Exosome Traceability and Cell Source Dependence on Composition and Cell-Cell Cross Talk
Abstract
:1. Introduction
2. Overview of Exosomes
2.1. Exosome Origins, Secretion Mechanics, and Structure
2.2. Known Exosome Functions
2.3. Mechanism of Uptake
2.4. Exosomal Applications
3. Exosome-Cell Interactions
3.1. The Interaction of Cancer Cell-Derived Exosomes with Tumor Cells and Normal Cells
3.2. The Interaction of Normal Cell-Derived Exosomes with Normal Cells
3.3. The Interaction of Normal Cell-Derived Exosomes with Tumor Cells
3.4. The Interaction between Exosomes and the Nervous System
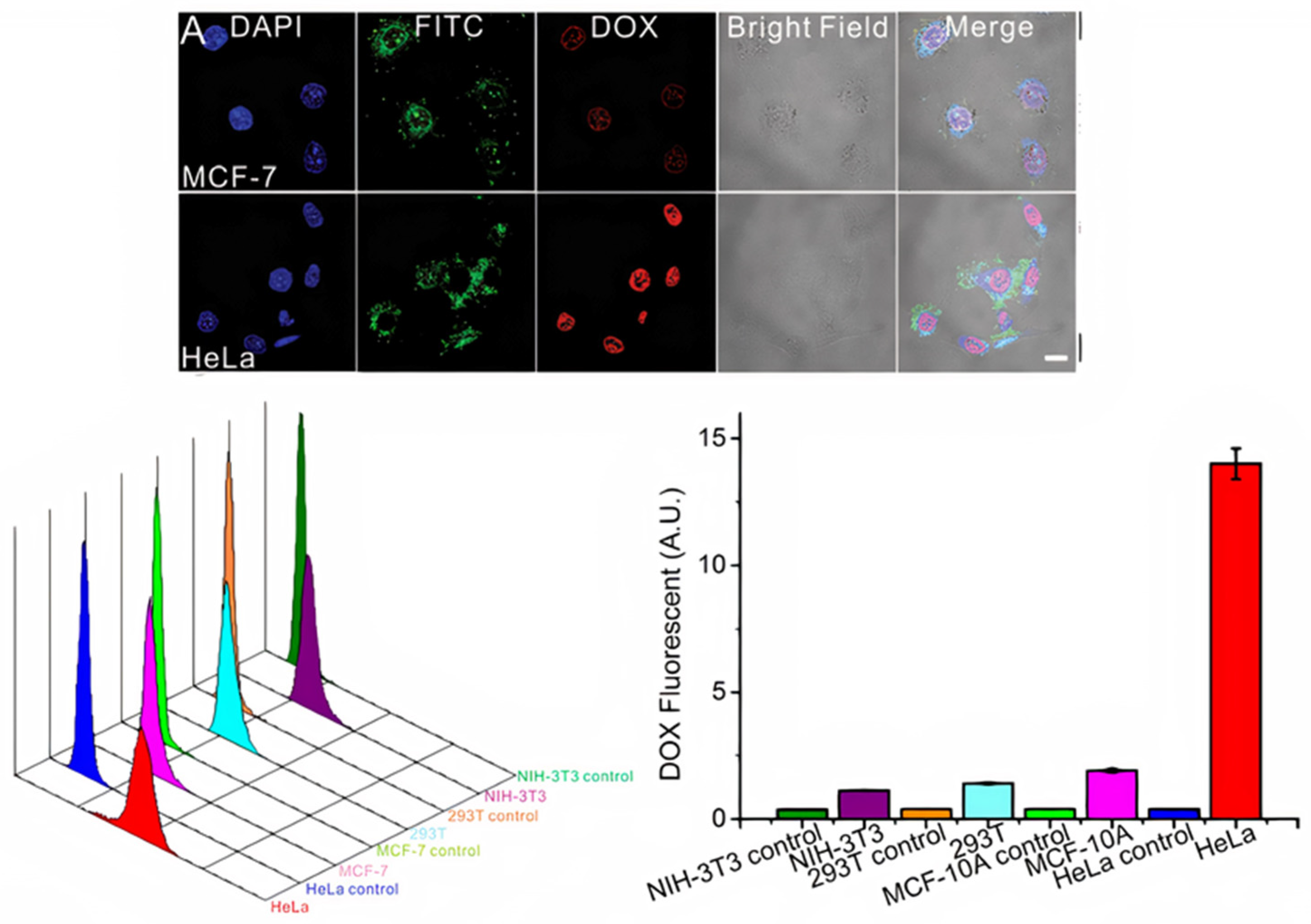
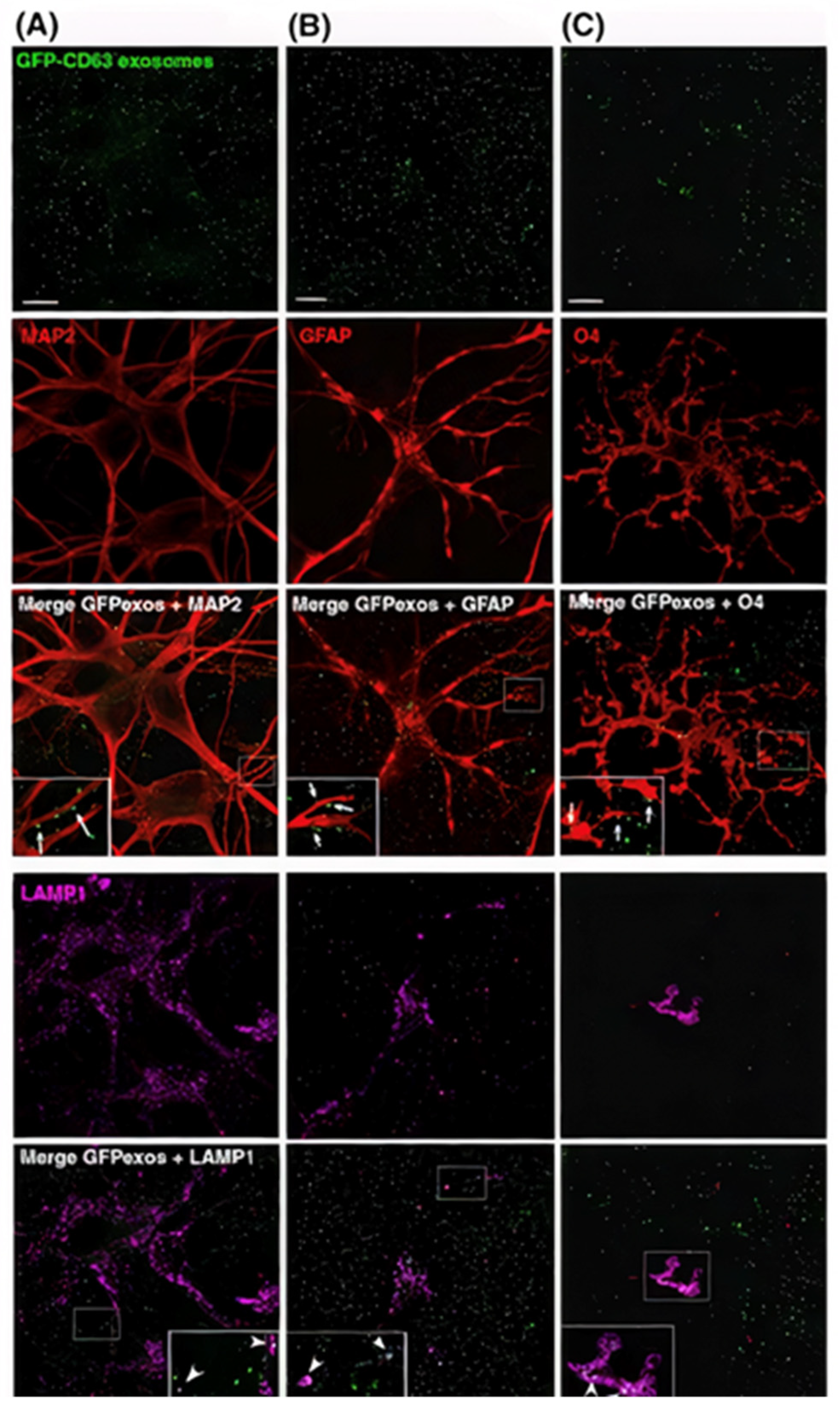
4. Exosome Labeling and Tracking: Bridging the Gap to Understand Exosome Functionality
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| NPS | Nanoparticles |
| CT | Computed tomography |
| MRI | Magnetic resonance imaging |
| PA | Photoacoustic microscopy |
| MVB | Multivesicular body membrane |
| ILVs | Intraluminal vesicles |
| ctDNA | Circulating tumor DNA |
| CTCs | Circulating tumor cells |
| cfDNA | Cell-free DNA |
| Exo | Exosomes |
| AST | Astrocyte cells |
| Oligo | Oligodendrocytes cells |
| CDE | Cancer cell-derived exosomes |
| U251-MG | Human brain cancer cells |
| MDA-MB-231 | Human breast cancer cells |
| HT-1080 | Human fibrosarcoma cells |
| EVs | Extracellular vesicles |
| DCs | Dendritic cells |
| MSC | Mesenchymal stem cell |
| 14DIV | Hippocampal cells |
| DRGs | Dorsal root ganglia cells |
| SCs | Schwann cells |
| HGNs | Hollow gold nanoparticle |
| BODIPY | Boron-dipyrromethene |
| PEG | Poly ethylene glycol |
| RGD | Arg-Gly-Asp |
| SEM | Scanning electron microscopy |
| TEM | Transmission electron microscopy |
| FC | Flow cytometry |
| GNRs | Gold nanorods |
| IONPs | Iron oxide nanoparticles |
References
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome Secretion: Molecular Mechanisms and Roles in Immune Responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Yang, H.; Zhang, X.; Xu, W. The emerging roles of exosomes in tumor–stroma interaction. J. Cancer Res. Clin. Oncol. 2016, 142, 1897–1907. [Google Scholar] [CrossRef] [PubMed]
- Yim, N.; Ryu, S.-W.; Choi, K.; Lee, K.R.; Lee, S.; Choi, H.; Kim, J.; Shaker, M.R.; Sun, M.R.S.W.; Park, J.-H.; et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein–protein interaction module. Nat. Commun. 2016, 7, 12277. [Google Scholar] [CrossRef]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and opportunities in exosome research—Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 3–21. [Google Scholar] [CrossRef] [Green Version]
- Maia, J.; Caja, S.; Strano, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Zech, D.; Rana, S.; Büchler, M.W.; Zöller, M. Tumor-exosomes and leukocyte activation: An ambivalent crosstalk. Cell Commun. Signal. 2012, 10, 1–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rana, S.; Yue, S.; Stadel, D.; Zöller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef]
- Othman, N.; Jamal, R.; Abu, N. Cancer-Derived Exosomes as Effectors of Key Inflammation-Related Players. Front. Immunol. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Chivet, M.; Javalet, C.; Laulagnier, K.; Blot, B.; Hemming, F.J.; Sadoul, R. Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. J. Extracell. Vesicles 2014, 3, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nat. Cell Biol. 2017, 546, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wang, G.; Hu, W.; Yao, Y.; Yu, X.-F. Emerging roles and therapeutic value of exosomes in cancer metastasis. Mol. Cancer 2019, 18, 1–11. [Google Scholar] [CrossRef]
- Vanni, I.; Alama, A.; Grossi, F.; Dal Bello, M.G.; Coco, S. Exosomes: A new horizon in lung cancer. Drug Discov. Today 2017, 22, 927–936. [Google Scholar] [CrossRef]
- Pan, J.; Ding, M.; Xu, K.; Yang, C.; Mao, L.-J. Exosomes in diagnosis and therapy of prostate cancer. Oncotarget 2017, 8, 97693–97700. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Fu, G.; Ming, L. Role of exosomes in pancreatic cancer (Review). Oncol. Lett. 2018, 15, 7479–7488. [Google Scholar] [CrossRef]
- Rodrigues, G.; Hoshino, A.; Kenific, C.M.; Matei, I.R.; Steiner, L.; Freitas, D.; Kim, H.S.; Oxley, P.R.; Scandariato, I.; Casanova-Salas, I.; et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019, 21, 1403–1412. [Google Scholar] [CrossRef]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome Delivered Anticancer Drugs Across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef]
- Lopez-Verrilli, M.A.; Picou, F.; Court, F.A. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia 2013, 61, 1795–1806. [Google Scholar] [CrossRef]
- He, F.; Ye, Z.-Y.; Zhao, L.-D.; Yin, B.-C.; Ye, B.-C. Probing exosome internalization pathways through confocal microscopy imaging. Chem. Commun. 2019, 55, 14015–14018. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.; Ashiq, K.A.; Phulpagar, P.; Singh, D.K.; Shiras, A. Effective Visualization and Easy Tracking of Extracellular Vesicles in Glioma Cells. Biol. Proced. Online 2019, 21, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Wu, T.; Zhang, K.; Meng, X.; Dai, W.; Wang, D.; Dong, H.; Zhang, X. Engineered Exosome-Mediated Near-Infrared-II Region V2C Quantum Dot Delivery for Nucleus-Target Low-Temperature Photothermal Therapy. ACS Nano 2019, 13, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wickline, S.A.; Hood, J.L. Magnetic resonance imaging of melanoma exosomes in lymph nodes. Magn. Reson. Med. 2015, 74, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Betzer, O.; Perets, N.; Angel, A.; Motiei, M.; Sadan, T.; Yadid, G.; Offen, D.; Popovtzer, R. In Vivo Neuroimaging of Exosomes Using Gold Nanoparticles. ACS Nano 2017, 11, 10883–10893. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular Vesicles in Cancer: Exosomes, Microvesicles and the Emerging Role of Large Oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Sahu, R.; Kaushik, S.; Clement, C.; Cannizzo, E.S.; Scharf, B.; Follenzi, A.; Potolicchio, I.; Nieves, E.; Cuervo, A.M.; Santambrogio, L. Microautophagy of Cytosolic Proteins by Late Endosomes. Dev. Cell 2011, 20, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Beer, K.B.; Wehman, A.M. Mechanisms and functions of extracellular vesicle release in vivo—What we can learn from flies and worms. Cell Adhes. Migr. 2017, 11, 135–150. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Hong, Y.; Cho, E.; Kim, G.B.; Kim, I.-S. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell. Vesicles 2018, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamzah, R.N.; Kore, R.A.; Alghazali, K.M.; Biris, A.S.; Griffin, R.J. Exosomes as Naturally Occurring, Abundant Nanoscale Soft Materials: Potential as Biomarkers and Delivery Vehicles for Solving Biomedical Problems. In Soft Matter and Biomaterials on the Nanoscale; Kim, J., Roper, D.K., Li, W., Eds.; World Scientific: Singapore, 2020; Volume 4, pp. 247–278. [Google Scholar]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Frydrychowicz, M.; Kolecka-Bednarczyk, A.; Madejczyk, M.; Yasar, S.; Dworacki, G. Exosomes-Structure, Biogenesis and Biological Role in Non-Small-Cell Lung Cancer. Scand. J. Immunol. 2015, 81, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-P.; Lin, Z.-X.; Jiang, X.-Y.; Yu, X.-Y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018, 39, 542–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, A.; Simons, M. Exosomes: Vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 2012, 352, 33–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilligan, K.E.; Dwyer, R.M. Engineering Exosomes for Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 1122. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Xu, Q. Functions and application of exosomes. Acta Pol. Pharm. Drug Res. 2014, 71, 537–543. [Google Scholar]
- Kore, R.A.; Henson, J.C.; Hamzah, R.N.; Griffin, R.J.; Tackett, A.J.; Ding, Z.; Mehta, J.L. Molecular events in MSC exosome mediated cytoprotection in cardiomyocytes. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Yanhua, T.; Suping, L.; Jian, S.; Tianjiao, J.; Motao, Z.; Gregory, J.A.; Jingyan, W.; Guangjun, N. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Pandanaboina, S.C.; Alghazali, K.M.; Nima, Z.A.; Alawajji, R.A.; Sharma, K.D.; Watanabe, F.; Saini, V.; Biris, A.S.; Srivatsan, M. Plasmonic nano surface for neuronal differentiation and manipulation. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102048. [Google Scholar] [CrossRef]
- Alghazali, K.M.; Newby, S.D.; Nima, Z.A.; Hamzah, R.N.; Watanabe, F.; Bourdo, S.E.; Masi, T.J.; Stephenson, S.M.; Anderson, D.E.; Dhar, M.S.; et al. Functionalized gold nanorod nanocomposite system to modulate differentiation of human mesenchymal stem cells into neural-like progenitors. Sci. Rep. 2017, 7, 16654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, H.; He, X.; Zheng, J. Exosomes and regenerative medicine: State of the art and perspectives. Transl. Res. J. Lab. Clin. Med. 2018, 196, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Miyaki, S.; Ishitobi, H.; Matsuyama, S.; Nakasa, T.; Kamei, N.; Akimoto, T.; Higashi, Y.; Ochi, M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015, 589, 1257–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuta, T.; Miyaki, S.; Ishitobi, H.; Ogura, T.; Kato, Y.; Kamei, N.; Miyado, K.; Higashi, Y.; Ochi, M. Mesenchymal Stem Cell-Derived Exosomes Promote Fracture Healing in a Mouse Model. Stem Cells Transl. Med. 2016, 5, 1620–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 1–15. [Google Scholar] [CrossRef]
- Mulcahy, L.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuyama, K.; Sun, H.; Mitsutake, S.; Igarashi, Y. Sphingolipid-modulated Exosome Secretion Promotes Clearance of Amyloid-β by Microglia. J. Biol. Chem. 2012, 287, 10977–10989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helming, L.; Gordon, S. The molecular basis of macrophage fusion. Immunobiology 2008, 212, 785–793. [Google Scholar] [CrossRef]
- Aplin, A.E.; Howe, A.; Alahari, S.K.; Juliano, R.L. Signal transduction and signal modulation by cell adhesion receptors: The role of integrins, cadherins, immunoglobulin-cell adhesion molecules, and selectins. Pharmacol. Rev. 1998, 50, 197–263. [Google Scholar]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef] [Green Version]
- van den Boorn, J.G.; Daßler, J.; Coch, C.; Schlee, M.; Hartmann, G. Exosomes as nucleic acid nanocarriers. Adv. Drug Deliv. Rev. 2013, 65, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.; Collinson, A.; Matthews, C.; Pointon, A.; Jenkinson, L.; Minter, R.R.; Vaughan, T.J.; Tigue, N.J. Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency. PLoS ONE 2019, 14, e0214545. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.-M.; Xu, Y.-M.; Huang, L.-F.; Wang, X.-Z. Exosomes: Novel Biomarkers for Clinical Diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef] [PubMed]
- Rabinowits, G.; Gerçel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Resnick, K.E.; Alder, H.; Hagan, J.P.; Richardson, D.L.; Croce, C.M.; Cohn, D.E. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 2009, 112, 55–59. [Google Scholar] [CrossRef]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef] [Green Version]
- Inamdar, S.; Nitiyanandan, R.; Rege, K. Emerging applications of exosomes in cancer therapeutics and diagnostics. Bioeng. Transl. Med. 2017, 2, 70–80. [Google Scholar] [CrossRef] [PubMed]
- De Jong, O.G.; Van Balkom, B.W.M.; Schiffelers, R.M.; Bouten, C.; Verhaar, M.C. Extracellular Vesicles: Potential Roles in Regenerative Medicine. Front. Immunol. 2014, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Richard, B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non–small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.; Enderle, D.; Noerholm, M.; Breakefield, X.; Skog, J. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Janku, F.; Zhan, Q.; Fan, J.-B. Accessing Genetic Information with Liquid Biopsies. Trends Genet. 2015, 31, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [Green Version]
- Keserű, J.S.; Soltész, B.; Lukács, J.; Márton, É.; Szilágyi-Bónizs, M.; Penyige, A.; Póka, R.; Nagy, B. Detection of cell-free, exosomal and whole blood mitochondrial DNA copy number in plasma or whole blood of patients with serous epithelial ovarian cancer. J. Biotechnol. 2019, 298, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Möhrmann, L.; Huang, H.J.; Hong, D.S.; Tsimberidou, A.M.; Funda, M.-B.; Piha-Paul, S.A.; Subbiah, V.; Karp, D.D.; Naing, A.; Krug, A.; et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin. Cancer Res. 2018, 24, 181–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tayoun, T.; Faugeroux, V.; Oulhen, M.; Aberlenc, A.; Pawlikowska, P.; Farace, F. CTC-Derived Models: A Window into the Seeding Capacity of Circulating Tumor Cells (CTCs). Cells 2019, 8, 1145. [Google Scholar] [CrossRef] [Green Version]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toda, Y.; Takata, K.; Nakagawa, Y.; Kawakami, H.; Fujioka, S.; Kobayashi, K.; Hattori, Y.; Kitamura, Y.; Akaji, K.; Ashihara, E. Effective internalization of U251-MG-secreted exosomes into cancer cells and characterization of their lipid components. Biochem. Biophys. Res. Commun. 2015, 456, 768–773. [Google Scholar] [CrossRef] [Green Version]
- Hellwinkel, J.E.; Redzic, J.S.; Harland, T.A.; Gunaydin, D.; Anchordoquy, T.J.; Graner, M.W. Glioma-derived extracellular vesicles selectively suppress immune responses. Neuro-Oncol. 2016, 18, 497–506. [Google Scholar] [CrossRef]
- Laura, M.; Epple, L.M.; Steve, G.; Griffiths, S.G.; Anjelika, M.; Dechkovskaia, A.M.; Nathaniel, L.; Dusto, N.L.; Jason White, J.; Rodney, J.; et al. Medulloblastoma Exosome Proteomics Yield Functional Roles for Extracellular Vesicles. PLoS ONE 2012, 7, e42064. [Google Scholar] [CrossRef] [Green Version]
- Oushy, S.; Hellwinkel, J.E.; Wang, M.; Nguyen, G.J.; Gunaydin, D.; Harland, T.A.; Anchordoquy, T.J.; Graner, M.W. Glioblastoma multiforme-derived extracellular vesicles drive normal astrocytes towards a tumour-enhancing phenotype. Philos. Trans. R. Soc. B Biol. Sci. 2017, 373, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Kowal, J.; Zucchetti, A.E.; Enserink, L.; Jouve, M.; Lankar, D.; Saitakis, M.; Martin-Jaular, L.; Théry, C. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 2017, 36, 3012–3028. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dong, Y.; Li, Y.; Li, W.; Cheng, K.; Qian, Y.; Xu, G.; Zhang, X.; Hu, L.; Chen, P.; et al. Designer Exosomes for Active Targeted Chemo-Photothermal Synergistic Tumor Therapy. Adv. Funct. Mater. 2018, 28, 1–14. [Google Scholar] [CrossRef]
- Sancho-Albero, M.; Navascués, N.; Mendoza, G.; Sebastián, V.; Arruebo, M.; Martín-Duque, P.; Santamaría, J. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J. Nanobiotechnol. 2019, 17, 1–13. [Google Scholar] [CrossRef]
- Liu, W.; Bai, X.; Zhang, A.; Huang, J.; Xu, S.; Zhang, J. Role of Exosomes in Central Nervous System Diseases. Front. Mol. Neurosci. 2019, 12, 240. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, Y.; Su, X.; He, J.; Bai, W.; He, X. MSCs-Derived Exosomes and Neuroinflammation, Neurogenesis and Therapy of Traumatic Brain Injury. Front. Cell. Neurosci. 2017, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Fröhlich, D.; Kuo, W.P.; Frühbeis, C.; Sun, J.-J.; Zehendner, C.M.; Luhmann, H.J.; Pinto, S.; Toedling, J.; Trotter, J.; Krämer-Albers, E.-M. Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130510. [Google Scholar] [CrossRef]
- Franzen, C.A.; Simms, P.E.; Huis, A.F.; Foreman, K.E.; Kuo, P.C.; Gopal, N.; Gupta, G.N. Characterization of Uptake and Internalization of Exosomes by Bladder Cancer Cells. Biomed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Zong, S.; Wang, Z.; Lu, J.; Zhu, D.; Zhang, Y.; Cui, Y. Imaging and Intracellular Tracking of Cancer-Derived Exosomes Using Single-Molecule Localization-Based Super-Resolution Microscope. ACS Appl. Mater. Interfaces 2016, 8, 25825–25833. [Google Scholar] [CrossRef]
- Pospíchalová, V.; Svoboda, J.; Dave, Z.; Kotrbová, A.; Kaiser, K.; Klemova, D.; Ilkovics, L.; Hampl, A.; Crha, I.; Jandakova, E.; et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [Google Scholar] [CrossRef]
- Nima, Z.A.; Alwbari, A.M.; Dantuluri, V.; Hamzah, R.N.; Sra, N.; Motwani, P.; Arnaoutakis, K.; Levy, R.A.; Bohliqa, A.F.; Nedosekin, D.; et al. Targeting nano drug delivery to cancer cells using tunable, multi-layer, silver-decorated gold nanorods. J. Appl. Toxicol. 2017, 37, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.P.; Cruz, M.A.E.; Tovani, C.B.; Ciancaglini, P. Biomedical applications of nanotechnology. Biophys. Rev. 2017, 9, 79–89. [Google Scholar] [CrossRef]
- Kedziorek, D.A.; Kraitchman, D.L. Superparamagnetic Iron Oxide Labeling of Stem Cells for MRI Tracking and Delivery in Cardiovascular Disease. Methods Mol. Biol. 2010, 660, 171–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin-Cheng, Y.; Liu, H.; Guo, X.; Yin, B.-C.; Ye, B.-C. Direct Exosome Quantification via Bivalent-Cholesterol-Labeled DNA Anchor for Signal Amplification. Anal. Chem. 2017, 89, 12968–12975. [Google Scholar] [CrossRef]
- Kang, C.; Han, P.; Lee, J.S.; Lee, D.; Kim, D. Anchor, Spacer, and Ligand-Modified Engineered Exosomes for Trackable Targeted Therapy. Bioconjugate Chem. 2020, 31, 2541–2552. [Google Scholar] [CrossRef]
- Gao, X.; Ran, N.; Dong, X.; Zuo, B.; Yang, R.; Zhou, Q.; Moulton, H.M.; Seow, Y.; Yin, H. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci. Transl. Med. 2018, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Deng, W.; Klinke, D.J., II. Exosomes: Improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst 2015, 140, 6631–6642. [Google Scholar] [CrossRef] [Green Version]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamzah, R.N.; Alghazali, K.M.; Biris, A.S.; Griffin, R.J. Exosome Traceability and Cell Source Dependence on Composition and Cell-Cell Cross Talk. Int. J. Mol. Sci. 2021, 22, 5346. https://doi.org/10.3390/ijms22105346
Hamzah RN, Alghazali KM, Biris AS, Griffin RJ. Exosome Traceability and Cell Source Dependence on Composition and Cell-Cell Cross Talk. International Journal of Molecular Sciences. 2021; 22(10):5346. https://doi.org/10.3390/ijms22105346
Chicago/Turabian StyleHamzah, Rabab N., Karrer M. Alghazali, Alexandru S. Biris, and Robert J. Griffin. 2021. "Exosome Traceability and Cell Source Dependence on Composition and Cell-Cell Cross Talk" International Journal of Molecular Sciences 22, no. 10: 5346. https://doi.org/10.3390/ijms22105346
APA StyleHamzah, R. N., Alghazali, K. M., Biris, A. S., & Griffin, R. J. (2021). Exosome Traceability and Cell Source Dependence on Composition and Cell-Cell Cross Talk. International Journal of Molecular Sciences, 22(10), 5346. https://doi.org/10.3390/ijms22105346





