tumor necrosis factor-related apoptosis-inducing ligand receptor |
| VACV | Vaccinia virus |
| VZV | Varicella Zoster Virus |
| VLPs | virus-like particles |
| WHO | World Health Organization |
References
- Agrawal, T.; Schu, P.; Medigeshi, G.R. Adaptor protein complexes-1 and 3 are involved at distinct stages of flavivirus life-cycle. Sci. Rep. 2013, 3, 1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehm, M.; Bonifacino, J.S. Adaptins. Mol. Biol. Cell 2001, 12, 2907–2920. [Google Scholar] [CrossRef]
- Hirst, J.; D. Barlow, L.; Francisco, G.C.; Sahlender, D.A.; Seaman, M.N.J.; Dacks, J.B.; Robinson, M.S. The Fifth Adaptor Protein Complex. PLoS Biol. 2011, 9, e1001170. [Google Scholar]
- Robinson, M.S.; Bonifacino, J.S. Adaptor-related proteins. Curr. Opin. Cell Biol. 2001, 13, 444–453. [Google Scholar] [CrossRef]
- Hirst, J.; Itzhak, D.N.; Antrobus, R.; Borner, G.H.H.; Robinson, M.S. Role of the AP-5 adaptor protein complex in late endosome-to-Golgi retrieval. PLoS Biol. 2018, 16, e2004411. [Google Scholar]
- Brodsky, F.M. Diversity of clathrin function: New tricks for an old protein. Annu. Rev. Cell Dev. Biol. 2012, 28, 309–336. [Google Scholar] [CrossRef]
- Yuan, S.; Chu, H.; Huang, J.; Zhao, X.; Ye, Z.W.; Lai, P.M.; Wen, L.; Cai, J.P.; Mo, Y.; Cao, J.; et al. Viruses harness YxxO motif to interact with host AP2M1 for replication: A vulnerable broad-spectrum antiviral target. Sci. Adv. 2020, 6, eaba7910. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traub, L.M. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim. Biophys. Acta 2005, 1744, 415–437. [Google Scholar]
- Collins, B.M.; McCoy, A.J.; Kent, H.M.; Evans, P.R.; Owen, D.J. Molecular architecture and functional model of the endocytic AP2 complex. Cell 2002, 109, 523–535. [Google Scholar] [CrossRef]
- Heldwein, E.E.; Macia, E.; Wang, J.; Yin, H.L.; Kirchhausen, T.; Harrison, S.C. Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl. Acad. Sci. USA 2004, 101, 14108–14113. [Google Scholar] [CrossRef] [Green Version]
- Edeling, M.A.; Mishra, S.K.; Keyel, P.A.; Steinhauser, A.L.; Collins, B.M.; Roth, R.; Heuser, J.E.; Owen, D.J.; Traub, L.M. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev. Cell. 2006, 10, 329–342. [Google Scholar] [CrossRef] [Green Version]
- Owen, D.J.; Vallis, Y.; Pearse, B.M.; McMahon, H.T.; Evans, P.R. The structure and function of the beta 2-adaptin appendage domain. EMBO J. 2000, 19, 4216–4227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zizioli, D.; Geumann, C.; Kratzke, M.; Mishra, R.; Borsani, G.; Finazzi, D.; Candiello, E.; Schu, P. gamma2 and gamma1AP-1 complexes: Different essential functions and regulatory mechanisms in clathrin-dependent protein sorting. Eur. J. Cell Biol. 2017, 96, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Zizioli, D.; Meyer, C.; Guhde, G.; Saftig, P.; von Figura, K.; Schu, P. Early embryonic death of mice deficient in gamma-adaptin. J. Biol. Chem. 1999, 274, 5385–5390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattera, R.; Boehm, M.; Chaudhuri, R.; Prabhu, Y.; Bonifacino, J.S. Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J. Biol. Chem. 2011, 286, 2022–2030. [Google Scholar] [CrossRef] [Green Version]
- Glyvuk, N.; Tsytsyura, Y.; Geumann, C.; D’Hooge, R.; Huve, J.; Kratzke, M.; Baltes, J.; Boening, D.; Klingauf, J.; Schu, P. AP-1/sigma1B-adaptin mediates endosomal synaptic vesicle recycling, learning and memory. EMBO J. 2010, 29, 1318–1330. [Google Scholar] [CrossRef] [Green Version]
- Ohno, H.; Tomemori, T.; Nakatsu, F.; Okazaki, Y.; Aguilar, R.C.; Foelsch, H.; Mellman, I.; Saito, T.; Shirasawa, T.; Bonifacino, J.S. Mu1B, a novel adaptor medium chain expressed in polarized epithelial cells. FEBS Lett. 1999, 449, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Traub, L.M. Tickets to ride: Selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 2009, 10, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Traub, L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003, 72, 395–447. [Google Scholar] [CrossRef] [Green Version]
- Baltes, J.; Larsen, J.V.; Radhakrishnan, K.; Geumann, C.; Kratzke, M.; Petersen, C.M.; Schu, P. Sigma1b adaptin regulates adipogenesis by mediating the sorting of sortilin in adipose tissue. J. Cell Sci. 2014, 127, 3477–3487. [Google Scholar] [CrossRef] [Green Version]
- Foote, C.; Nothwehr, S.F. The clathrin adaptor complex 1 directly binds to a sorting signal in Ste13p to reduce the rate of its trafficking to the late endosome of yeast. J. Cell Biol. 2006, 173, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Dib, K.; Tikhonova, I.G.; Ivetic, A.; Schu, P. The cytoplasmic tail of L-selectin interacts with the adaptor-protein complex AP-1 subunit μ1A via a novel basic binding motif. J. Biol. Chem. 2017, 292, 6703–6714. [Google Scholar] [CrossRef] [Green Version]
- Orzech, E.; Schlessinger, K.; Weiss, A.; Okamoto, C.T.; Aroeti, B. Interactions of the AP-1 Golgi Adaptor with the Polymeric Immunoglobulin Receptor and Their Possible Role in Mediating Brefeldin A-sensitive Basolateral Targeting from the trans-Golgi Network. J. Biol. Chem. 1999, 274, 2201–2215. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, P.; Kornfeld, S. AP-1 binding to sorting signals and release from clathrin-coated vesicles is regulated by phosphorylation. J. Cell Biol. 2003, 160, 699–708. [Google Scholar] [CrossRef] [Green Version]
- Ricotta, D.; Conner, S.D.; Schmid, S.L.; von Figura, K.; Honing, S. Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 2002, 156, 791–795. [Google Scholar] [CrossRef] [Green Version]
- Conner, S.D.; Schroter, T.; Schmid, S.L. AAK1-mediated micro2 phosphorylation is stimulated by assembled clathrin. Traffic 2003, 4, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Banting, G. CK2 and GAK/auxilin2 are major protein kinases in clathrin-coated vesicles. Traffic 2002, 3, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.P.; Kelly, B.T.; McCoy, A.J.; Gaffry, T.; James, L.C.; Collins, B.M.; Honing, S.; Evans, P.R.; Owen, D.J. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 2010, 141, 1220–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, X.; Farias, G.G.; Canagarajah, B.J.; Bonifacino, J.S.; Hurley, J.H. Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell 2013, 152, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Fölsch, H. Role of the epithelial cell-specific clathrin adaptor complex AP-1B in cell polarity. Cell Logist. 2015, 5, e1074331. [Google Scholar] [CrossRef] [Green Version]
- Eskelinen, E.L.; Meyer, C.; Ohno, H.; von Figura, K.; Schu, P. The polarized epithelia-specific mu 1B-adaptin complements mu 1A-deficiency in fibroblasts. EMBO Rep. 2002, 3, 471–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dohgu, S.; Ryerse, J.S.; Robinson, S.M.; Banks, W.A. Human immunodeficiency virus-1 uses the mannose-6-phosphate receptor to cross the blood-brain barrier. PLoS ONE 2012, 7, e39565. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Salinas, M.A.; Casorla, L.A.; Lopez, T.; Lopez, S.; Arias, C.F. Most rotavirus strains require the cation-independent mannose-6-phosphate receptor, sortilin-1, and cathepsins to enter cells. Virus Res. 2018, 245, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Salinas, M.A.; Silva-Ayala, D.; Lopez, S.; Arias, C.F. Rotaviruses reach late endosomes and require the cation-dependent mannose-6-phosphate receptor and the activity of cathepsin proteases to enter the cell. J. Virol. 2014, 88, 4389–4402. [Google Scholar] [CrossRef] [Green Version]
- Girsch, J.H.; Jackson, W.; Carpenter, J.E.; Moninger, T.O.; Jarosinski, K.W.; Grose, C. Exocytosis of Progeny Infectious Varicella-Zoster Virus Particles via a Mannose-6-Phosphate Receptor Pathway without Xenophagy following Secondary Envelopment. J. Virol. 2020, 94, e00800-20. [Google Scholar] [CrossRef]
- Chen, J.J.; Zhu, Z.; Gershon, A.A.; Gershon, M.D. Mannose 6-phosphate receptor dependence of varicella zoster virus infection in vitro and in the epidermis during varicella and zoster. Cell 2004, 119, 915–926. [Google Scholar] [CrossRef] [Green Version]
- Schmid, E.M.; McMahon, H.T. Integrating molecular and network biology to decode endocytosis. Nature 2007, 448, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Ruckert, C.; Saenger, W.; Haucke, V. The early steps of endocytosis: From cargo selection to membrane deformation. Eur. J. Cell Biol. 2012, 91, 226–233. [Google Scholar] [CrossRef]
- Pascolutti, R.; Algisi, V.; Conte, A.; Raimondi, A.; Pasham, M.; Upadhyayula, S.; Gaudin, R.; Maritzen, T.; Barbieri, E.; Caldieri, G.; et al. Molecularly Distinct Clathrin-Coated Pits Differentially Impact EGFR Fate and Signaling. Cell Rep. 2019, 27, 3049–3061.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyers, L.; Zwickl, H.; Blaas, D. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. J. Virol. 2003, 77, 5360–5369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharyya, S.; Warfield, K.L.; Ruthel, G.; Bavari, S.; Aman, M.J.; Hope, T.J. Ebola virus uses clathrin-mediated endocytosis as an entry pathway. Virology 2010, 401, 18–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharyya, S.; Hope, T.J.; Young, J.A. Differential requirements for clathrin endocytic pathway components in cellular entry by Ebola and Marburg glycoprotein pseudovirions. Virology 2011, 419, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Peden, A.A.; Oorschot, V.; Hesser, B.A.; Austin, C.D.; Scheller, R.H.; Klumperman, J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 2004, 164, 1065–1076. [Google Scholar] [CrossRef]
- Dell’Angelica, E.C.; Shotelersuk, V.; Aguilar, R.C.; Gahl, W.A.; Bonifacino, J.S. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol. Cell. 1999, 3, 11–21. [Google Scholar] [CrossRef]
- Reusch, U.; Bernhard, O.; Koszinowski, U.; Schu, P. AP-1A and AP-3A Lysosomal Sorting Functions. Traffic 2002, 3, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Kimpler, L.A.; Glosson, N.L.; Downs, D.; Gonyo, P.; May, N.A.; Hudson, A.W. Adaptor Protein Complexes AP-1 and AP-3 Are Required by the HHV-7 Immunoevasin U21 for Rerouting of Class I MHC Molecules to the Lysosomal Compartment. PLoS ONE 2014, 9, e99139. [Google Scholar] [CrossRef] [Green Version]
- Hudson, A.W.; Howley, P.M.; Ploegh, H.L. A Human Herpesvirus 7 Glycoprotein, U21, Diverts Major Histocompatibility Complex Class I Molecules to Lysosomes. J. Virol. 2001, 75, 12347–12358. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Li, H.; Derdowski, A.; Ding, L.; Burnett, A.; Chen, X.; Peters, T.R.; Dermody, T.S.; Woodruff, E.; Wang, J.J.; et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 2005, 120, 663–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, N.; Plutner, H.; Hahn, K.; Balch, W.E. The delta subunit of AP-3 is required for efficient transport of VSV-G from the trans-Golgi network to the cell surface. Proc. Natl. Acad. Sci. USA 2002, 99, 6755–6760. [Google Scholar] [CrossRef] [Green Version]
- Ross, B.H.; Lin, Y.; Corales, E.A.; Burgos, P.V.; Mardones, G.A. Structural and functional characterization of cargo-binding sites on the mu4-subunit of adaptor protein complex 4. PLoS ONE 2014, 9, e88147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmen, T.; Honing, S.; Icking, A.; Tikkanen, R.; Hunziker, W. AP-4 binds basolateral signals and participates in basolateral sorting in epithelial MDCK cells. Nat. Cell Biol. 2002, 4, 154–159. [Google Scholar] [CrossRef]
- Matsuda, S.; Yuzaki, M. How polarized sorting to neuronal dendrites is achieved: A newly discovered AP4-based system. Tanpakushitsu Kakusan Koso. 2008, 53 (Suppl. S16), 2214–2219. [Google Scholar]
- Mattera, R.; Park, S.Y.; De Pace, R.; Guardia, C.M.; Bonifacino, J.S. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc. Natl. Acad. Sci. USA 2017, 114, E10697–E10706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivankovic, D.; Drew, J.; Lesept, F.; White, I.J.; Lopez Domenech, G.; Tooze, S.A.; Kittler, J.T. Axonal autophagosome maturation defect through failure of ATG9A sorting underpins pathology in AP-4 deficiency syndrome. Autophagy 2020, 16, 391–407. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Wang, S.; Barouch-Bentov, R.; Neveu, G.; Pu, S.; Beer, M.; Schor, S.; Kumar, S.; Nicolaescu, V.; Lindenbach, B.D.; et al. Interactions between the Hepatitis C Virus Nonstructural 2 Protein and Host Adaptor Proteins 1 and 4 Orchestrate Virus Release. mBio 2018, 9, e02233-17. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Palefsky, J.M.; Herrera, R.; Berline, J.; Tugizov, S.M. EBV BMRF-2 facilitates cell-to-cell spread of virus within polarized oral epithelial cells. Virology 2009, 388, 335–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montecchiani, C.; Pedace, L.; Lo Giudice, T.; Casella, A.; Mearini, M.; Gaudiello, F.; Pedroso, J.L.; Terracciano, C.; Caltagirone, C.; Massa, R.; et al. ALS5/SPG11/KIAA1840 mutations cause autosomal recessive axonal Charcot-Marie-Tooth disease. Brain 2016, 139 Pt 1, 73–85. [Google Scholar] [CrossRef] [Green Version]
- Khundadze, M.; Ribaudo, F.; Hussain, A.; Rosentreter, J.; Nietzsche, S.; Thelen, M.; Winter, D.; Hoffmann, B.; Afzal, M.A.; Hermann, T.; et al. A mouse model for SPG48 reveals a block of autophagic flux upon disruption of adaptor protein complex five. Neurobiol. Dis. 2019, 127, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Lee, S.; Blackstone, C. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J. Clin. Investig. 2014, 124, 5249–5262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alford, J.E.; Marongiu, M.; Watkins, G.L.; Anderson, E.C. Human Immunodeficiency Virus Type 2 (HIV-2) Gag Is Trafficked in an AP-3 and AP-5 Dependent Manner. PLoS ONE 2016, 11, e0158941. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Gonzalez, J.M.; Gravotta, D.; Mattera, R.; Diaz, F.; Perez Bay, A.; Roman, A.C.; Schreiner, R.P.; Thuenauer, R.; Bonifacino, J.S.; Rodriguez-Boulan, E. Basolateral sorting of the coxsackie and adenovirus receptor through interaction of a canonical YXXPhi motif with the clathrin adaptors AP-1A and AP-1B. Proc. Natl. Acad. Sci. USA 2012, 109, 3820–3825. [Google Scholar] [CrossRef] [Green Version]
- Cianciola, N.L.; Crooks, D.; Shah, A.H.; Carlin, C. A tyrosine-based signal plays a critical role in the targeting and function of adenovirus RIDalpha protein. J. Virol. 2007, 81, 10437–10450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flatt, J.W.; Butcher, S.J. Adenovirus flow in host cell networks. Open Biol. 2019, 9, 190012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J. Exp. Med. 2004, 199, 1331–1341. [CrossRef] [Green Version]
- Johnson, D.C.; Webb, M.; Wisner, T.W.; Brunetti, C. Herpes Simplex Virus gE/gI Sorts Nascent Virions to Epithelial Cell Junctions, Promoting Virus Spread. J. Virol. 2001, 75, 821–833. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Gallo, R.M.; Duffy, C.; Brutkiewicz, R.R. A VP22-Null HSV-1 Is Impaired in Inhibiting CD1d-Mediated Antigen Presentation. Viral Immunol. 2016, 29, 409–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Minnebruggen, G.; Favoreel, H.W.; Nauwynck, H.J. Internalization of Pseudorabies Virus Glycoprotein B Is Mediated by an Interaction between the YQRL Motif in Its Cytoplasmic Domain and the Clathrin-Associated AP-2 Adaptor Complex. J. Virol. 2004, 78, 8852–8859. [Google Scholar] [CrossRef] [Green Version]
- Lebrun, M.; Lambert, J.; Riva, L.; Thelen, N.; Rambout, X.; Blondeau, C.; Thiry, M.; Snoeck, R.; Twizere, J.-C.; Dequiedt, F.; et al. Varicella-Zoster Virus ORF9p Binding to Cellular Adaptor Protein Complex 1 Is Important for Viral Infectivity. J. Virol. 2018, 92, e00295-18. [Google Scholar] [CrossRef] [Green Version]
- Strazic Geljic, I.; Kucan Brlic, P.; Angulo, G.; Brizic, I.; Lisnic, B.; Jenus, T.; Juranic Lisnic, V.; Pietri, G.P.; Engel, P.; Kaynan, N.; et al. Cytomegalovirus protein m154 perturbs the adaptor protein-1 compartment mediating broad-spectrum immune evasion. eLife 2020, 9, e50803. [Google Scholar] [CrossRef]
- Fink, A.; Blaum, F.; Babic Cac, M.; Ebert, S.; Lemmermann, N.A.; Reddehase, M.J. An endocytic YXXPhi (YRRF) cargo sorting motif in the cytoplasmic tail of murine cytomegalovirus AP2 ’adapter adapter’ protein m04/gp34 antagonizes virus evasion of natural killer cells. Med. Microbiol. Immunol. 2015, 204, 383–394. [Google Scholar] [CrossRef]
- Jelcic, I.; Reichel, J.; Schlude, C.; Treutler, E.; Sinzger, C.; Steinle, A. The Polymorphic HCMV Glycoprotein UL20 Is Targeted for Lysosomal Degradation by Multiple Cytoplasmic Dileucine Motifs. Traffic 2011, 12, 1444–1456. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Quinn, L.L.; Tamblyn, J.; Thomas, W.A.; Feederle, R.; Delecluse, H.-J.; Hislop, A.D.; Rowe, M. The Epstein-Barr Virus-Encoded BILF1 Protein Modulates Immune Recognition of Endogenously Processed Antigen by Targeting Major Histocompatibility Complex Class I Molecules Trafficking on both the Exocytic and Endocytic Pathways. J. Virol. 2011, 85, 1604–1614. [Google Scholar] [CrossRef] [Green Version]
- Veettil, M.; Bandyopadhyay, C.; Dutta, D.; Chandran, B. Interaction of KSHV with Host Cell Surface Receptors and Cell Entry. Viruses 2014, 6, 4024–4046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Orba, Y.; Makino, Y.; Okada, Y.; Sunden, Y.; Hasegawa, H.; Hall, W.W.; Sawa, H. Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc. Natl. Acad. Sci. USA 2013, 110, 18668–18673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husain, M.; Moss, B. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J. Virol. 2003, 77, 11754–11766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, B.M.; Moss, B. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J. Virol. 2000, 74, 3771–3780. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.G.; Tang, D.J.; Hua, Z.; Wang, Z.; An, J. Sunitinib reduces the infection of SARS-CoV, MERS-CoV and SARS-CoV-2 partially by inhibiting AP2M1 phosphorylation. Cell Discov. 2020, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Eifart, P.; Ludwig, K.; Bottcher, C.; de Haan, C.A.; Rottier, P.J.; Korte, T.; Herrmann, A. Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J. Virol. 2007, 81, 10758–10768. [Google Scholar] [CrossRef] [Green Version]
- Pu, Y.; Zhang, X. Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J. Virol. 2008, 82, 8112–8123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, F.; Wong, A.P.; Ng, M.M.; Chu, J.J. Small interference RNA profiling reveals the essential role of human membrane trafficking genes in mediating the infectious entry of dengue virus. Virol. J. 2010, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Yasamut, U.; Tongmuang, N.; Yenchitsomanus, P.T.; Junking, M.; Noisakran, S.; Puttikhunt, C.; Chu, J.J.; Limjindaporn, T. Adaptor Protein 1A Facilitates Dengue Virus Replication. PLoS ONE 2015, 10, e0130065. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Niu, Y.; Cheng, M.; Chi, X.; Liu, X.; Yang, W. AP1S3 is required for hepatitis C virus infection by stabilizing E2 protein. Antiviral Res. 2016, 131, 26–34. [Google Scholar] [CrossRef]
- Neveu, G.; Barouch-Bentov, R.; Ziv-Av, A.; Gerber, D.; Jacob, Y.; Einav, S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012, 8, e1002845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.; Schor, S.; Barouch-Bentov, R.; Einav, S. Viral journeys on the intracellular highways. Cell Mol Life Sci. 2018, 75, 3693–3714. [Google Scholar] [CrossRef]
- Bekerman, E.; Neveu, G.; Shulla, A.; Brannan, J.; Pu, S.Y.; Wang, S.; Xiao, F.; Barouch-Bentov, R.; Bakken, R.R.; Mateo, R.; et al. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Investig. 2017, 127, 1338–1352. [Google Scholar] [CrossRef]
- Garrison, A.R.; Radoshitzky, S.R.; Kota, K.P.; Pegoraro, G.; Ruthel, G.; Kuhn, J.H.; Altamura, L.A.; Kwilas, S.A.; Bavari, S.; Haucke, V.; et al. Crimean-Congo hemorrhagic fever virus utilizes a clathrin- and early endosome-dependent entry pathway. Virology 2013, 444, 45–54. [Google Scholar] [CrossRef] [Green Version]
- Keren, T.; Roth, M.G.; Henis, Y.I. Internalization-competent influenza hemagglutinin mutants form complexes with clathrin-deficient multivalent AP-2 oligomers in live cells. J. Biol. Chem. 2001, 276, 28356–28363. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Jiang, L.; Wang, J.; Zhang, J.; Kong, F.; Li, Q.; Yan, Y.; Huang, S.; Zhao, Y.; Liang, L.; et al. The G Protein-Coupled Receptor FFAR2 Promotes Internalization during Influenza A Virus Entry. J. Virol. 2020, 94, e01707-19. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; McCrory, T.S.; Khaw, W.Y.; Petzing, S.; Myers, T.; Schmitt, A.P. Matrix proteins of Nipah and Hendra viruses interact with beta subunits of AP-3 complexes. J. Virol. 2014, 88, 13099–13110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattera, R.; Farias, G.G.; Mardones, G.A.; Bonifacino, J.S. Co-assembly of viral envelope glycoproteins regulates their polarized sorting in neurons. PLoS Pathog. 2014, 10, e1004107. [Google Scholar] [CrossRef] [Green Version]
- Kolokoltsov, A.A.; Deniger, D.; Fleming, E.H.; Roberts, N.J., Jr.; Karpilow, J.M.; Davey, R.A. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J. Virol. 2007, 81, 7786–7800. [Google Scholar] [CrossRef] [Green Version]
- Krzyzaniak, M.A.; Zumstein, M.T.; Gerez, J.A.; Picotti, P.; Helenius, A. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog. 2013, 9, e1003309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, C.; Maselko, M.; Lupfer, C.; Prescott, M.; Pastey, M.K. Interaction of the Human Respiratory Syncytial Virus matrix protein with cellular adaptor protein complex 3 plays a critical role in trafficking. PLoS ONE 2017, 12, e0184629. [Google Scholar] [CrossRef] [Green Version]
- Cureton, D.K.; Massol, R.H.; Saffarian, S.; Kirchhausen, T.L.; Whelan, S.P. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009, 5, e1000394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujisawa, R.; Masuda, M. Ecotropic murine leukemia virus envelope protein affects interaction of cationic amino acid transporter 1 with clathrin adaptor protein complexes, leading to receptor downregulation. Virology 2007, 368, 342–350. [Google Scholar] [CrossRef] [Green Version]
- Buffalo, C.Z.; Iwamoto, Y.; Hurley, J.H.; Ren, X. How HIV Nef Proteins Hijack Membrane Traffic To Promote Infection. J. Virol. 2019, 93, e01322-19. [Google Scholar] [CrossRef]
- Pereira, E.A.; daSilva, L.L. HIV-1 Nef: Taking Control of Protein Trafficking. Traffic 2016, 17, 976–996. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.; Weber, E.; Tokarev, A.; Lewinski, M.; Rizk, M.; Suarez, M.; Guatelli, J.; Xiong, Y. Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. Elife 2014, 3, e02362. [Google Scholar] [CrossRef]
- Chu, H.; Wang, J.J.; Spearman, P. Human immunodeficiency virus type-1 gag and host vesicular trafficking pathways. Curr. Top. Microbiol. Immunol. 2009, 339, 67–84. [Google Scholar] [PubMed]
- Hirao, K.; Andrews, S.; Kuroki, K.; Kusaka, H.; Tadokoro, T.; Kita, S.; Ose, T.; Rowland-Jones, S.L.; Maenaka, K. Structure of HIV-2 Nef Reveals Features Distinct from HIV-1 Involved in Immune Regulation. iScience 2020, 23, 100758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noble, B.; Abada, P.; Nunez-Iglesias, J.; Cannon, P.M. Recruitment of the adaptor protein 2 complex by the human immunodeficiency virus type 2 envelope protein is necessary for high levels of virus release. J. Virol. 2006, 80, 2924–2932. [Google Scholar] [CrossRef] [Green Version]
- Cooper, A.; Shaul, Y. Clathrin-mediated endocytosis and lysosomal cleavage of hepatitis B virus capsid-like core particles. J. Biol. Chem. 2006, 281, 16563–16569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.C.; Chen, C.C.; Chang, W.C.; Tao, M.H.; Huang, C. Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J. Virol. 2012, 86, 9443–9453. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, M.; Saso, W.; Nishioka, K.; Ohashi, H.; Sugiyama, R.; Ryo, A.; Ohki, M.; Yun, J.H.; Park, S.Y.; Ohshima, T.; et al. The machinery for endocytosis of epidermal growth factor receptor coordinates the transport of incoming hepatitis B virus to the endosomal network. J. Biol. Chem. 2020, 295, 800–807. [Google Scholar] [CrossRef]
- Wold, W.S.; Toth, K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther. 2013, 13, 421–433. [Google Scholar] [CrossRef]
- Majhen, D.; Calderon, H.; Chandra, N.; Fajardo, C.A.; Rajan, A.; Alemany, R.; Custers, J. Adenovirus-based vaccines for fighting infectious diseases and cancer: Progress in the field. Hum. Gene Ther. 2014, 25, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Segerman, A.; Atkinson, J.P.; Marttila, M.; Dennerquist, V.; Wadell, G.; Arnberg, N. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 2003, 77, 9183–9191. [Google Scholar] [CrossRef] [Green Version]
- Meier, O.; Greber, U.F. Adenovirus endocytosis. J. Gene Med. 2004, 6 (Suppl. S1), S152–S163. [Google Scholar] [CrossRef]
- Diaz, F.; Gravotta, D.; Deora, A.; Schreiner, R.; Schoggins, J.; Falck-Pedersen, E.; Rodriguez-Boulan, E. Clathrin adaptor AP1B controls adenovirus infectivity of epithelial cells. Proc. Natl. Acad. Sci. USA 2009, 106, 11143–11148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichtenstein, D.L.; Toth, K.; Doronin, K.; Tollefson, A.E.; Wold, W.S. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 2004, 23, 75–111. [Google Scholar] [CrossRef]
- Tollefson, A.E.; Stewart, A.R.; Yei, S.P.; Saha, S.K.; Wold, W.S. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus form a complex and function together to down-regulate the epidermal growth factor receptor. J. Virol. 1991, 65, 3095–3105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlin, C.R.; Tollefson, A.E.; Brady, H.A.; Hoffman, B.L.; Wold, W.S. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell 1989, 57, 135–144. [Google Scholar] [CrossRef]
- Elsing, A.; Burgert, H.G. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. USA 1998, 95, 10072–10077. [Google Scholar] [PubMed] [Green Version]
- Tollefson, A.E.; Hermiston, T.W.; Lichtenstein, D.L.; Colle, C.F.; Tripp, R.A.; Dimitrov, T.; Toth, K.; Wells, C.E.; Doherty, P.C.; Wold, W.S. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 1998, 392, 726–730. [Google Scholar] [CrossRef]
- Tollefson, A.E.; Toth, K.; Doronin, K.; Kuppuswamy, M.; Doronina, O.A.; Lichtenstein, D.L.; Hermiston, T.W.; Smith, C.A.; Wold, W.S. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J. Virol. 2001, 75, 8875–8887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilgendorf, A.; Lindberg, J.; Ruzsics, Z.; Honing, S.; Elsing, A.; Lofqvist, M.; Engelmann, H.; Burgert, H.G. Two distinct transport motifs in the adenovirus E3/10.4-14.5 proteins act in concert to down-modulate apoptosis receptors and the epidermal growth factor receptor. J. Biol. Chem. 2003, 278, 51872–51884. [Google Scholar]
- Farnsworth, A.; Johnson, D.C. Herpes Simplex Virus gE/gI Must Accumulate in the trans-Golgi Network at Early Times and Then Redistribute to Cell Junctions To Promote Cell-Cell Spread. J. Virol. 2006, 80, 3167–3179. [Google Scholar] [CrossRef] [Green Version]
- Mcmillan, T.N.; Johnson, D.C. Cytoplasmic Domain of Herpes Simplex Virus gE Causes Accumulation in the trans-Golgi Network, a Site of Virus Envelopment and Sorting of Virions to Cell Junctions. J. Virol. 2001, 75, 1928–1940. [Google Scholar] [CrossRef] [Green Version]
- Favoreel, H.W.; Nauwynck, H.J.; Halewyck, H.M.; Van Oostveldt, P.; Mettenleiter, T.C.; Pensaert, M.B. Antibody-induced endocytosis of viral glycoproteins and major histocompatibility complex class I on pseudorabies virus-infected monocytes. J. Gen. Virol. 1999, 80, 1283–1291. [Google Scholar] [CrossRef] [Green Version]
- Favoreel, H.W.; Van Minnebruggen, G.; Nauwynck, H.J.; Enquist, L.W.; Pensaert, M.B. A Tyrosine-Based Motif in the Cytoplasmic Tail of Pseudorabies Virus Glycoprotein B Is Important for both Antibody-Induced Internalization of Viral Glycoproteins and Efficient Cell-to-Cell Spread. J. Virol. 2002, 76, 6845–6851. [Google Scholar] [CrossRef] [Green Version]
- Heineman, T.C.; Hall, S.L. VZV gB Endocytosis and Golgi Localization Are Mediated by YXXφ Motifs in Its Cytoplasmic Domain. Virology 2001, 285, 42–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, J.K.; Grose, C. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: Internalization mediated by a YXXL motif in the cytoplasmic tail. J. Virol. 1997, 71, 4042–4054. [Google Scholar] [CrossRef] [Green Version]
- Frank, I.; Friedman, H.M. A novel function of the herpes simplex virus type 1 Fc receptor: Participation in bipolar bridging of antiviral immunoglobulin G. J. Virol. 1989, 63, 4479–4488. [Google Scholar] [CrossRef] [Green Version]
- Ndjamen, B.; Farley, A.H.; Lee, T.; Fraser, S.E.; Bjorkman, P.J. The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface. PLoS Pathog. 2014, 10, e1003961. [Google Scholar] [CrossRef]
- Van De Walle, G.R.; Favoreel, H.W.; Nauwynck, H.J.; Pensaert, M.B. Antibody-induced internalization of viral glycoproteins and gE–gI Fc receptor activity protect pseudorabies virus-infected monocytes from efficient complement-mediated lysis. J. Gen. Virol. 2003, 84, 939–947. [Google Scholar] [CrossRef]
- Favoreel, H.W.; Nauwynck, H.J.; Van Oostveldt, P.; Pensaert, M.B. Role of Anti-gB and -gD Antibodies in Antibody-Induced Endocytosis of Viral and Cellular Cell Surface Glycoproteins Expressed on Pseudorabies Virus-Infected Monocytes. Virology 2000, 267, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Lenac Rovis, T.; Kucan Brlic, P.; Kaynan, N.; Juranic Lisnic, V.; Brizic, I.; Jordan, S.; Tomic, A.; Kvestak, D.; Babic, M.; Tsukerman, P.; et al. Inflammatory monocytes and NK cells play a crucial role in DNAM-1-dependent control of cytomegalovirus infection. J. Exp. Med. 2016, 213, 1835–1850. [Google Scholar] [CrossRef]
- Zarama, A.; Perez-Carmona, N.; Farre, D.; Tomic, A.; Borst, E.M.; Messerle, M.; Jonjic, S.; Engel, P.; Angulo, A. Cytomegalovirus m154 hinders CD48 cell-surface expression and promotes viral escape from host natural killer cell control. PLoS Pathog. 2014, 10, e1004000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, J.; Currin, A.; Griffin, B.D.; Shannon-Lowe, C.; Thomas, W.A.; Ressing, M.E.; Wiertz, E.J.H.J.; Rowe, M. The Epstein-Barr Virus G-Protein-Coupled Receptor Contributes to Immune Evasion by Targeting MHC Class I Molecules for Degradation. PLoS Pathog. 2009, 5, e1000255. [Google Scholar] [CrossRef] [Green Version]
- Coscoy, L.; Ganem, D. Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 2000, 97, 8051–8056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishido, S.; Wang, C.; Lee, B.-S.; Cohen, G.B.; Jung, J.U. Downregulation of Major Histocompatibility Complex Class I Molecules by Kaposi’s Sarcoma-Associated Herpesvirus K3 and K5 Proteins. J. Virol. 2000, 74, 5300–5309. [Google Scholar] [CrossRef] [PubMed]
- Coscoy, L.; Sanchez, D.J.; Ganem, D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 2001, 155, 1265–1274. [Google Scholar] [CrossRef] [Green Version]
- Assetta, B.; Atwood, W.J. The biology of JC polyomavirus. Biol. Chem. 2017, 398, 839–855. [Google Scholar] [CrossRef]
- Querbes, W.; O’Hara, B.A.; Williams, G.; Atwood, W.J. Invasion of host cells by JC virus identifies a novel role for caveolae in endosomal sorting of noncaveolar ligands. J. Virol. 2006, 80, 9402–9413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Orba, Y.; Okada, Y.; Sunden, Y.; Kimura, T.; Tanaka, S.; Nagashima, K.; Hall, W.W.; Sawa, H. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog. 2010, 6, e1000801. [Google Scholar] [CrossRef] [Green Version]
- Moss, B. Poxviridae: The viruses and their replication. In Fields Virology, 4th ed.; Knie, D.M., Howley, P.M., Eds.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2001; pp. 2849–2883. [Google Scholar]
- Ichihashi, Y. Extracellular enveloped vaccinia virus escapes neutralization. Virology 1996, 217, 478–485. [Google Scholar] [CrossRef]
- Bengali, Z.; Townsley, A.C.; Moss, B. Vaccinia virus strain differences in cell attachment and entry. Virology 2009, 389, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Husain, M.; Moss, B. Role of receptor-mediated endocytosis in the formation of vaccinia virus extracellular enveloped particles. J. Virol. 2005, 79, 4080–4089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husain, M.; Moss, B. Intracellular trafficking of a palmitoylated membrane-associated protein component of enveloped vaccinia virus. J. Virol. 2003, 77, 9008–9019. [Google Scholar] [CrossRef] [Green Version]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [PubMed] [Green Version]
- Wang, S.; Guo, F.; Liu, K.; Wang, H.; Rao, S.; Yang, P.; Jiang, C. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008, 136, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells following viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Van der Schaar, H.M.; Rust, M.J.; Chen, C.; van der Ende-Metselaar, H.; Wilschut, J.; Zhuang, X.; Smit, J.M. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008, 4, e1000244. [Google Scholar] [CrossRef] [Green Version]
- Smit, J.M.; Moesker, B.; Rodenhuis-Zybert, I.; Wilschut, J. Flavivirus cell entry and membrane fusion. Viruses 2011, 3, 160–171. [Google Scholar] [CrossRef] [Green Version]
- Pu, S.; Schor, S.; Karim, M.; Saul, S.; Robinson, M.; Kumar, S.; Prugar, L.I.; Dorosky, D.E.; Brannan, J.; Dye, J.M.; et al. BIKE regulates dengue virus infection and is a cellular target for broad-spectrum antivirals. Antiviral Res. 2020, 184, 104966. [Google Scholar] [CrossRef]
- Pu, S.Y.; Wouters, R.; Schor, S.; Rozenski, J.; Barouch-Bentov, R.; Prugar, L.I.; O’Brien, C.M.; Brannan, J.M.; Dye, J.M.; Herdewijn, P.; et al. Optimization of Isothiazolo[4,3- b]pyridine-Based Inhibitors of Cyclin G Associated Kinase (GAK) with Broad-Spectrum Antiviral Activity. J. Med. Chem. 2018, 61, 6178–6192. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Da Costa, D.; Baumert, T.F. Opening the door for hepatitis C virus infection in genetically humanized mice. Hepatology 2011, 54, 1873–1875. [Google Scholar] [CrossRef]
- Neveu, G.; Ziv-Av, A.; Barouch-Bentov, R.; Berkerman, E.; Mulholland, J.; Einav, S. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J. Virol. 2015, 89, 4387–4404. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, S.E.; Lamson, D.M.; St George, K.; Walsh, T.J. Human rhinoviruses. Clin. Microbiol. Rev. 2013, 26, 135–162. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, R.; Blaas, D. Productive entry pathways of human rhinoviruses. Adv. Virol. 2012, 2012, 826301. [Google Scholar]
- Kuhn, J.H.; Amarasinghe, G.K.; Basler, C.F.; Bavari, S.; Bukreyev, A.; Chandran, K.; Crozier, I.; Dolnik, O.; Dye, J.M.; Formenty, P.B.H.; et al. ICTV Virus Taxonomy Profile: Filoviridae. J. Gen. Virol. 2019, 100, 911–912. [Google Scholar] [CrossRef]
- Schnittler, H.J.; Feldmann, H. Viral hemorrhagic fever--a vascular disease? Thromb. Haemost. 2003, 89, 967–972. [Google Scholar]
- Bell, B.P.; Damon, I.K.; Jernigan, D.B.; Kenyon, T.A.; Nichol, S.T.; O’Connor, J.P.; Tappero, J.W. Overview, Control Strategies, and Lessons Learned in the CDC Response to the 2014-2016 Ebola Epidemic. MMWR Suppl. 2016, 65, 4–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aleksandrowicz, P.; Marzi, A.; Biedenkopf, N.; Beimforde, N.; Becker, S.; Hoenen, T.; Feldmann, H.; Schnittler, H.J. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J. Infect. Dis. 2011, 204 (Suppl. S3), S957–S967. [Google Scholar] [CrossRef] [Green Version]
- Nanbo, A.; Imai, M.; Watanabe, S.; Noda, T.; Takahashi, K.; Neumann, G.; Halfmann, P.; Kawaoka, Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010, 6, e1001121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, M.F.; Kolokoltsov, A.A.; Albrecht, T.; Davey, R.A. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010, 6, e1001110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitehouse, C.A. Crimean-Congo hemorrhagic fever. Antiviral Res. 2004, 64, 145–160. [Google Scholar] [CrossRef]
- Simon, M.; Johansson, C.; Mirazimi, A. Crimean-Congo hemorrhagic fever virus entry and replication is clathrin-, pH- and cholesterol-dependent. J. Gen. Virol. 2009, 90, 210–215. [Google Scholar] [CrossRef]
- Von Kleist, L.; Stahlschmidt, W.; Bulut, H.; Gromova, K.; Puchkov, D.; Robertson, M.J.; MacGregor, K.A.; Tomilin, N.; Pechstein, A.; Chau, N.; et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 2011, 146, 471–484. [Google Scholar] [CrossRef] [Green Version]
- Simpson, C.; Yamauchi, Y. Microtubules in Influenza Virus Entry and Egress. Viruses 2020, 12, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edinger, T.O.; Pohl, M.O.; Stertz, S. Entry of influenza A virus: Host factors and antiviral targets. J. Gen. Virol. 2014, 95 Pt 2, 263–277. [Google Scholar] [CrossRef]
- Rust, M.J.; Lakadamyali, M.; Zhang, F.; Zhuang, X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 2004, 11, 567–573. [Google Scholar] [CrossRef] [Green Version]
- Mercer, J.; Helenius, A. Virus entry by macropinocytosis. Nat. Cell Biol. 2009, 11, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Rossman, J.S.; Leser, G.P.; Lamb, R.A. Filamentous influenza virus enters cells via macropinocytosis. J. Virol. 2012, 86, 10950–10960. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Zhuang, X. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc. Natl. Acad. Sci. USA 2008, 105, 11790–11795. [Google Scholar] [CrossRef] [Green Version]
- Mair, C.M.; Ludwig, K.; Herrmann, A.; Sieben, C. Receptor binding and pH stability - how influenza A virus hemagglutinin affects host-specific virus infection. Biochim. Biophys. Acta 2014, 1838, 1153–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, M.K.; Feldmann, F.; Gary, J.M.; Jordan, R.; Bannister, R.; Cronin, J.; Patel, N.R.; Klena, J.D.; Nichol, S.T.; Cihlar, T.; et al. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci. Transl. Med. 2019, 11, eaau9242. [Google Scholar] [CrossRef]
- Diederich, S.; Maisner, A. Molecular characteristics of the Nipah virus glycoproteins. Ann. N. Y. Acad. Sci. 2007, 1102, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Lee, B. Envelope-receptor interactions in Nipah virus pathobiology. Ann. N. Y. Acad. Sci. 2007, 1102, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Ataman, Z.A. Modes of paramyxovirus fusion: A Henipavirus perspective. Trends Microbiol. 2011, 19, 389–399. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, C.M.; Roby, J.A.; Scott, C.J.; Raidal, S.R.; Forwood, J.K. The Structural Features of Henipavirus Matrix Protein Driving Intracellular Trafficking. Viral. Immunol. 2021, 34, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Borchers, A.T.; Chang, C.; Gershwin, M.E.; Gershwin, L.J. Respiratory syncytial virus--a comprehensive review. Clin. Rev. Allergy Immunol. 2013, 45, 331–379. [Google Scholar] [CrossRef] [PubMed]
- Rozo-Lopez, P.; Drolet, B.S.; Londono-Renteria, B. Vesicular Stomatitis Virus Transmission: A Comparison of Incriminated Vectors. Insects 2018, 9, 190. [Google Scholar] [CrossRef] [Green Version]
- Johannsdottir, H.K.; Mancini, R.; Kartenbeck, J.; Amato, L.; Helenius, A. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 2009, 83, 440–453. [Google Scholar] [CrossRef] [Green Version]
- Rein, A.; Datta, S.A.; Jones, C.P.; Musier-Forsyth, K. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 2011, 36, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Cingoz, O.; Paprotka, T.; Delviks-Frankenberry, K.A.; Wildt, S.; Hu, W.S.; Pathak, V.K.; Coffin, J.M. Characterization, mapping, and distribution of the two XMRV parental proviruses. J. Virol. 2012, 86, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Jager, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV-human protein complexes. Nature 2011, 481, 365–370. [Google Scholar] [CrossRef]
- Craig, H.M.; Pandori, M.W.; Guatelli, J.C. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc. Natl. Acad. Sci. USA 1998, 95, 11229–11234. [Google Scholar] [CrossRef] [Green Version]
- Gondim, M.V.; Wiltzer-Bach, L.; Maurer, B.; Banning, C.; Arganaraz, E.; Schindler, M. AP-2 Is the Crucial Clathrin Adaptor Protein for CD4 Downmodulation by HIV-1 Nef in Infected Primary CD4+ T Cells. J. Virol. 2015, 89, 12518–12524. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, R.; Mattera, R.; Lindwasser, O.W.; Robinson, M.S.; Bonifacino, J.S. A basic patch on alpha-adaptin is required for binding of human immunodeficiency virus type 1 Nef and cooperative assembly of a CD4-Nef-AP-2 complex. J. Virol. 2009, 83, 2518–2530. [Google Scholar] [CrossRef] [Green Version]
- Kelly, B.T.; McCoy, A.J.; Spate, K.; Miller, S.E.; Evans, P.R.; Honing, S.; Owen, D.J. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 2008, 456, 976–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, Y.; Kaake, R.M.; Echeverria, I.; Suarez, M.; Karimian Shamsabadi, M.; Stoneham, C.; Ramirez, P.W.; Kress, J.; Singh, R.; Sali, A.; et al. Structural basis of CD4 downregulation by HIV-1 Nef. Nat. Struct. Mol. Biol. 2020, 27, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Doray, B.; Lee, I.; Knisely, J.; Bu, G.; Kornfeld, S. The gamma/sigma1 and alpha/sigma2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol Biol Cell. 2007, 18, 1887–1896. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Park, S.Y.; Bonifacino, J.S.; Hurley, J.H. How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. Elife 2014, 3, e01754. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.A.; Johnson, A.L.; Pawlak, E.N.; Dirk, B.S.; Van Nynatten, L.R.; Haeryfar, S.M.M.; Dikeakos, J.D. The interaction between HIV-1 Nef and adaptor protein-2 reduces Nef-mediated CD4(+) T cell apoptosis. Virology 2017, 509, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, O.; Marechal, V.; Le Gall, S.; Lemonnier, F.; Heard, J.M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 1996, 2, 338–342. [Google Scholar] [CrossRef]
- Roeth, J.F.; Williams, M.; Kasper, M.R.; Filzen, T.M.; Collins, K.L. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J. Cell Biol. 2004, 167, 903–913. [Google Scholar] [CrossRef]
- Le Gall, S.; Erdtmann, L.; Benichou, S.; Berlioz-Torrent, C.; Liu, L.; Benarous, R.; Heard, J.M.; Schwartz, O. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 1998, 8, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Cohen, G.B.; Gandhi, R.T.; Davis, D.M.; Mandelboim, O.; Chen, B.K.; Strominger, J.L.; Baltimore, D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999, 10, 661–671. [Google Scholar] [CrossRef]
- Wonderlich, E.R.; Williams, M.; Collins, K.L. The tyrosine binding pocket in the adaptor protein 1 (AP-1) mu1 subunit is necessary for Nef to recruit AP-1 to the major histocompatibility complex class I cytoplasmic tail. J. Biol. Chem. 2008, 283, 3011–3022. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.; Singh, R.; Homann, S.; Yang, H.; Guatelli, J.; Xiong, Y. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat. Struct. Mol. Biol. 2012, 19, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Noviello, C.M.; Benichou, S.; Guatelli, J.C. Cooperative binding of the class I major histocompatibility complex cytoplasmic domain and human immunodeficiency virus type 1 Nef to the endosomal AP-1 complex via its mu subunit. J. Virol. 2008, 82, 1249–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulpa, D.A.; Del Cid, N.; Peterson, K.A.; Collins, K.L. Adaptor protein 1 promotes cross-presentation through the same tyrosine signal in major histocompatibility complex class I as that targeted by HIV-1. J. Virol. 2013, 87, 8085–8098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willard-Gallo, K.E.; Furtado, M.; Burny, A.; Wolinsky, S.M. Down-modulation of TCR/CD3 surface complexes after HIV-1 infection is associated with differential expression of the viral regulatory genes. Eur. J. Immunol. 2001, 31, 969–979. [Google Scholar] [CrossRef] [Green Version]
- Rosa, A.; Chande, A.; Ziglio, S.; De Sanctis, V.; Bertorelli, R.; Goh, S.L.; McCauley, S.M.; Nowosielska, A.; Antonarakis, S.E.; Luban, J.; et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 2015, 526, 212–217. [Google Scholar] [CrossRef] [Green Version]
- Stove, V.; Van de Walle, I.; Naessens, E.; Coene, E.; Stove, C.; Plum, J.; Verhasselt, B. Human immunodeficiency virus Nef induces rapid internalization of the T-cell coreceptor CD8alphabeta. J. Virol. 2005, 79, 11422–11433. [Google Scholar] [CrossRef] [Green Version]
- Swigut, T.; Shohdy, N.; Skowronski, J. Mechanism for down-regulation of CD28 by Nef. EMBO J. 2001, 20, 1593–1604. [Google Scholar] [CrossRef] [Green Version]
- Usami, Y.; Wu, Y.; Gottlinger, H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Morris, K.L.; Buffalo, C.Z.; Sturzel, C.M.; Heusinger, E.; Kirchhoff, F.; Ren, X.; Hurley, J.H. HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation. Cell 2018, 174, 659–671.e14. [Google Scholar] [CrossRef] [Green Version]
- Coleman, S.H.; Hitchin, D.; Noviello, C.M.; Guatelli, J.C. HIV-1 Nef stabilizes AP-1 on membranes without inducing ARF1-independent de novo attachment. Virology 2006, 345, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Janvier, K.; Craig, H.; Hitchin, D.; Madrid, R.; Sol-Foulon, N.; Renault, L.; Cherfils, J.; Cassel, D.; Benichou, S.; Guatelli, J. HIV-1 Nef stabilizes the association of adaptor protein complexes with membranes. J. Biol. Chem. 2003, 278, 8725–8732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wonderlich, E.R.; Leonard, J.A.; Kulpa, D.A.; Leopold, K.E.; Norman, J.M.; Collins, K.L. ADP ribosylation factor 1 activity is required to recruit AP-1 to the major histocompatibility complex class I (MHC-I) cytoplasmic tail and disrupt MHC-I trafficking in HIV-1-infected primary T cells. J. Virol. 2011, 85, 12216–12226. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.T.; Ren, X.; Zhang, R.; Lee, I.H.; Hurley, J.H. HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Science 2015, 350, aac5137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahauad-Fernandez, W.D.; Okeoma, C.M. The role of BST-2/Tetherin in host protection and disease manifestation. Immun. Inflamm. Dis. 2016, 4, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Perez-Caballero, D.; Zang, T.; Ebrahimi, A.; McNatt, M.W.; Gregory, D.A.; Johnson, M.C.; Bieniasz, P.D. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 2009, 139, 499–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neil, S.J.; Zang, T.; Bieniasz, P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, N.; Goff, D.; Katsura, C.; Jorgenson, R.L.; Mitchell, R.; Johnson, M.C.; Stephens, E.B.; Guatelli, J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 2008, 3, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skasko, M.; Wang, Y.; Tian, Y.; Tokarev, A.; Munguia, J.; Ruiz, A.; Stephens, E.B.; Opella, S.J.; Guatelli, J. HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J. Biol. Chem. 2012, 287, 58–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kueck, T.; Foster, T.L.; Weinelt, J.; Sumner, J.C.; Pickering, S.; Neil, S.J. Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors. PLoS Pathog. 2015, 11, e1005141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, E.; Nikolic, D.S.; Piguet, V. HIV-1 replication in dendritic cells occurs through a tetraspanin-containing compartment enriched in AP-3. Traffic 2008, 9, 200–214. [Google Scholar] [CrossRef]
- Kyere, S.K.; Mercredi, P.Y.; Dong, X.; Spearman, P.; Summers, M.F. The HIV-1 matrix protein does not interact directly with the protein interactive domain of AP-3delta. Virus Res. 2012, 169, 411–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camus, G.; Segura-Morales, C.; Molle, D.; Lopez-Verges, S.; Begon-Pescia, C.; Cazevieille, C.; Schu, P.; Bertrand, E.; Berlioz-Torrent, C.; Basyuk, E. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol. Biol. Cell. 2007, 18, 3193–3203. [Google Scholar] [CrossRef]
- Batonick, M.; Favre, M.; Boge, M.; Spearman, P.; Honing, S.; Thali, M. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology 2005, 342, 190–200. [Google Scholar] [CrossRef] [Green Version]
- Byland, R.; Vance, P.J.; Hoxie, J.A.; Marsh, M. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol. Biol. Cell. 2007, 18, 414–425. [Google Scholar] [CrossRef] [Green Version]
- Ohno, H.; Aguilar, R.C.; Fournier, M.C.; Hennecke, S.; Cosson, P.; Bonifacino, J.S. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 1997, 238, 305–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyss, S.; Berlioz-Torrent, C.; Boge, M.; Blot, G.; Honing, S.; Benarous, R.; Thali, M. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor [correction of adapter]. J. Virol. 2001, 75, 2982–2992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell-Yesufu, O.T.; Gandhi, R.T. Update on human immunodeficiency virus (HIV)-2 infection. Clin. Infect. Dis. 2011, 52, 780–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyamweya, S.; Hegedus, A.; Jaye, A.; Rowland-Jones, S.; Flanagan, K.L.; Macallan, D.C. Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev. Med. Virol. 2013, 23, 221–240. [Google Scholar] [CrossRef] [PubMed]
- Schindler, M.; Munch, J.; Kutsch, O.; Li, H.; Santiago, M.L.; Bibollet-Ruche, F.; Muller-Trutwin, M.C.; Novembre, F.J.; Peeters, M.; Courgnaud, V.; et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 2006, 125, 1055–1067. [Google Scholar] [CrossRef] [Green Version]
- Herrscher, C.; Pastor, F.; Burlaud-Gaillard, J.; Dumans, A.; Seigneuret, F.; Moreau, A.; Patient, R.; Eymieux, S.; de Rocquigny, H.; Hourioux, C.; et al. Hepatitis B virus entry into HepG2-NTCP cells requires clathrin-mediated endocytosis. Cell Microbiol. 2020, 22, e13205. [Google Scholar] [CrossRef]
- Trepo, C. A brief history of hepatitis milestones. Liver Int. 2014, 34 (Suppl. S1), 29–37. [Google Scholar] [CrossRef]
- Hayes, C.N.; Zhang, Y.; Makokha, G.N.; Hasan, M.Z.; Omokoko, M.D.; Chayama, K. Early events in hepatitis B virus infection: From the cell surface to the nucleus. J. Gastroenterol. Hepatol. 2016, 31, 302–309. [Google Scholar] [CrossRef] [Green Version]
- Schulze, A.; Gripon, P.; Urban, S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 2007, 46, 1759–1768. [Google Scholar] [CrossRef]
- Verrier, E.R.; Colpitts, C.C.; Bach, C.; Heydmann, L.; Weiss, A.; Renaud, M.; Durand, S.C.; Habersetzer, F.; Durantel, D.; Abou-Jaoude, G.; et al. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology 2016, 63, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Zhang, F.; Duan, L.; Wang, B.; Ye, Y.; Li, P.; Li, D.; Yang, S.; Zhou, L.; Chen, W. E-cadherin Plays a Role in Hepatitis B Virus Entry Through Affecting Glycosylated Sodium-Taurocholate Cotransporting Polypeptide Distribution. Front Cell Infect Microbiol. 2020, 10, 74. [Google Scholar] [CrossRef] [Green Version]
- Umetsu, T.; Inoue, J.; Kogure, T.; Kakazu, E.; Ninomiya, M.; Iwata, T.; Takai, S.; Nakamura, T.; Sano, A.; Shimosegawa, T. Inhibitory effect of silibinin on hepatitis B virus entry. Biochem. Biophys. Rep. 2018, 14, 20–25. [Google Scholar] [CrossRef]
- Ren, S.; Ding, C.; Sun, Y. Morphology Remodeling and Selective Autophagy of Intracellular Organelles during Viral Infections. Int. J. Mol. Sci. 2020, 21, 3689. [Google Scholar] [CrossRef]
- Helenius, A.; Kartenbeck, J.; Simons, K.; Fries, E. On the entry of Semliki forest virus into BHK-21 cells. J. Cell Biol. 1980, 84, 404–420. [Google Scholar] [CrossRef] [Green Version]


| Group | Viral Family Members included in the Review | Viral Protein Involved | AP Complex Involved | References | ||
|---|---|---|---|---|---|---|
| dsDNA Viruses | Adenoviridae | Human Adenoviruses (HAdV, HAdV2, HAdV5) | RIDα, RIDβ | AP-1, AP-2 | [62,63,64] | |
| Herpesviridae | α-herpesvirinae | Herpes simplex virus 1 and 2 (HSV-1, HSV-2) | gE/gI, VP22 | AP-1 | [65,66,67] | |
| Pseudorabiesvirus (PRV) | gB, gE | AP-1, AP-2 | [68] | |||
| Varicella zoster virus (VZV) | ORF9p | AP-1 | [69] | |||
| β-herpesvirinae | Murine Cytomegalovirus (MCMV) | m154, gp48, m04 | AP-1, AP-2, AP-3 | [46,70,71] | ||
| Human Cytomegalovirus (HCMV) | UL20 | AP-1 | [72] | |||
| Human herpes virus 6, -7 (HHV-6, -7) | U21 | AP-1, AP-3 | [47] | |||
| γ-herpesvirinae | Epstein-Barr virus (EBV) | BILF1, BMRF, | AP-1, AP-4 | [57,73] | ||
| Kaposi’s sarcoma-associated herpesvirus (KSHV) | unknown | AP-2 | [74] | |||
| Polyomaviridae | Human polyomavirus (JCV) | agnoprotein | AP-2, AP-3 | [75] | ||
| Poxviridae | Vaccinia virus (VACV) | VACV F13, VACV A33 | AP-2 | [76,77] | ||
| ssDNA viruses (+ strand or “sense”) | not identified | / | / | / | / | |
| dsRNA viruses | not identified | / | / | / | / | |
| (+) ssRNA viruses (+ strand or sense) | Coronavirdae | Severe acute respiratory syndrome coronavirus (SARS-CoV, SARS-CoV-2) | unknown | AP-2 | [7,78,79] | |
| Murine hepatitis virus (MHV) | unknown | AP-2 | [80,81] | |||
| Flaviviridae | Dengue virus (DENV) | unknown | AP-1, AP-2 | [82,83] | ||
| Hepatitis C virus (HCV) | NS2, NS5A, core | AP-1, AP-2, AP-4 | [26,29,56,84,85,86] | |||
| Rhinoviridae | Human rhinovirus (hRV) | LDLR (minor group) | AP-2 | [41] | ||
| (−) ssRNA viruses (− strand or antisense) RNA | Filoviridae | Zaire ebolavirus (EBOV) | unknown | AP-1, AP-2 | [43,87] | |
| Nairoviriade | Crimean-congo hemorragic fever (CCHFV) | unknown | AP-2 | [88] | ||
| Orthomyxoviridae | Influenza A (IAV) | Hemagglutinin | AP-2 | [7,89,90] | ||
| Paramyxoviridae | Hendra virus (HeV) | M-protein | AP-3 | [91] | ||
| Nipah virus (NiV) | NiV-F, M-protein | AP-1, AP-3 | [91,92] | |||
| Pneumoviridae | Human respiratory syncytial virus (RSV) | M-protein | AP-2, AP-3 | [93,94,95] | ||
| Rhabdoviridae | Vesicular stomatitis Indiana virus (VSV) | VSV-G | AP-2, AP-3 | [50,96] | ||
| ssRNA-RT viruses (+ strand or sense) RNA with DNA intermediate in life-cycle | Retroviridae | Ecotropic murine leukemia virus (eMuLV) | Env | AP-3? | [97] | |
| Human immunodeficiency virus 1 (HIV-1) | Nef, Gag, Vpu | AP-1, AP-2, AP-3 | [98,99,100,101] | |||
| Human immunodeficiency virus 2 (HIV-2) | Nef, Env, Gag | AP-1, AP-2, AP-3, AP-5 | [61,102,103] | |||
| dsDNA-RT viruses DNA with RNA intermediate in life-cycle | Hepadnaviridae | Hepatitis B virus (HVB) | preS1 | AP-2 | [104,105,106] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strazic Geljic, I.; Kucan Brlic, P.; Musak, L.; Karner, D.; Ambriović-Ristov, A.; Jonjic, S.; Schu, P.; Rovis, T.L. Viral Interactions with Adaptor-Protein Complexes: A Ubiquitous Trait among Viral Species. Int. J. Mol. Sci. 2021, 22, 5274. https://doi.org/10.3390/ijms22105274
Strazic Geljic I, Kucan Brlic P, Musak L, Karner D, Ambriović-Ristov A, Jonjic S, Schu P, Rovis TL. Viral Interactions with Adaptor-Protein Complexes: A Ubiquitous Trait among Viral Species. International Journal of Molecular Sciences. 2021; 22(10):5274. https://doi.org/10.3390/ijms22105274
Chicago/Turabian StyleStrazic Geljic, Ivana, Paola Kucan Brlic, Lucija Musak, Dubravka Karner, Andreja Ambriović-Ristov, Stipan Jonjic, Peter Schu, and Tihana Lenac Rovis. 2021. "Viral Interactions with Adaptor-Protein Complexes: A Ubiquitous Trait among Viral Species" International Journal of Molecular Sciences 22, no. 10: 5274. https://doi.org/10.3390/ijms22105274
APA StyleStrazic Geljic, I., Kucan Brlic, P., Musak, L., Karner, D., Ambriović-Ristov, A., Jonjic, S., Schu, P., & Rovis, T. L. (2021). Viral Interactions with Adaptor-Protein Complexes: A Ubiquitous Trait among Viral Species. International Journal of Molecular Sciences, 22(10), 5274. https://doi.org/10.3390/ijms22105274








