Ubiquitin-Specific Protease 3 Deubiquitinates and Stabilizes Oct4 Protein in Human Embryonic Stem Cells
Abstract
:1. Introduction
2. Results
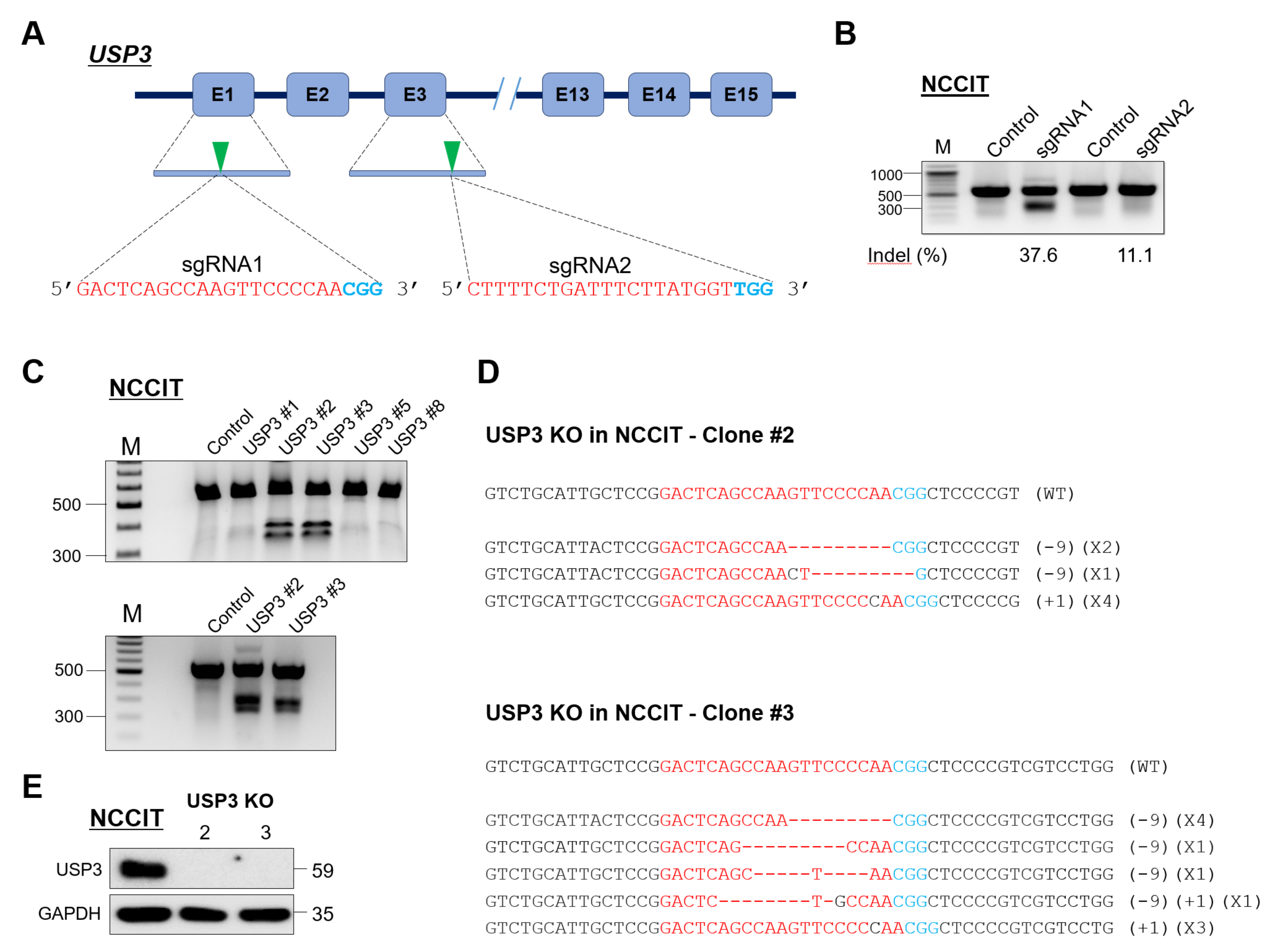
2.1. Generation of Single-Cell-Derived USP3 Gene Knockout Clones in Human Embryonic Carcinoma Stem Cells
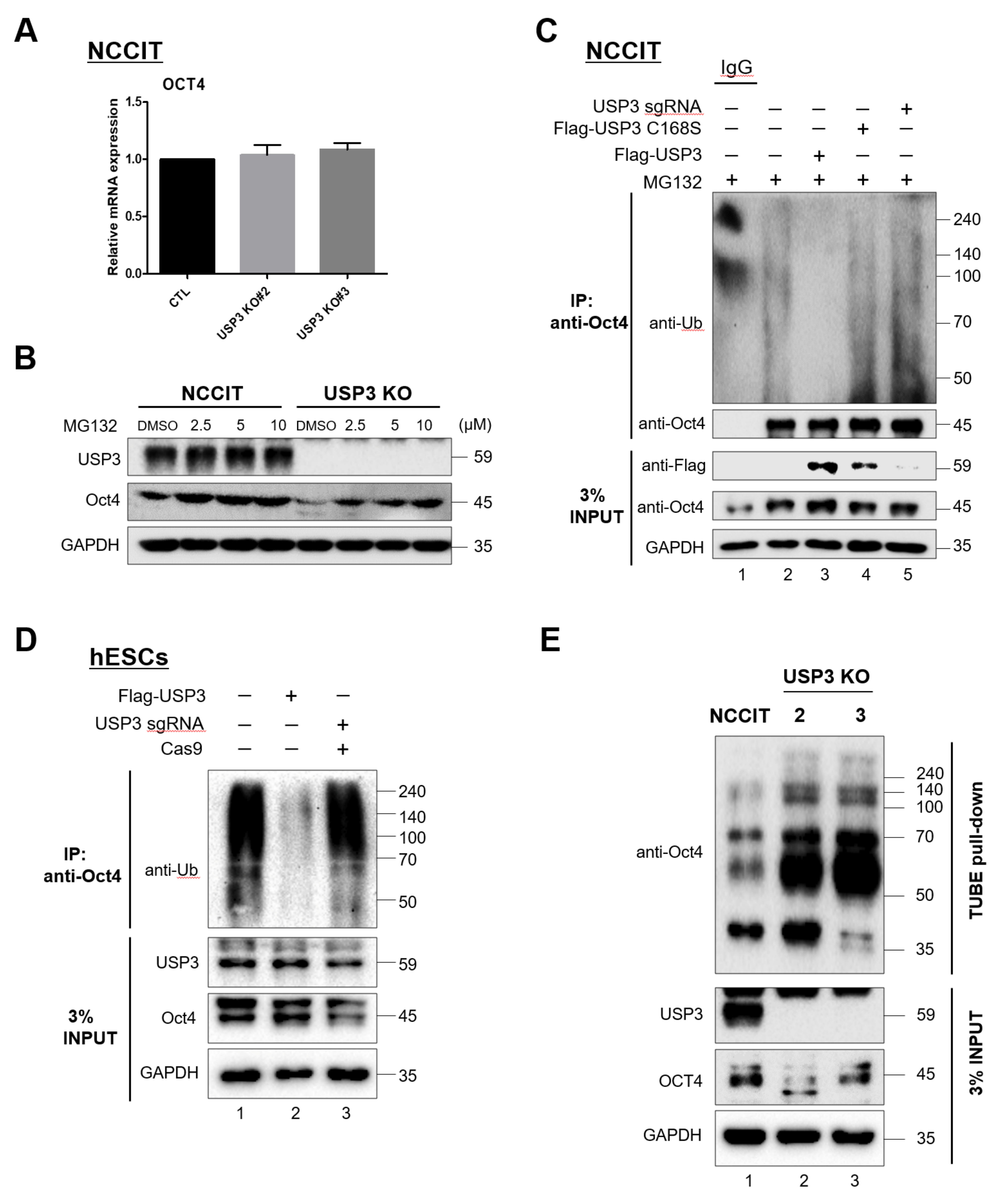
2.2. USP3 Regulated Oct4 Protein Stability and Half-Life
2.3. USP3 Interacted with Oct4
2.4. Deubiquitination of Oct4 by USP3
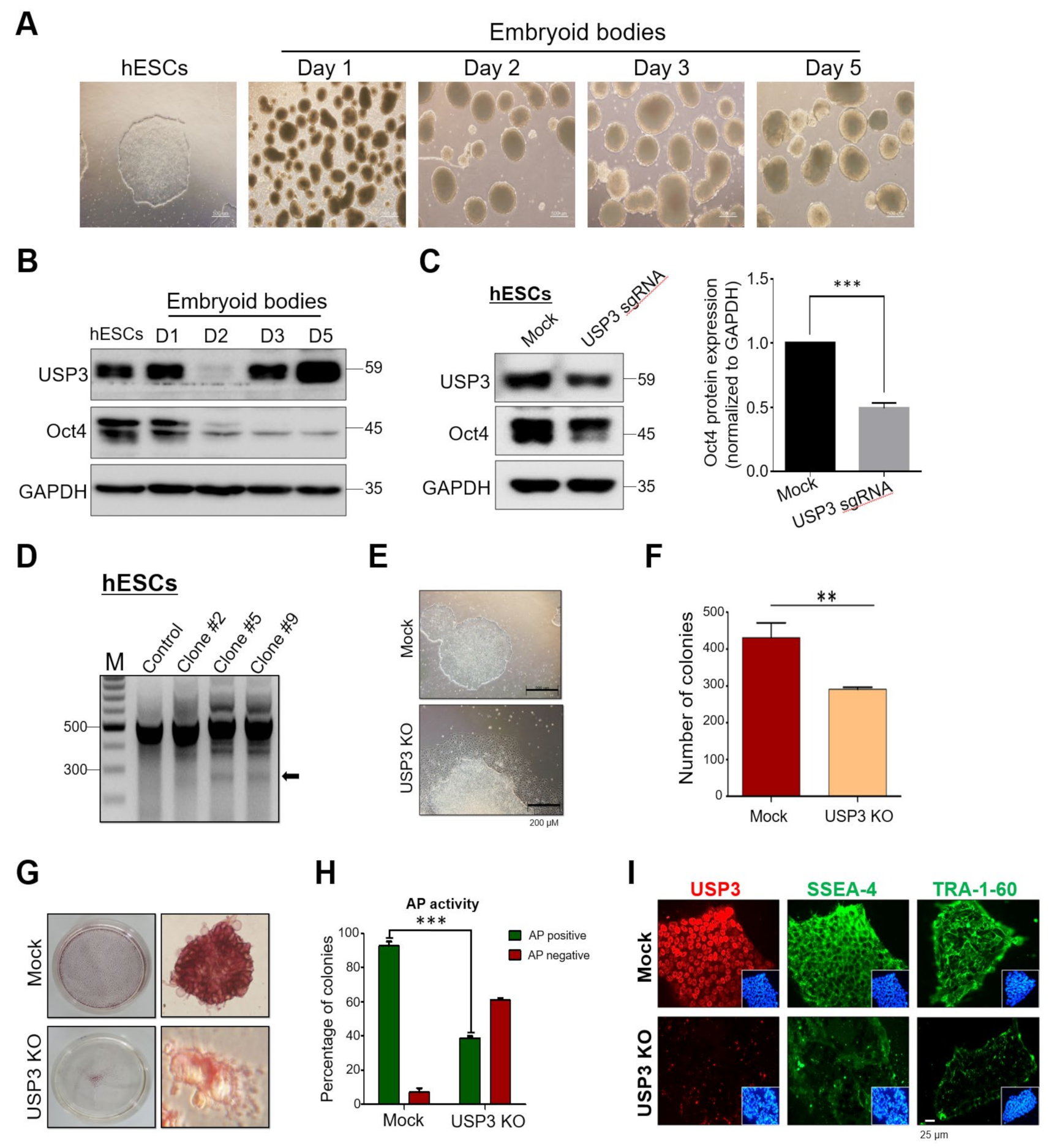
2.5. Loss of USP3 Affected Self-Renewal of hESCs
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Antibodies and Reagents
4.3. Cell Culture and Treatments
4.4. Embryoid Body (EB) Differentiation
4.5. Cas9 and sgRNA Constructs
4.6. T7E1 Assay
4.7. Generation of DUB Knockout Single-Cell-Derived Clones
4.8. Immunoprecipitation
4.9. Deubiquitination Assay
4.10. Immunofluorescence
4.11. qRT-PCR
4.12. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckley, S.M.; Aranda-Orgilles, B.; Strikoudis, A.; Apostolou, E.; Loizou, E.; Moran-Crusio, K.; Farnsworth, C.L.; Koller, A.A.; Dasgupta, R.; Silva, J.C.; et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 2012, 11, 783–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okita, Y.; Nakayama, K.I. UPS delivers pluripotency. Cell Stem Cell 2012, 11, 728–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatetsu, H.; Kong, N.R.; Chong, G.; Amabile, G.; Tenen, D.G.; Chai, L. SALL4, the missing link between stem cells, development and cancer. Gene 2016, 584, 111–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Suresh, B.; Lee, J.; Kim, H.; Ramakrishna, S. Regulation of pluripotency and differentiation by deubiquitinating enzymes. Cell Death Differ. 2016, 23, 1257–1264. [Google Scholar] [CrossRef]
- Wolford, J.L.; Chishti, Y.; Jin, Q.; Ward, J.; Chen, L.; Vogt, S.; Finney, L. Loss of pluripotency in human embryonic stem cells directly correlates with an increase in nuclear zinc. PLoS ONE 2010, 5, e12308. [Google Scholar] [CrossRef]
- Chen, L.; Daley, G.Q. Molecular basis of pluripotency. Hum. Mol. Genet. 2008, 17, R23–R27. [Google Scholar] [CrossRef] [Green Version]
- Niwa, H.; Miyazaki, J.; Smith, A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000, 24, 372–376. [Google Scholar] [CrossRef]
- Nichols, J.; Zevnik, B.; Anastassiadis, K.; Niwa, H.; Klewe-Nebenius, D.; Chambers, I.; Schöler, H.; Smith, A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998, 95, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Saxe, J.P.; Tomilin, A.; Schöler, H.R.; Plath, K.; Huang, J. Post-translational regulation of Oct4 transcriptional activity. PLoS ONE 2009, 4, e4467. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.M.; Liao, B.; Zhang, Q.J.; Wang, B.B.; Li, H.; Zhong, X.M.; Sheng, H.Z.; Zhao, Y.X.; Zhao, Y.M.; Jin, Y. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J. Biol. Chem. 2004, 279, 23495–23503. [Google Scholar] [CrossRef] [Green Version]
- Wei, F.; Schöler, H.R.; Atchison, M.L. Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J. Biol. Chem. 2007, 282, 21551–21560. [Google Scholar] [CrossRef] [Green Version]
- Tolkunova, E.; Malashicheva, A.; Parfenov, V.N.; Sustmann, C.; Grosschedl, R.; Tomilin, A. PIAS proteins as repressors of Oct4 function. J. Mol. Biol. 2007, 374, 1200–1212. [Google Scholar] [CrossRef]
- Jang, H.; Kim, T.W.; Yoon, S.; Choi, S.-Y.; Kang, T.-W.; Kim, S.-Y.; Kwon, Y.-W.; Cho, E.-J.; Youn, H.-D. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 2012, 11, 62–74. [Google Scholar] [CrossRef] [Green Version]
- Suresh, B.; Lee, J.; Kim, K.-S.; Ramakrishna, S. The Importance of Ubiquitination and Deubiquitination in Cellular Reprogramming. Stem Cells Int. 2016, 2016, 6705927. [Google Scholar] [CrossRef] [Green Version]
- Amm, I.; Sommer, T.; Wolf, D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 2014, 1843, 182–196. [Google Scholar] [CrossRef] [Green Version]
- Liao, B.; Zhong, X.; Xu, H.; Xiao, F.; Fang, Z.; Gu, J.; Chen, Y.; Zhao, Y.; Jin, Y. Itch, an E3 ligase of Oct4, is required for embryonic stem cell self-renewal and pluripotency induction. J. Cell Physiol. 2013, 228, 1443–1451. [Google Scholar] [CrossRef]
- Liao, B.; Jin, Y. Wwp2 mediates Oct4 ubiquitination and its own auto-ubiquitination in a dosage-dependent manner. Cell Res. 2010, 20, 332–344. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, D.; Shen, Y.; Tao, X.; Liu, L.; Zhong, Y.; Fang, S. DPF2 regulates OCT4 protein level and nuclear distribution. Biochim. Biophys. Acta 2015, 1853, 3279–3293. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Kang, H.G.; Kim, S.J.; Lee, S.; Jee, S.; Ahn, S.G.; Kang, M.J.; Song, J.S.; Chung, J.Y.; Yi, E.C.; et al. Post-translational modification of OCT4 in breast cancer tumorigenesis. Cell Death Differ. 2018, 25, 1781–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushal, K.; Antao, A.M.; Kim, K.-S.; Ramakrishna, S. Deubiquitinating enzymes in cancer stem cells: Functions and targeted inhibition for cancer therapy. Drug Discov. Today 2018, 23, 1974–1982. [Google Scholar] [CrossRef]
- Wei, X.; Guo, J.; Li, Q.; Jia, Q.; Jing, Q.; Li, Y.; Zhou, B.; Chen, J.; Gao, S.; Zhang, X.; et al. Bach1 regulates self-renewal and impedes mesendodermal differentiation of human embryonic stem cells. Sci. Adv. 2019, 5, eaau7887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Chandrasekaran, A.P.; Suresh, B.; Haq, S.; Kang, J.H.; Lee, S.J.; Kim, J.; Kim, J.; Lee, S.; Kim, H.H.; et al. Genome-scale screening of deubiquitinase subfamily identifies USP3 as a stabilizer of Cdc25A regulating cell cycle in cancer. Cell Death Differ. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Neganova, I.; Przyborski, S.; Yang, C.; Cooke, M.; Atkinson, S.P.; Anyfantis, G.; Fenyk, S.; Keith, W.N.; Hoare, S.F.; et al. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J. Cell Biol. 2009, 184, 67–82. [Google Scholar] [CrossRef] [PubMed]
- van der Laan, S.; Tsanov, N.; Crozet, C.; Maiorano, D. High Dub3 expression in mouse ESCs couples the G1/S checkpoint to pluripotency. Mol. Cell 2013, 52, 366–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bárta, T.; Vinarský, V.; Holubcová, Z.; Doležalová, D.; Verner, J.; Pospíšilová, Š.; Dvořák, P.; Hampl, A. Human Embryonic Stem Cells Are Capable of Executing G1/S Checkpoint Activation. Stem Cells 2010, 28, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.A.; Perez-Iratxeta, C.; Andrade-Navarro, M.A.; Rudnicki, M.A. Oct4 targets regulatory nodes to modulate stem cell function. PLoS ONE 2007, 2, e553. [Google Scholar] [CrossRef]
- Wu, Y.; Qin, J.; Li, F.; Yang, C.; Li, Z.; Zhou, Z.; Zhang, H.; Li, Y.; Wang, X.; Liu, R.; et al. USP3 promotes breast cancer cell proliferation by deubiquitinating KLF5. J. Biol. Chem. 2019, 294, 17837–17847. [Google Scholar] [CrossRef]
- Wu, X.; Liu, M.; Zhu, H.; Wang, J.; Dai, W.; Li, J.; Zhu, D.; Tang, W.; Xiao, Y.; Lin, J.; et al. Ubiquitin-specific protease 3 promotes cell migration and invasion by interacting with and deubiquitinating SUZ12 in gastric cancer. J. Exp. Clin. Cancer Res. 2019, 38, 277. [Google Scholar] [CrossRef] [Green Version]
- You, J.S.; Kang, J.K.; Seo, D.W.; Park, J.H.; Park, J.W.; Lee, J.C.; Jeon, Y.J.; Cho, E.J.; Han, J.W. Depletion of embryonic stem cell signature by histone deacetylase inhibitor in NCCIT cells: Involvement of Nanog suppression. Cancer Res. 2009, 69, 5716–5725. [Google Scholar] [CrossRef] [Green Version]
- Cauffman, G.; Liebaers, I.; Van Steirteghem, A.; Van de Velde, H. POU5F1 Isoforms Show Different Expression Patterns in Human Embryonic Stem Cells and Preimplantation Embryos. Stem Cells 2006, 24, 2685–2691. [Google Scholar] [CrossRef]
- Verneri, P.; Vazquez Echegaray, C.; Oses, C.; Stortz, M.; Guberman, A.; Levi, V. Dynamical reorganization of the pluripotency transcription factors Oct4 and Sox2 during early differentiation of embryonic stem cells. Sci. Rep. 2020, 10, 5195. [Google Scholar] [CrossRef]
- Cai, N.; Li, M.; Qu, J.; Liu, G.H.; Izpisua Belmonte, J.C. Post-translational modulation of pluripotency. J. Mol. Cell Biol. 2012, 4, 262–265. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, A.P.; Suresh, B.; Kim, H.H.; Kim, K.S.; Ramakrishna, S. Concise Review: Fate Determination of Stem Cells by Deubiquitinating Enzymes. Stem Cells 2017, 35, 9–16. [Google Scholar] [CrossRef]
- Saez, I.; Koyuncu, S.; Gutierrez-Garcia, R.; Dieterich, C.; Vilchez, D. Insights into the ubiquitin-proteasome system of human embryonic stem cells. Sci. Rep. 2018, 8, 4092. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Michowski, W.; Kolodziejczyk, A.; Sicinski, P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 2019, 21, 1060–1067. [Google Scholar] [CrossRef]
- Shin, J.; Kim, T.W.; Kim, H.; Kim, H.J.; Suh, M.Y.; Lee, S.; Lee, H.T.; Kwak, S.; Lee, S.E.; Lee, J.H.; et al. Aurkb/PP1-mediated resetting of Oct4 during the cell cycle determines the identity of embryonic stem cells. eLife 2016, 5, e10877. [Google Scholar] [CrossRef]
- Wu, G.; Schöler, H.R. Role of Oct4 in the early embryo development. Cell Regen. 2014, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishna, S.; Kwaku Dad, A.-B.; Beloor, J.; Gopalappa, R.; Lee, S.-K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Lee, H.J.; Kim, H.; Cho, S.W.; Kim, J.-S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009, 19, 1279–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene | sgRNA | Direction | Sequence (5′ to 3′) | |
|---|---|---|---|---|
| Oligonucleotides used for sgRNA plasmid construction | ||||
| USP3 | sgRNA1 | FP | GACTCAGCCAAGTTCCCCAA | |
| RP | TTGGGGAACTTGGCTGAGTC | |||
| sgRNA2 | FP | AGTTCAGCACACAGTATGTA | ||
| RP | TACATACTGTGTGCTGAACT | |||
| Oligonucleotides used for amplify the PCR target for the T7E1 assay | ||||
| USP3 | sgRNA1 | I PCR | FP | TCGGAGTTACACGTTCTACGG |
| RP | CTGCGGAGAAGCGCGG | |||
| II PCR | FP | TCGGAGTTACACGTTCTACGG | ||
| RP | GCCTCGGGAAACAAAGGA | |||
| sgRNA2 | I PCR | FP | CTTGCCTGAGCCTACTCTTGT | |
| RP | AGGATGGATGAGGAACGGGA | |||
| II PCR | FP | TCACTACATGATTGACTGCTGTT | ||
| RP | AGGATGGATGAGGAACGGGA | |||
| Gene | sgRNA | PCR Size | Cleavage Size | Orientation |
|---|---|---|---|---|
| USP3 | sgRNA1 | 507 | 280 + 227 | Sense |
| sgRNA2 | 504 | 274 + 230 | Sense |
| Gene | Direction | Sequence (5′ to 3′) |
|---|---|---|
| Oct4 | FP | GCTGGATGTCAGGGCTCTTT |
| RP | TCAAGAGATTTATCGAGCACCTTCT | |
| GAPDH | FP | GTCATCCCTGAGCTGAACGG |
| RP | CCACCTGGTGCTCAGTGTAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rhie, B.-H.; Antao, A.M.; Karapurkar, J.K.; Kim, M.-S.; Jo, W.-J.; Ramakrishna, S.; Kim, K.-S. Ubiquitin-Specific Protease 3 Deubiquitinates and Stabilizes Oct4 Protein in Human Embryonic Stem Cells. Int. J. Mol. Sci. 2021, 22, 5584. https://doi.org/10.3390/ijms22115584
Rhie B-H, Antao AM, Karapurkar JK, Kim M-S, Jo W-J, Ramakrishna S, Kim K-S. Ubiquitin-Specific Protease 3 Deubiquitinates and Stabilizes Oct4 Protein in Human Embryonic Stem Cells. International Journal of Molecular Sciences. 2021; 22(11):5584. https://doi.org/10.3390/ijms22115584
Chicago/Turabian StyleRhie, Byung-Ho, Ainsley Mike Antao, Janardhan Keshav Karapurkar, Min-Seong Kim, Won-Jun Jo, Suresh Ramakrishna, and Kye-Seong Kim. 2021. "Ubiquitin-Specific Protease 3 Deubiquitinates and Stabilizes Oct4 Protein in Human Embryonic Stem Cells" International Journal of Molecular Sciences 22, no. 11: 5584. https://doi.org/10.3390/ijms22115584







