Genome-Wide Analysis of RAV Transcription Factors and Functional Characterization of Anthocyanin-Biosynthesis-Related RAV Genes in Pear
Abstract
:1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of RAV TFs in Pear
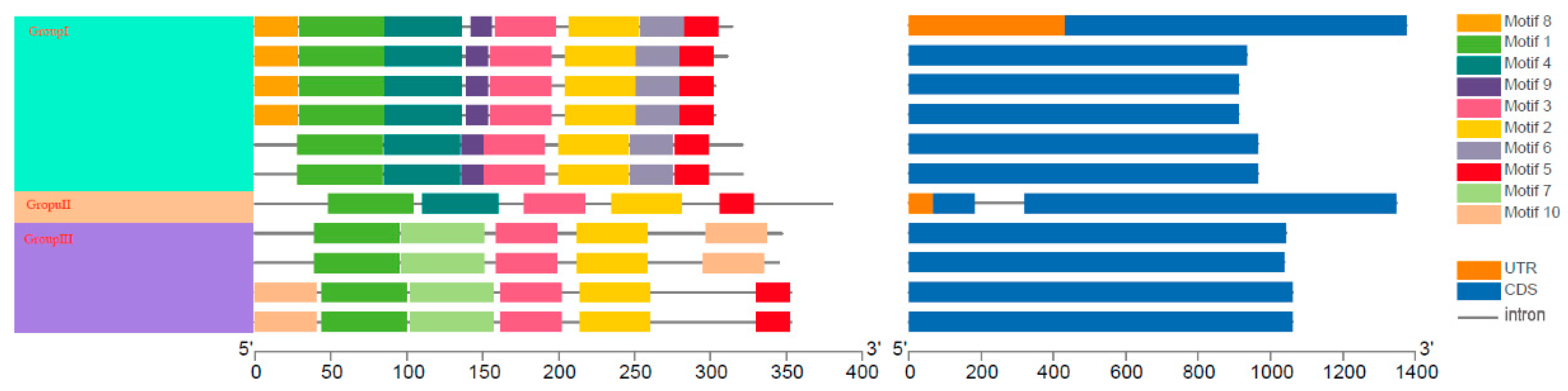
2.2. Conserved Motifs and Gene Structure of PbRAV TFs
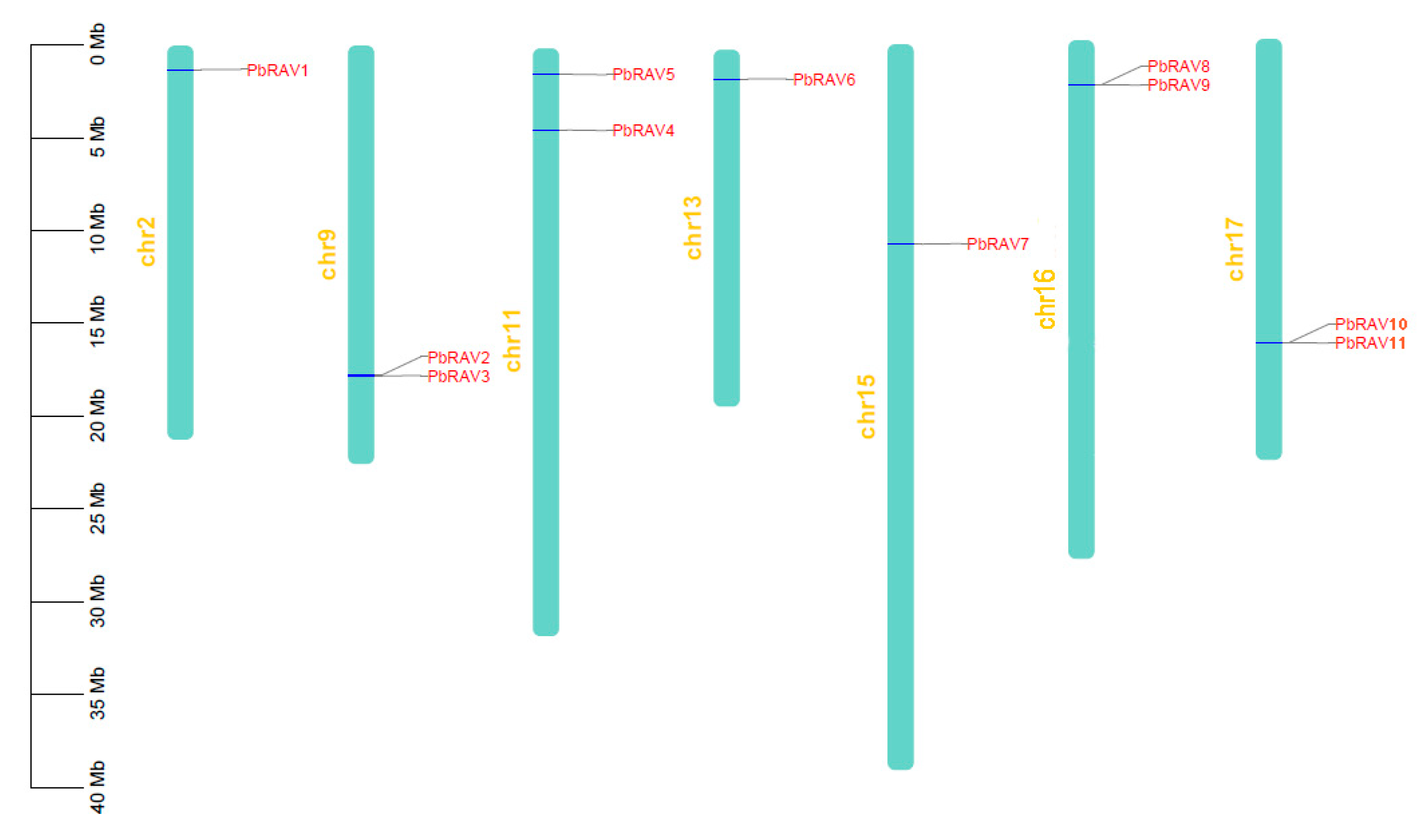
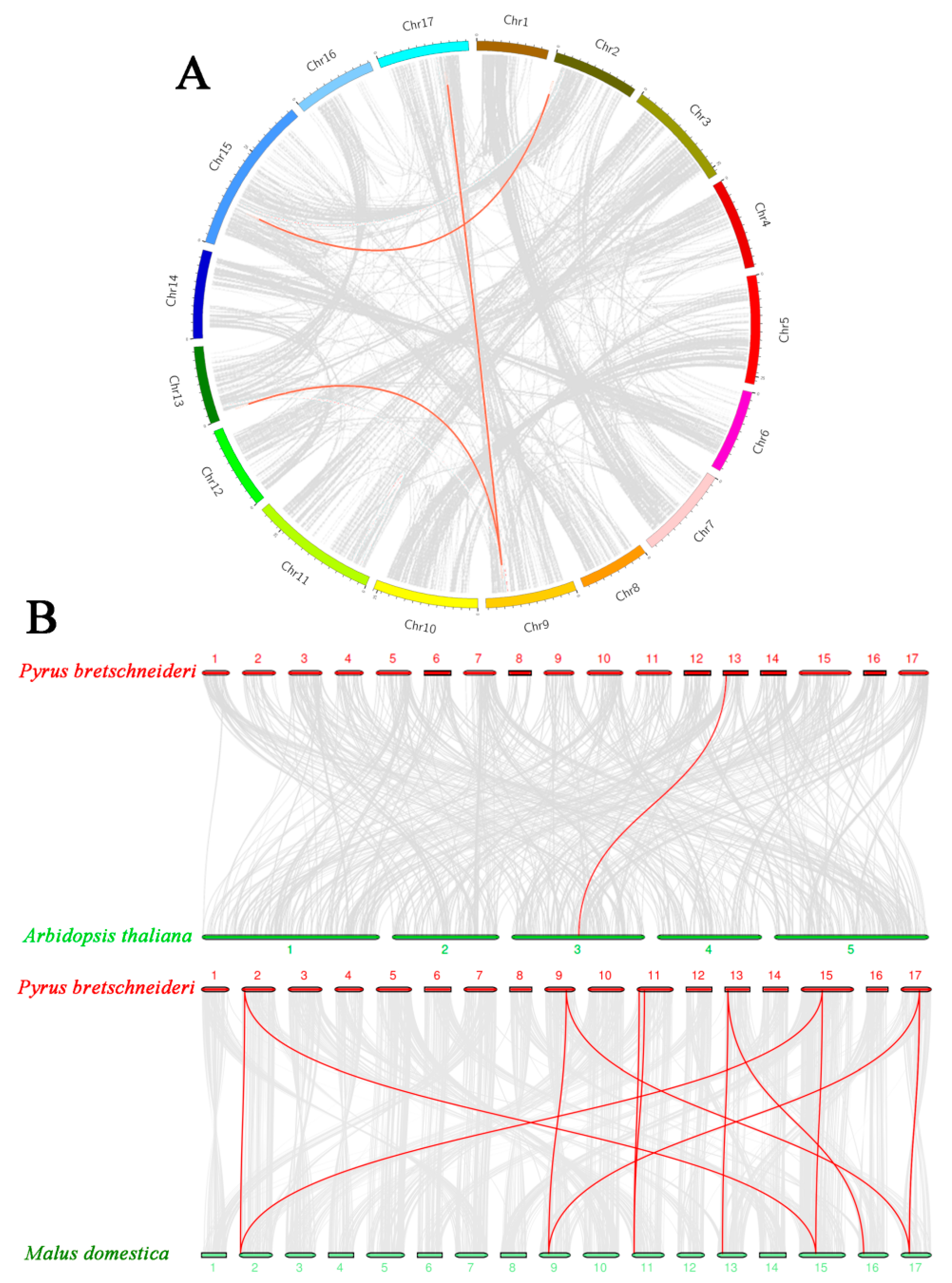
2.3. Chromosomal Distribution and Orthologous Relationships of PbRAV Genes in P. bretschneideri, Arabidopsis, and M. domestica
2.4. Expression Pattern of RAV Genes during Pear Development
2.5. Expression of PbRAV Genes in Response to Abiotic and Biotic Stress
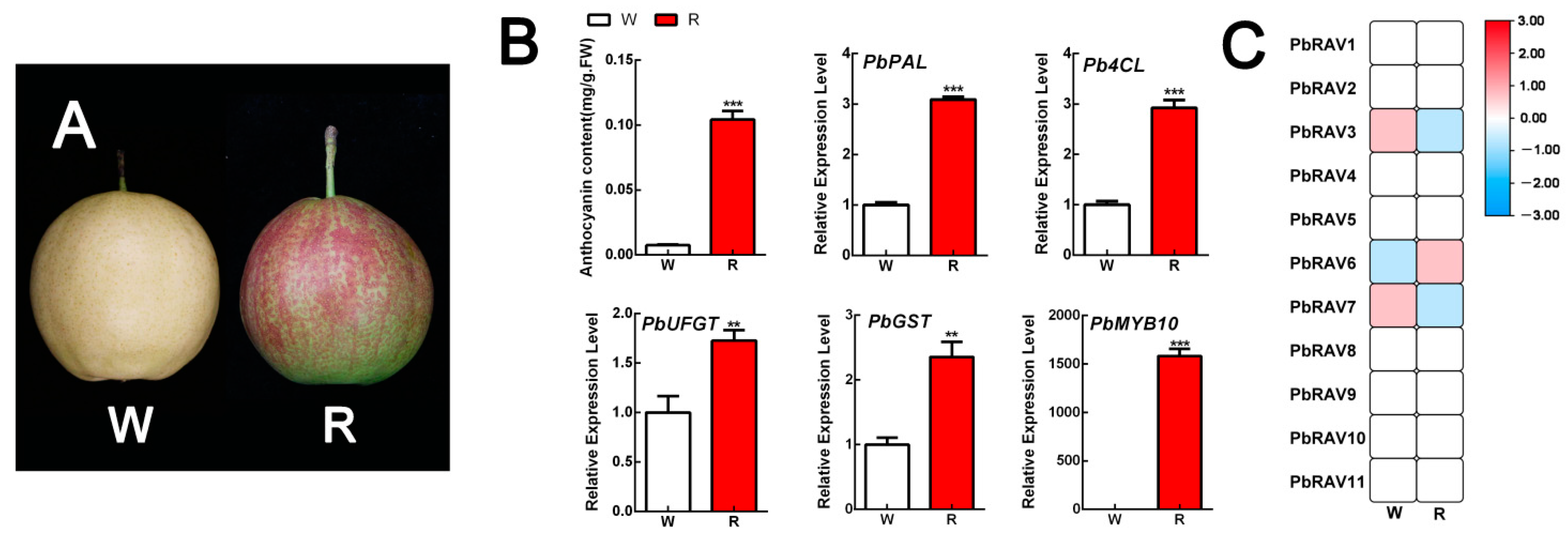
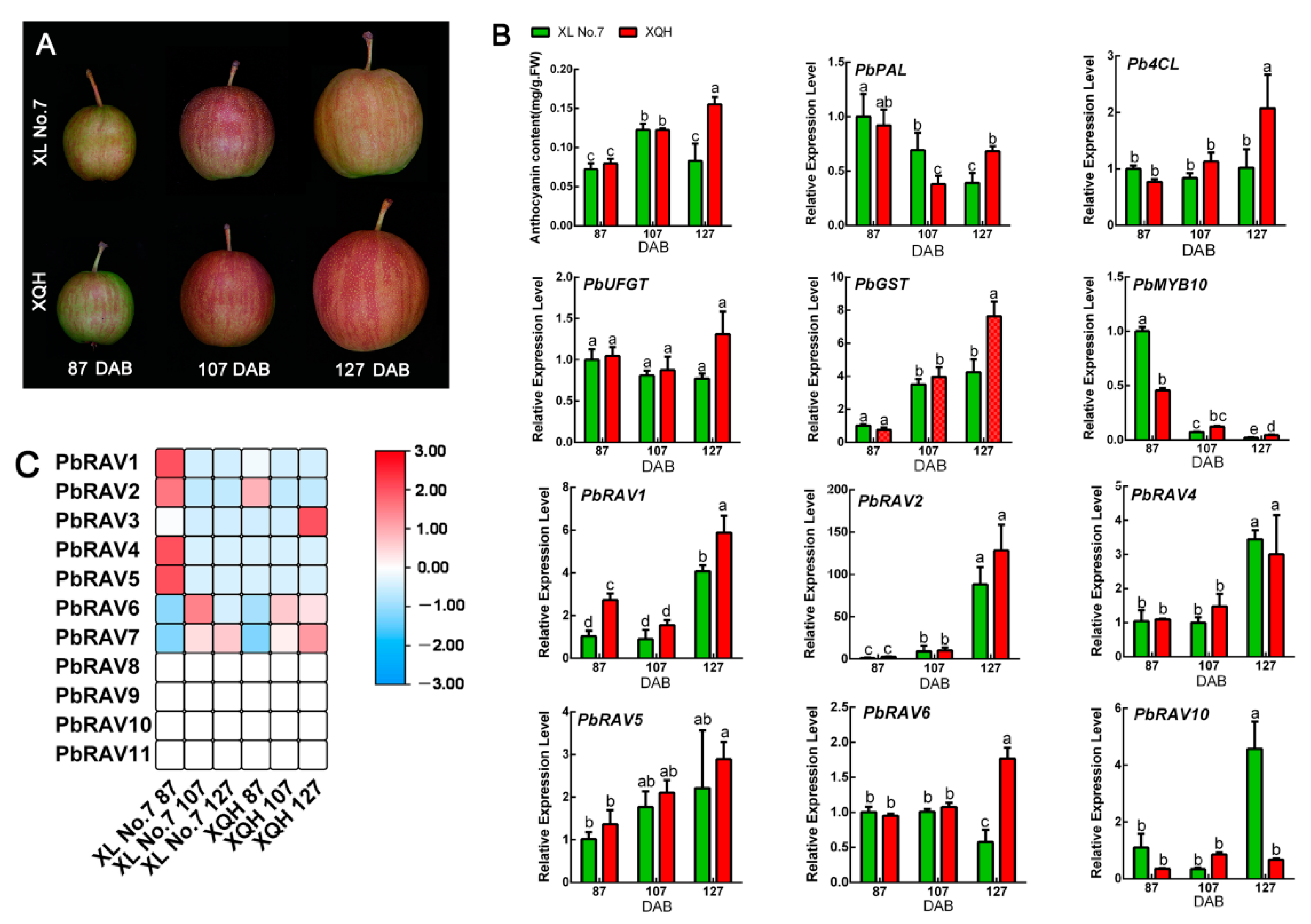
2.6. Expression of PbRAV Genes during Anthocyanin Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Database Search and Sequence Retrieval
4.2. Chromosomal Mapping, Intron/Exon Distribution, and Conserved Domain Analysis
4.3. Phylogenetic Comparison of RAV Proteins in P. bretschneideri, M. domestica, and A. thaliana
4.4. Expression Patterns of PbRAVs in Various Tissues of Pear at Different Developmental Stages
4.5. Expression Patterns of PbRAVs under Different Stress Conditions
4.5.1. Salt Stress
4.5.2. Drought Stress
4.5.3. Cork Spots
4.5.4. Pear Black Spot Disease
4.6. Plant Growth Conditions and Quantitative Real-Time (qRT)-PCR Analysis
4.7. Measurement of Anthocyanin Content in Peel
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Pear. Available online: http://www.fao.org/faostat/en/#data/QC Faq 2018 (accessed on 26 April 2021).
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions of the ERF transcription factor family in plants. Botany 2008, 86, 969–977. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Tsujimoto, Y.; Naito, Y.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the Arabidopsis and rice ERF gene families. Plant Cell Physiol. 2004, 45, S204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Li, F.; Liu, C.; Zhang, C.; Wu, Z.; Chen, Y. Isolation and characterization of an ERF transcription factor gene from cotton (Gossypium barbadense L.). Plant Mol. Biol. Rep. 2010, 28, 176–183. [Google Scholar] [CrossRef]
- Chen, N.; Yang, Q.; Su, M.; Pan, L.J.; Chi, X.Y.; Chen, M.N.; He, Y.A.; Yang, Z.; Wang, T.; Wang, M.A.; et al. Cloning of six ERF family transcription factor genes from peanut and analysis of their expression during abiotic stress. Plant Mol. Biol. Rep. 2012, 30, 1415–1425. [Google Scholar] [CrossRef]
- Liu, D.; Chen, X.; Liu, J.; Ye, J.; Guo, Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 2012, 63, 3899–3911. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Cao, X.L.; Bai, R.; Yao, N.; Li, L.B.; He, C.F. Isolation and characterization of the cold-induced Phyllostachys edulis AP2/ERF family transcription factor, peDREB1. Plant Mol. Biol. Rep. 2012, 30, 679–689. [Google Scholar] [CrossRef]
- Du, D.; Hao, R.; Cheng, T.; Pan, H.; Yang, W.; Wang, J.; Zhang, Q. Genome-wide analysis of the AP2/ERF gene family in Prunus mume. Plant Mol. Biol. Rep. 2013, 31, 741–750. [Google Scholar] [CrossRef]
- Girardi, C.L.; Rombaldi, C.V.; Dal Cero, J.; Nobile, P.M.; Laurens, F.; Bouzayen, M.; Quecini, V. Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis. Sci. Hortic. 2013, 151, 112–121. [Google Scholar] [CrossRef] [Green Version]
- Cao, P.B.; Azar, S.; SanClemente, H.; Mounet, F.; Dunand, C.; Marque, G.; Teulieres, C. Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis: An intriguing over-representation of stress-responsive DREB1/CBF genes. PLoS ONE 2015, 10, 0121041. [Google Scholar] [CrossRef] [PubMed]
- Charfeddine, M.; Sa€ıdi, M.N.; Charfeddine, S.; Hammami, A.; Bouzid, R.G. Genome-wide analysis and expression profiling of the ERF transcription factor family in potato (Solanum tuberosum L.). Mol. Biotechnol 2015, 57, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Lakhwani, D.; Pandey, A.; Dhar, Y.V.; Bag, S.K.; Trivedi, P.K.; Asif, M.H. Genome-wide analysis of the AP2/ERF family in Musa species reveals divergence and neofunctionalisation during evolution. Sci. Rep. 2016, 6, 18878. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Tao, S.; Wei, S.; Ming, M.; Huang, X.; Zhang, S.; Wu, J. The mining and evolutionary investigation of AP2/ERF genes in pear (Pyrus). BMC Plant Biol. 2018, 18, 46. [Google Scholar] [CrossRef] [Green Version]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchishinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.T.; Wu, X.; Wang, R.Z.; Chang, Y.J.; Qi, K.J.; Zhang, S.L.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, T.; Sato, F. The function of ETHYLENE RESPONSE FACTOR genes in the light-induced anthocyanin production of Arabidopsis thaliana leaves. Plant Biotechnol. 2018, 35, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xu, H.; Wang, N.; Jiang, S.; Fang, H.; Zhang, Z.; Yang, G.; Wang, Y.; Su, M.; Chen, X. The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol. Biol. 2018, 98, 205–218. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Li, Y.Y.; Song, L.Q.; Zhao, L.L.; You, C.X.; Hao, Y.J. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018, 178, 808–823. [Google Scholar] [CrossRef]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.J.; Kang, H.K.; Son, S.H.; Kim, S.K.; Nam, K.H. A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 2014, 55, 1892–1904. [Google Scholar] [CrossRef] [Green Version]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.X.; Lin, Z.; Yang, X.; Li, M.M.; Sun, J.Z.; Wang, K.; Li, Y.H.; Zheng, Y.H.; Yao, Y.H.; Li, W.B. GmRAV1 regulates regeneration of roots and adventitious buds by the cytokinin signaling pathway in Arabidopsis and soybean. Physiol. Plant. 2018, 165, 814–829. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.Z.; Yun, C.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2015, 80, 654–668. [Google Scholar] [CrossRef]
- Gao, Y.; Han, D.; Jia, W.; Ma, X.H.; Yang, Y.X.; Xu, Z.C. Molecular characterization and systematic analysis of NtAP2/ERF in tobacco and functional determination of NtRAV-4 under drought stress. Plant Physiol. Biochem. 2020, 156, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, F.; Liu, B.; Yang, S.L.; Xiong, X.; Hassani, D.; Zhang, Y.D. CmRAV1 shows differential expression in two melon (Cucumis melo L.) cultivars and enhances salt tolerance in transgenic Arabidopsis plants. Acta Biochim. Biophys. Sin. 2019, 51, 1123–1133. [Google Scholar] [CrossRef]
- Li, X.J.; Li, M.; Zhou, Y.; Hu, S.; Hu, R.; Chen, Y.; Li, X.B. Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS ONE 2015, 10, 0118056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Chang, Y.; Zeng, H.; Liu, G.Y.; He, C.Z.; Shi, H.T. RAV transcription factors are essential for disease resistance against cassava bacterial blight via activation of melatonin biosynthesis genes. J. Pineal Res. 2018, 64, e12454. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. Plant TFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Chen, Y.; Jin, X.F.; Cai, Y.X.; Yuan, Y.Y.; Fu, C.H.; Yu, L.J. New different origins and evolutionary processes of AP2/EREBP transcription factors in Taxus chinensis. BMC Plant Biol. 2019, 19, 413. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.X.; Zhang, T.Z. Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genom. 2017, 18, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Ramamoorthy, R.; Jiang, S.Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef]
- Rose, A.B. Intron-mediated regulation of gene expression. Curr. Top. Microbiol. Immunol. 2008, 326, 277–290. [Google Scholar]
- Li, H.; Wang, Y.; Wu, M.; Li, L.; Li, C.; Han, Z.; Yuan, J.; Chen, C.; Song, W.; Wang, C. Genome-wide identification of AP2/ERF transcription factors in Cauliflower and expression profiling of the ERF family under salt and drought stresses. Front. Plant Sci. 2017, 8, 946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Shi, Y.N.; Ma, Y.C.; Yang, X.F.; Yin, X.R.; Zhang, Y.Y.; Xiao, Y.W.; Liu, W.L.; Li, Y.D.; Li, S.J.; et al. The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 2020, 11, 2267–2279. [Google Scholar] [CrossRef] [Green Version]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, W1, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Qiao, X.; Yin, H.; Zhou, Y.; Dong, H.; Qi, K.; Li, L.; Zhang, S. Unbiased subgenome evolution following a recent whole-genome duplication in pear (Pyrus bretschneideri Rehd.). Hortic. Res. 2019, 6, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Wang, Z.; Liu, J.; Liu, F.; Zhai, R.; Zhu, C.; Wang, H.; Ma, F.; Xu, L. Histological, hormonal and transcriptomic reveal the changes upon gibberellininduced parthenocarpy in pear fruit. Hortic. Res. 2018, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Wei, Y.; Pang, H.; Xu, J.; Li, Y.; Zhang, H.; Zhang, J.; Zhang, Y. Genome-wide identification of the PEBP genes in pears and the putative role of PbFT in flower bud differentiation. PeerJ 2020, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.Y.; Xue, C.; Xu, L.; Sun, H.; Qin, M.F.; Zhang, S.; Wu, J. Distinct transcriptome profiles reveal gene expression patterns during fruit development and maturation in five main cultivated species of pear (Pyrus L.). Sci. Rep. 2016, 6, 28130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.Q.; Xu, X.Y.; Huang, X.S. Identification of differentially expressed genes related to dehydration resistance in a highly drought-tolerant pear, Pyrus betulaefolia, as through RNA-Seq. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hu, H.; Yu, D.; Sun, Z.; He, X.; Zhang, J.; Chen, Q.; Tian, R.; Fan, J. Candidate resistant genes of sand pear (Pyrus pyrifolia Nakai) to Alternaria alternate revealed by transcriptome sequencing. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circ. Calif. Agric. Expt. Stn. 1950, 347, 32. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xin, G.; Zhang, B.; Yang, J.Y. Optimization of extraction technique of anthocyanin from red peel of ‘Nanguo’ pear. Food Sci. 2009, 30, 97–100. [Google Scholar]



| Gene Name | Gene ID Number 1 | Chromosome | Start Site | Termination Site | Amino Acid Residues | MW (Da) | pI | Hydrophilicity | Subcellular Localization 2 |
|---|---|---|---|---|---|---|---|---|---|
| PbRAV1 | rna20334 | 2 | 1254332 | 1255369 | 345 | 39,656.68 | 8.75 | −0.663 | Nuclear |
| PbRAV2 | rna11028 | 9 | 17713298 | 17714715 | 314 | 35,670.50 | 8.61 | −0.525 | Cytoplasmic |
| PbRAV3 | rna11030 | 9 | 17751768 | 17752703 | 311 | 35,287.23 | 8.98 | −0.497 | Cytoplasmic |
| PbRAV4 | rna18153 | 11 | 4367928 | 4368989 | 353 | 40,901.03 | 6.34 | −0.635 | Cytoplasmic |
| PbRAV5 | rna51022 | 11 | 1362857 | 1363918 | 353 | 40,901.03 | 6.34 | −0.635 | Nuclear |
| PbRAV6 | rna37631 | 13 | 1545457 | 1547183 | 380 | 41,708.85 | 9.14 | −0.553 | Cytoplasmic |
| PbRAV7 | rna12487 | 15 | 10731607 | 10732650 | 347 | 40,063.17 | 8.87 | −0.631 | Nuclear |
| PbRAV8 | rna50871 | 16 | 2581667 | 2582926 | 313 | 35,191.35 | 8.79 | −0.491 | Cytoplasmic |
| PbRAV9 | rna50870 | 16 | 2568673 | 2569996 | 331 | 37,772.82 | 8.42 | −0.568 | Cytoplasmic |
| PbRAV10 | rna25443 | 17 | 16315324 | 16316296 | 303 | 34,391.24 | 8.89 | −0.491 | Cytoplasmic |
| PbRAV11 | rna25444 | 17 | 16328353 | 16329416 | 321 | 36,732.82 | 8.71 | −0.568 | Cytoplasmic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Deng, Z.; Liang, C.; Sun, H.; Li, D.; Song, J.; Zhang, S.; Wang, R. Genome-Wide Analysis of RAV Transcription Factors and Functional Characterization of Anthocyanin-Biosynthesis-Related RAV Genes in Pear. Int. J. Mol. Sci. 2021, 22, 5567. https://doi.org/10.3390/ijms22115567
Liu J, Deng Z, Liang C, Sun H, Li D, Song J, Zhang S, Wang R. Genome-Wide Analysis of RAV Transcription Factors and Functional Characterization of Anthocyanin-Biosynthesis-Related RAV Genes in Pear. International Journal of Molecular Sciences. 2021; 22(11):5567. https://doi.org/10.3390/ijms22115567
Chicago/Turabian StyleLiu, Jianlong, Zhiwei Deng, Chenglin Liang, Hongwei Sun, Dingli Li, Jiankun Song, Shaoling Zhang, and Ran Wang. 2021. "Genome-Wide Analysis of RAV Transcription Factors and Functional Characterization of Anthocyanin-Biosynthesis-Related RAV Genes in Pear" International Journal of Molecular Sciences 22, no. 11: 5567. https://doi.org/10.3390/ijms22115567





