Plastic Degradation by Extremophilic Bacteria
Abstract
1. General Features of Plastic Degradation
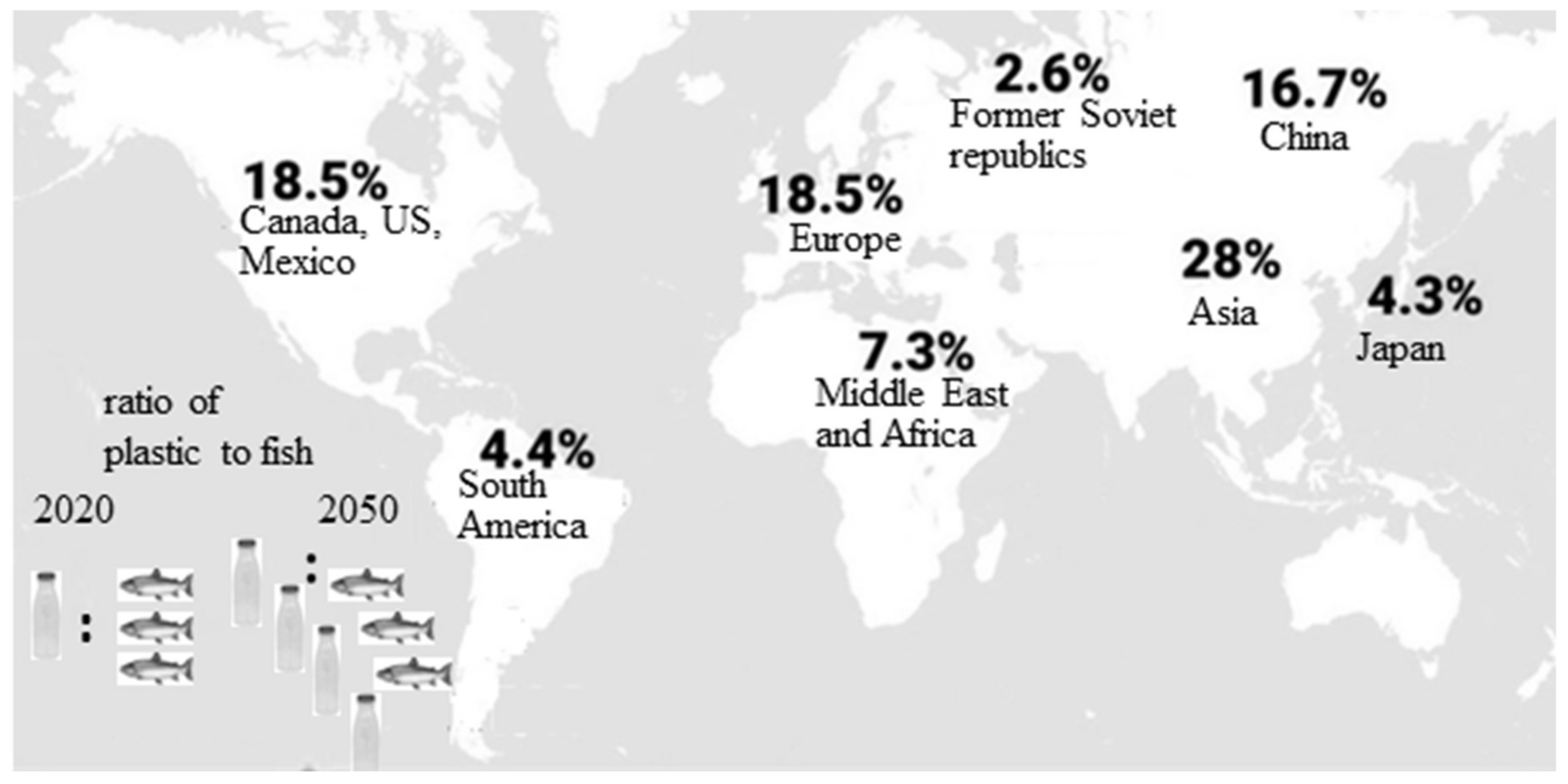
1.1. Plastics—Unavoidable Part of Our Daily Life. Negative Consequences from Plastic Accumulation and Slow Degradation
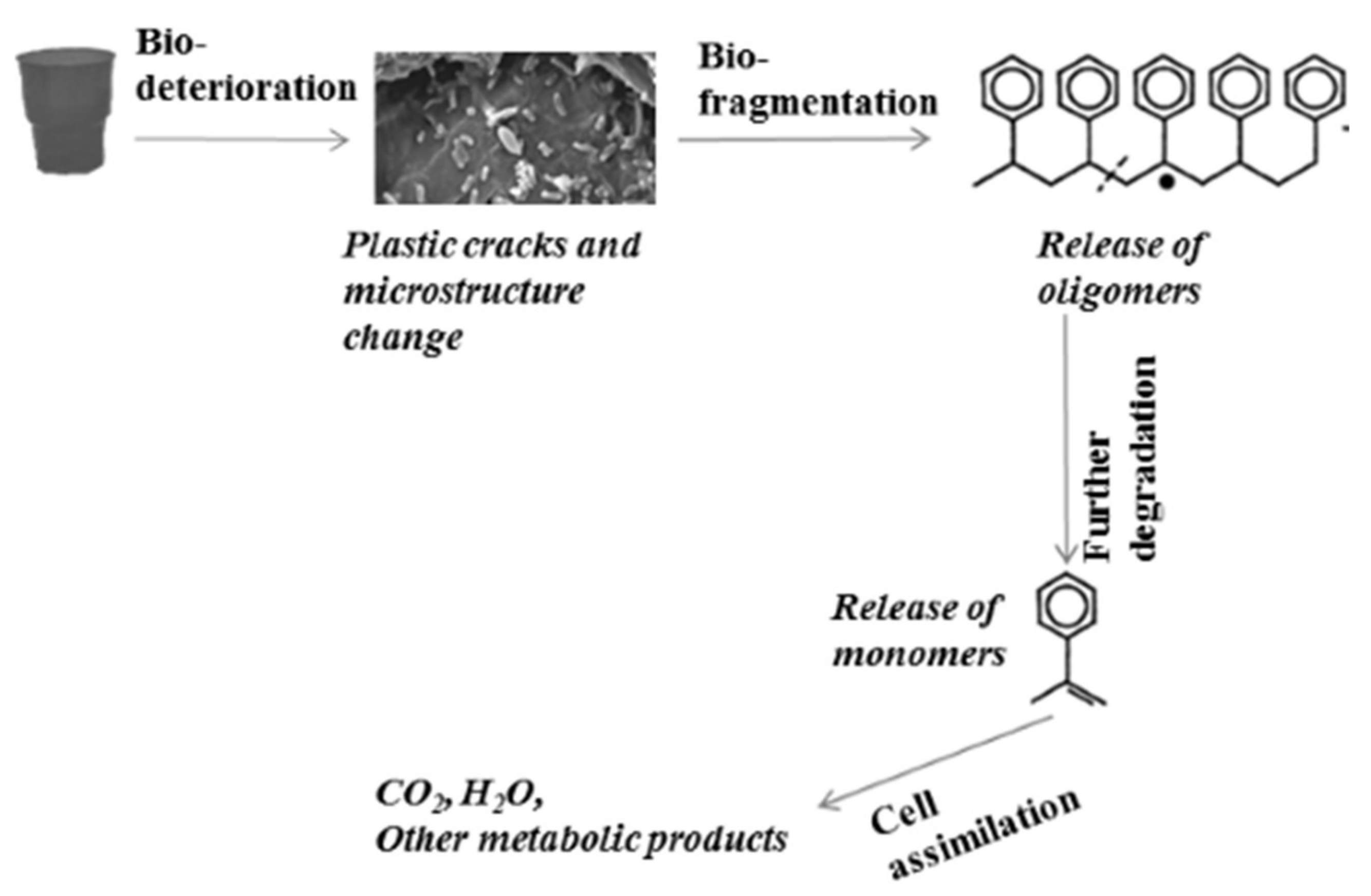
1.2. Microbial Degradation of Plastics
- -
- Bio-deterioration—microbial metabolic activity provokes plastic cracks and aggravates physical properties or changes the microstructure of the matrix by pH change as a result of the released acid or biofilm formation.
- -
- Bio-fragmentation of the long polymer chain—the activity of enzymes produced by microorganisms leads to oligomer release.
- -
- Degradation of oligomers to monomers—oligomers enter inside the cells, and secondary degraders assimilate them as a carbon source, thus increasing the microbial biomass.
- -
- Assimilation of oligomers and excretion of completely oxidized metabolites to H2O, CO2, N2, and CH4.
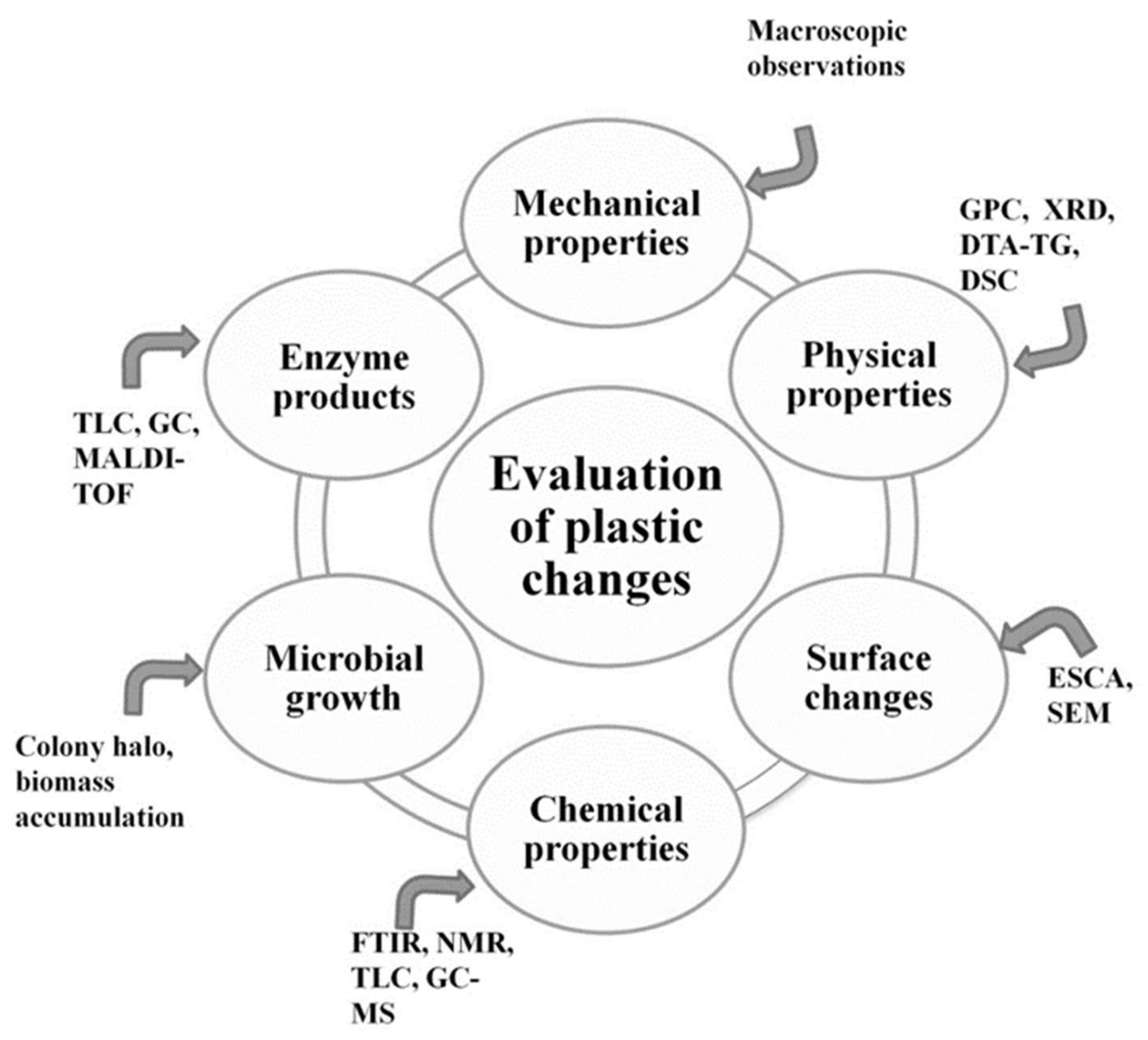
1.3. Standard Testing Methods
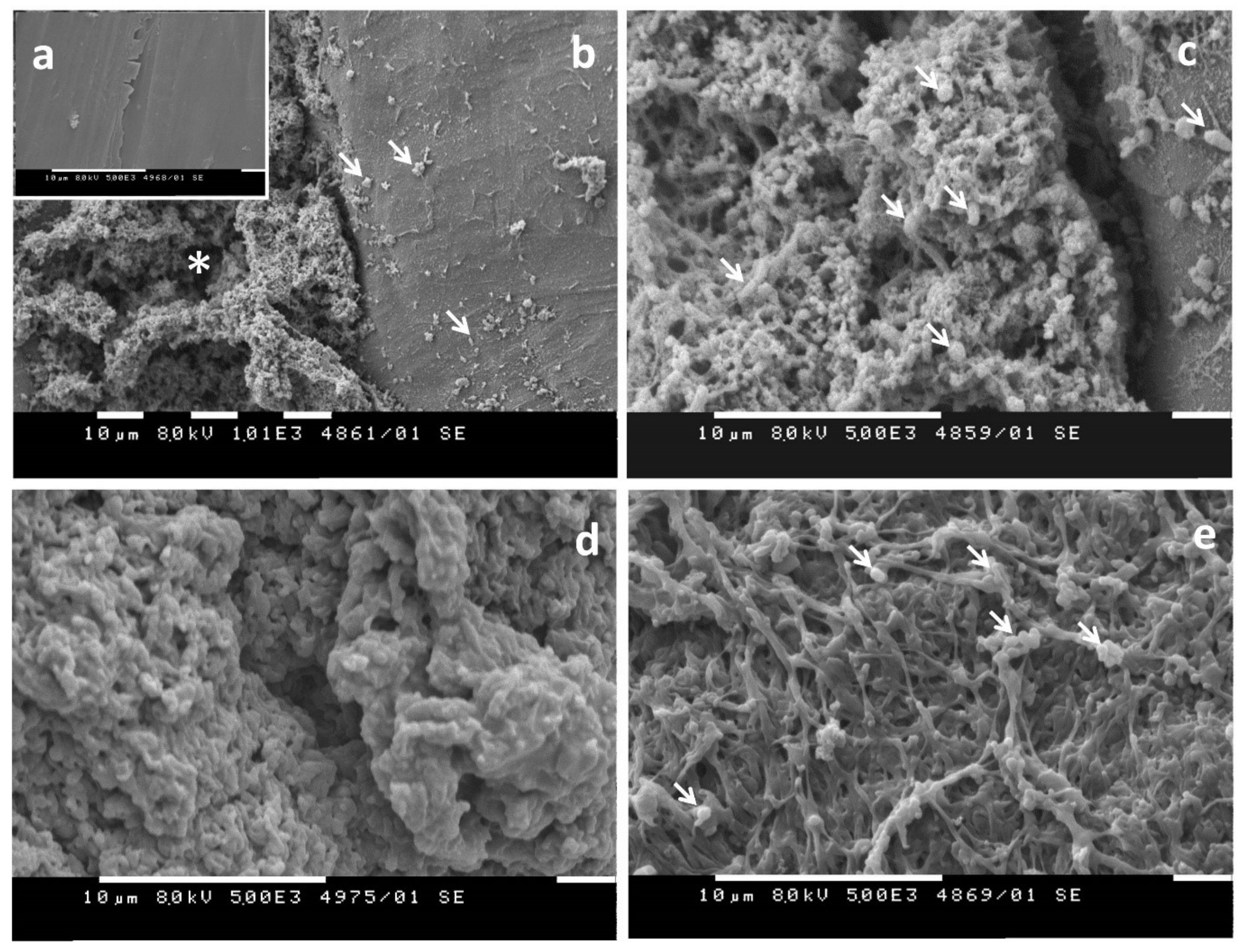
- Evaluation of visible changes in plastics such as appearance of holes or cracks, changes in surface view or color, or formation of biofilms. Scanning electron microscopy (SEM) and atomic force microscopy (AFM) represent good approaches for more sophisticated observations.
- Registering of changes in physical polymer properties, such as mass loss and tensile strength, and chemical properties, such as molecular weight.
- Measurement of utilized carbon dioxide and oxygen consumption rate.
- Evaluation of growth by:
- -
- measurement of the accumulated biomass, usually in minimal media with a polymer as the sole carbon source;
- -
- formation of a clear halo around the colonies that depolymerize the polymer.
- Enzyme assay for detection and characterization of the depolymerization products.
1.3.1. Biofilm Formation on the Plastic Surface
1.3.2. Enzymes Participating in Plastic Degradation
2. Extreme Environments and Extremophiles
3. Plastic-Degrading Thermophilic Bacteria
4. Alkaliphilic Degraders
5. Halophilic Degraders
| Plastic Degradation Type | Polymer | Microorganism | Isolation Source | Physico-Chemical Parameters of Environment | Effectiveness of Degradation | Reference |
|---|---|---|---|---|---|---|
| Non-biodegradable | Polyethylene (72.2%) + PP (18.0%) and PS (2.8%) | Cyanobacteria (Calothrix, Pleurocapsa, Phormidium), Erythrobacter | Western Mediterranean Sea | 3.87% salinity | not reported | [68] |
| Polystyrene | Community, Erythrobacter | Black Sea water | 1.86% salinity | not reported | [70] | |
| Polyurethane | Community | Baltic Sea | 10 °C, pH 8.0, 1.86% salinity | 19% weight loss for PU-A, 4% weight loss for PU-B after 12 months | [75] | |
| Biodegradable | Polycaprolactone | Shewanella, Moritella, Psychrobacter, and Pseudomonas | Kurile and Japan Trenches | depth of 5000–7000 m, 4 °C | not reported | [76] |
6. Psychrophilic Degraders
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Plastics Europe. Plastics-The Facts 2020: An Analysis of European Plastics Production, Demand and Waste Data. Available online: https://www.plasticseurope.org/application/files/8016/1125/2189/AF_Plastics_the_facts-WEB-2020-ING_FINAL.pdf (accessed on 23 March 2021).
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.; Wagner, M. Microplastics Are Contaminants of Emerging Concern in Freshwater Environments: An Overview; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2018; pp. 1–23. [Google Scholar]
- Lebreton, L.; Andrady, A. Future scenarios of global plastic waste generation and disposal. Palgrave Commun. 2019, 5, 6. [Google Scholar] [CrossRef]
- Sumathi, T.; Viswanath, B.; Lakshmi, A.S.; SaiGopal, D.V.R. Production of Laccase byCochliobolussp. Isolated from Plastic Dumped Soils and Their Ability to Degrade Low Molecular Weight PVC. Biochem. Res. Int. 2016, 2016, 9519527. [Google Scholar] [CrossRef] [PubMed]
- World Economic Forum. The Future of the Plastics Industry Is Green. 2021. Available online: https://www.weforum.org/agenda/2021/03/future-plastics-industry-green (accessed on 23 March 2021).
- Geyer, R.; Jambeck, J.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed]
- Jambeck, J.R.; Geyer, R.; Wilcox, C.; Siegler, T.R.; Perryman, M.; Andrady, A.; Narayan, R.; Law, K.L. Plastic waste inputs from land into the ocean. Science 2015, 347, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Ragaert, K.; Delva, L.; Van Geem, K. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017, 69, 24–58. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Zimmermann, W. Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb. Biotechnol. 2017, 10, 1302–1307. [Google Scholar] [CrossRef] [PubMed]
- Webb, H.K.; Arnott, J.; Crawford, R.J.; Ivanova, E.P. Plastic Degradation and Its Environmental Implications with Special Reference to Poly(ethylene terephthalate). Polymers 2012, 5, 1–18. [Google Scholar] [CrossRef]
- Mir, S.; Asghar, B.; Khan, A.K.; Rashid, R.; Shaikh, A.; Khan, R.A.; Murtaza, G. The effects of nanoclay on thermal, mechanical and rheological properties of LLDPE/chitosan blend. J. Polym. Eng. 2017, 37, 143–149. [Google Scholar] [CrossRef]
- Tokiwa, Y.; Calabia, B.P.; Ugwu, C.U.; Aiba, S. Biodegradability of Plastics. Int. J. Mol. Sci. 2009, 10, 3722–3742. [Google Scholar] [CrossRef]
- Ahmed, T.; Shahid, M.; Azeem, F.; Rasul, I.; Shah, A.A.; Noman, M.; Hameed, A.; Manzoor, N.; Manzoor, I.; Muhammad, S. Biodegradation of plastics: Current scenario and future prospects for environmental safety. Environ. Sci. Pollut. Res. 2018, 25, 7287–7298. [Google Scholar] [CrossRef] [PubMed]
- Dussud, C.; Ghiglione, J.F. Bacterial degradation of synthetic plastics. CIESM Workshop Monogr. 2014, 46, 49–54. Available online: https://oceans.taraexpeditions.org/en/m/science/news/bacterial-degradation-of-synthetic-plastics (accessed on 25 March 2021).
- Debroas, D.; Mone, A.; Ter Halle, A. Plastics in the North Atlantic garbage patch: A boat-microbe for hitchhikers and plastic degraders. Sci. Total. Environ. 2017, 599–600, 1222–1232. [Google Scholar] [CrossRef]
- Vijaya, C.; Reddy, R.M. Impact of soil composting using municipal solid waste on biodegradation of plastics. Indian J. Biotechnol. 2008, 7, 235–239. [Google Scholar]
- Muhamad, W.N.A.W.; Othman, R.; Shaharuddin, R.I.; Irani, M.S. Microorganism as plastic biodegradation agent towards sustainable environment. Adv. Environ. Biol. 2015, 9, 8–13. [Google Scholar]
- Ganesh, K.A.; Anjana, K.; Hinduja, M.; Sujitha, K.; Dharani, G. Review on plastic wastes in marine environment–Biodegradation and biotechnological solutions. Mar. Pollut. Bull. 2020, 150, 110733. [Google Scholar]
- Ghosh, S.; Qureshi, A.; Purohit, H. Microbial degradation of plastics: Biofilms and degradation pathways. In Contaminants in Agriculture and Environment: Health Risks and Remediation; Agro Environ Media: Haridwar, India, 2019; Volume 1, pp. 184–199. [Google Scholar]
- Jacquin, J.; Cheng, J.; Odobel, C.; Pandin, C.; Conan, P.; Pujo-Pay, M.; Barbe, V.; Meistertzheim, A.-L.; Ghiglione, J.-F. Microbial Ecotoxicology of Marine Plastic Debris: A Review on Colonization and Biodegradation by the “Plastisphere”. Front. Microbiol. 2019, 10, 865. [Google Scholar] [CrossRef]
- Pinto, M.; Langer, T.M.; Hüffer, T.; Hofmann, T.; Herndl, G.J. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS ONE 2019, 14, e0217165. [Google Scholar] [CrossRef]
- Awasthi, S.; Srivastava, P.; Singh, P.; Tiwary, D.; Mishra, P.K. Biodegradation of thermally treated high-density polyethylene (HDPE) by Klebsiella pneumoniae CH001. Biotech 2017, 7, 332. [Google Scholar] [CrossRef]
- Gilan, I.; Hadar, Y.; Sivan, A. Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Appl. Microbiol. Biotechnol. 2004, 65, 97–104. [Google Scholar] [CrossRef]
- Giacomucci, L.; Raddadi, N.; Soccio, M.; Lotti, N.; Fava, F. Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. New Biotechnol. 2019, 52, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Demirkan, E.; Guller, B.E.; Sevgi, T. Analysis by scanning electron microscopy of polyethylene terephthalate and nylon biodegradation abilities of Bacillus sp. strains isolated from soil. J. Biol. Environ. Sci. 2020, 14, 107–114. [Google Scholar]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.-J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.-D. Enzymatic Degradation of Poly(ethylene terephthalate): Rapid Hydrolyse using a Hydrolase from T. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Rowe, L.; Howard, G.T. Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme. Int. Biodeterior. Biodegrad. 2002, 50, 33–40. [Google Scholar] [CrossRef]
- Oceguera-Cervantes, A.; Carrillo-García, A.; López, N.; Bolaños-Nuñez, S.; Cruz-Gómez, M.J.; Wacher, C.; Loza-Tavera, H. Characterization of the Polyurethanolytic Activity of Two Alicycliphilus sp. Strains Able to Degrade Polyurethane and N-Methylpyrrolidone. Appl. Environ. Microbiol. 2007, 73, 6214–6223. [Google Scholar] [CrossRef]
- Magnin, A.; Pollet, E.; Perrin, R.; Ullmann, C.; Persillon, C.; Phalip, V.; Avérous, L. Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 2019, 85, 141–150. [Google Scholar] [CrossRef]
- Tomita, K.; Hayashi, N.; Ikeda, N.; Kikuchi, Y. Isolation of a thermophilic bacterium degrading some nylons. Polym. Degrad. Stab. 2003, 81, 511–514. [Google Scholar] [CrossRef]
- Almeida, E.L.; Rincón, A.F.C.; Jackson, S.A.; Dobson, A.D.W. In silico Screening and Heterologous Expression of a Polyethylene Terephthalate Hydrolase (PETase)-Like Enzyme (SM14est) With Polycaprolactone (PCL)-Degrading Activity, From the Marine Sponge-Derived Strain Streptomyces sp. SM14. Front. Microbiol. 2019, 10, 2187. [Google Scholar] [CrossRef]
- Oda, Y.; Oida, N.; Urakami, T.; Tonomura, K. Polycaprolactone depolymerase produced by the bacterium Alcaligenes faecalis. FEMS Microbiol. Lett. 2006, 152, 339–343. [Google Scholar] [CrossRef]
- Negoro, S. Biodegradation of nylon oligomers. Appl. Microbiol. Biotechnol. 2000, 54, 461–466. [Google Scholar] [CrossRef]
- Hirota-Mamoto, R.; Nagai, R.; Tachibana, S.; Yasuda, M.; Tani, A.; Kimbara, K.; Kawai, F. Cloning and expression of the gene for periplasmic poly(vinyl alcohol) dehydrogenase from Sphingomonas sp. strain 113P3, a novel-type quinohaemoprotein alcohol dehydrogenase. Microbiology 2006, 152, 1941–1949. [Google Scholar] [CrossRef]
- Yoon, M.G.; Jeon, H.J. Biodegradation of Polyethylene by a Soil Bacterium and AlkB Cloned Recombinant Cell. J. Bioremediation Biodegrad. 2012, 3, 145. [Google Scholar] [CrossRef]
- Santo, M.; Weitsman, R.; Sivan, A. The role of the copper-binding enzyme–laccase–in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegradation 2013, 84, 204–210. [Google Scholar] [CrossRef]
- Takei, D.; Washio, K.; Morikawa, M. Identification of alkane hydroxylase genes in Rhodococcus sp. strain TMP2 that degrades a branched alkane. Biotechnol. Lett. 2008, 30, 1447–1452. [Google Scholar] [CrossRef]
- Iakovlev, V.V.; Guelcher, S.A.; Bendavid, R. Degradation of polypropylenein vivo: A microscopic analysis of meshes explanted from patients. J. Biomed. Mater. Res. Part B Appl. Biomater. 2015, 105, 237–248. [Google Scholar] [CrossRef]
- Krueger, M.C.; Hofmann, U.; Moeder, M.; Schlosser, D. Potential of Wood-Rotting Fungi to Attack Polystyrene Sulfonate and Its Depolymerisation by Gloeophyllum trabeum via Hydroquinone-Driven Fenton Chemistry. PLoS ONE 2015, 10, e0131773. [Google Scholar] [CrossRef]
- Kour, D.; Rana, K.L.; Kaur, T.; Singh, B.; Chauhan, V.S.; Kumar, A.; Rastegari, A.A.; Yadav, N.; Yadav, A.N.; Gupta, V.K. Extremophiles for Hydrolytic Enzymes Productions: Biodiversity and Potential Biotechnological Applications. In Bioprocessing for Biomolecules Production; Wiley &Sons: Hoboken, NJ, USA, 2019; pp. 321–372. [Google Scholar]
- Takai, K.; Nakamura, K.; Toki, T.; Tsunogai, U.; Miyazaki, M.; Miyazaki, J.; Hirayama, H.; Nakagawa, S.; Nunoura, T.; Horikoshi, K. Cell proliferation at 122 C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. USA 2008, 105, 10949–10954. [Google Scholar] [CrossRef]
- Berenguer, J. Thermophile. In Encyclopedia of Astrobiology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2011; pp. 1666–1667. [Google Scholar]
- Orellana, R.; Macaya, C.; Bravo, G.; Dorochesi, F.; Cumsille, A.; Valencia, R.; Rojas, C.; Seeger, M. Living at the Frontiers of Life: Extremophiles in Chile and Their Potential for Bioremediation. Front. Microbiol. 2018, 9, 2309. [Google Scholar] [CrossRef]
- Preiss, L.; Hicks, D.B.; Suzuki, S.; Meier, T.; Krulwich, T.A. Alkaliphilic Bacteria with Impact on Industrial Applications, Concepts of Early Life Forms, and Bioenergetics of ATP Synthesis. Front. Bioeng. Biotechnol. 2015, 3, 75. [Google Scholar] [CrossRef]
- Clarke, A.; Morris, G.J.; Fonseca, F.; Murray, B.J.; Acton, E.; Price, H.C. A Low Temperature Limit for Life on Earth. PLoS ONE 2013, 8, e66207. [Google Scholar] [CrossRef] [PubMed]
- Nogi, Y. Taxonomy of Psychrophiles. In Extremophiles Handbook; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2011; pp. 777–792. [Google Scholar]
- Ventosa, A.; de la Haba, R.R.; Sánchez-Porro, C.; Papke, R.T. Microbial diversity of hypersaline environments: A metagenomic approach. Curr. Opin. Microbiol. 2015, 25, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Hadad, D.; Geresh, S.; Sivan, A. Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J. Appl. Microbiol. 2005, 98, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.C.H.; Nguyen, D.T.; Thai, H.; Nguyen, T.C.; Tran, T.T.H.; Le, V.H.; Nguyen, V.H.; Tran, X.B.; Pham, T.P.T.; Nguyen, T.G.; et al. Plastic degradation by thermophilic Bacillus sp. BCBT21 isolated from composting agricultural residual in Vietnam. Adv. Nat. Sci. Nanosci. Nanotechnol. 2018, 9, 015014. [Google Scholar] [CrossRef]
- Yan, F.; Wei, R.; Cui, Q.; Bornscheuer, U.T.; Liu, Y. Thermophilic whole-cell degradation of polyethylene terephthalate using engineered Clostridium thermocellum. Microb. Biotechnol. 2021, 14, 374–385. [Google Scholar] [CrossRef]
- Mahdi, M.S.; Ameen, R.S.; Ibrahim, H.K. Study on degradation of nylon 6 by thermophilic bacteria Anoxybacillus rupiensis Ir3 (JQ912241). International Journal of Advanced Research in Biological Sciences (IJARBS). Int. J. Adv. Res. Biol. Sci. 2016, 3, 200–209. [Google Scholar] [CrossRef]
- Nakasaki, K.; Matsuura, H.; Tanaka, H.; Sakai, T. Synergy of two thermophiles enables decomposition of poly-É›-caprolactone under composting conditions. FEMS Microbiol. Ecol. 2006, 58, 373–383. [Google Scholar] [CrossRef]
- Chua, T.-K.; Tseng, M.; Yang, M.-K. Degradation of Poly(ε-caprolactone) by thermophilic Streptomyces thermoviolaceus subsp. thermoviolaceus 76T-2. AMB Express 2013, 3, 8. [Google Scholar] [CrossRef]
- Tseng, M.; Hoang, K.-C.; Yang, M.-K.; Yang, S.-F.; Chu, W.S. Polyester-degrading thermophilic actinomycetes isolated from different environment in Taiwan. Biodegradation 2007, 18, 579–583. [Google Scholar] [CrossRef]
- Ru, J.; Huo, Y.; Yang, Y. Microbial Degradation and Valorization of Plastic Wastes. Front. Microbiol. 2020, 11, 442. [Google Scholar] [CrossRef]
- Kawai, F.; Kawabata, T.; Oda, M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019, 103, 4253–4268. [Google Scholar] [CrossRef]
- Hu, X.; Thumarat, U.; Zhang, X.; Tang, M.; Kawai, F. Diversity of polyester-degrading bacteria in compost and molecular analysis of a thermoactive esterase from Thermobifida alba AHK119. Appl. Microbiol. Biotechnol. 2010, 87, 771–779. [Google Scholar] [CrossRef]
- Ribitsch, D.; Acero, E.H.; Greimel, K.; Dellacher, A.; Zitzenbacher, S.; Marold, A.; Rodriguez, R.D.; Steinkellner, G.; Gruber, K.; Schwab, H.; et al. A New Esterase from Thermobifida halotolerans Hydrolyses Polyethylene Terephthalate (PET) and Polylactic Acid (PLA). Polymers 2012, 4, 617–629. [Google Scholar] [CrossRef]
- Wei, R.; Oeser, T.; Then, J.; Kühn, N.; Barth, M.; Schmidt, J.; Zimmermann, W. Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata. AMB Express 2014, 4, 44. [Google Scholar] [CrossRef]
- Wei, R.; Breite, D.; Song, C.; Gräsing, D.; Ploss, T.; Hille, P.; Schwerdtfeger, R.; Matysik, J.; Schulze, A.; Zimmermann, W. Biocatalytic Degradation Efficiency of Postconsumer Polyethylene Terephthalate Packaging Determined by Their Polymer Microstructures. Adv. Sci. 2019, 6, 1900491. [Google Scholar] [CrossRef]
- Chen, S.; Su, L.; Billig, S.; Zimmermann, W.; Chen, J.; Wu, J. Biochemical characterization of the cutinases from Thermobifida fusca. J. Mol. Catal. B Enzym. 2010, 63, 121–127. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.-J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a Novel Cutinase Homolog with Polyethylene Terephthalate-Degrading Activity from Leaf-Branch Compost by Using a Metagenomic Approach. Appl. Environ. Microbiol. 2011, 78, 1556–1562. [Google Scholar] [CrossRef]
- Morales-Gámez, L.; Soto, D.; Franco, L.; Puiggalí, J. Brill transition and melt crystallization of nylon 56: An odd–even polyamide with two hydrogen-bonding directions. Polymers 2010, 51, 5788–5798. [Google Scholar] [CrossRef]
- Torre, D.Y.Z.D.; Santos, L.A.D.; Reyes, M.L.C.; Baculi, R.Q. Biodegradation of low-density polyethylene by bacteria isolated from serpentinization-driven alkaline spring. Philipp. Sci. Lett. 2018, 11, 1–12. [Google Scholar]
- Cada, E.J.G.; Muyot, M.L.C.; Sison, J.M.C.; Baculi, R.Q. Enhanced in vitro biodegradation of low-density polyethylene using alkaliphilic bacterial consortium supplemented with iron oxide nanoparticles. Philipp. Sci. Lett. 2019, 12, 55–69. [Google Scholar]
- Dussud, C.; Meistertzheim, A.; Conan, P.; Pujo-Pay, M.; George, M.; Fabre, P.; Coudane, J.; Higgs, P.; Elineau, A.; Pedrotti, M.; et al. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ. Pollut. 2018, 236, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; De Philippis, R. Role of Cyanobacterial Exopolysaccharides in Phototrophic Biofilms and in Complex Microbial Mats. Life 2015, 5, 1218–1238. [Google Scholar] [CrossRef] [PubMed]
- Tourova, T.; Sokolova, D.; Nazina, T.; Grouzdev, D.; Kurshev, E.; Laptev, A. Biodiversity of Microorganisms Colonizing the Surface of Polystyrene Samples Exposed to Different Aqueous Environments. Sustainability 2020, 12, 3624. [Google Scholar] [CrossRef]
- Nagpal, S.; Haque, M.M.; Singh, R.; Mande, S.S. iVikodak—A Platform and Standard Workflow for Inferring, Analyzing, Comparing, and Visualizing the Functional Potential of Microbial Communities. Front. Microbiol. 2019, 9, 3336. [Google Scholar] [CrossRef]
- Schmitt, G.; Arndt, F.; Kahnt, J.; Heider, J. Adaptations to a Loss-of-Function Mutation in the Betaproteobacterium Aromatoleum aromaticum: Recruitment of Alternative Enzymes for Anaerobic Phenylalanine Degradation. J. Bacteriol. 2017, 199, e00383-17. [Google Scholar] [CrossRef]
- Tourova, T.P.; Sokolova, D.S.; Nazina, T.N.; Gruzdev, D.S.; Laptev, A.B. Phylogenetic Diversity of Microbial Communities from the Surface of Polyethylene Terephthalate Materials Exposed to Different Water Environments. Microbiology 2020, 89, 96–106. [Google Scholar] [CrossRef]
- Paunova-Krasteva, T.; Atanasova, N.; Radchenkova, N.; Boyadzhieva, I.; Stoitsova, S.; Kambourova, M.; Taktarova, Y.; Malakhova, D.; Tsavkelova, E.; Egorova, M.; et al. Plastic Degradation by Extremophilic Microbial Communities. In Proceedings of the International Seminar of Ecology, Sofia, Bulgaria, 23–24 April 2020; Available online: https://2e1fb47a-6580-419b-ad7b-abb2f84bb78c.filesusr.com/ugd/16129b_68d8df1e28b94b54832f02f64033eefb.pdf (accessed on 28 March 2021).
- Heimowska, A.; Rutkowska, M. Environmental Degradability of Polyurethanes. In Thermoplastic Elastomers-Synthesis and Applications; IntechOpen: London, UK, 2015; pp. 75–94. [Google Scholar]
- Sekiguchi, T.; Sato, T.; Enoki, M.; Kanehiro, H.; Uematsu, K.; Kato, C. Isolation and characterization of biodegradable plastic degrading bacteria from deep-sea environments. JAMSTEC Rep. Res. Dev. 2011, 11, 33–41. [Google Scholar] [CrossRef]
- Moyer, C.L.; Morita, R.Y. Psychrophiles and Psychrotrophs. In eLS; Wiley &Sons: Hoboken, NJ, USA, 2007; pp. 1–5. [Google Scholar]
- Urbanek, A.K.; Rymowicz, W.; Mirończuk, A.M. Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl. Microbiol. Biotechnol. 2018, 102, 7669–7678. [Google Scholar] [CrossRef]
- UNEP. Plastic Debris in the Ocean. UNEP Year Book 2014 Emerging Issues Update. Available online: http://www.unep.org/yearbook/2014/PDF/UNEP_YearBook_2014.pdf (accessed on 30 March 2021).
- Oliveira, M.M.; Proenca, A.M.; Moreira-Silva, E.; de Castro, A.M.; dos Santos, F.M.; Marconatto, L.; Medina-Silva, R. Biofilms of Pseudomonas and Lysinibacillus Marine Strains on High-Density Polyethylene. Microb. Ecol. 2021, 81, 833–846. [Google Scholar] [CrossRef]

| Enzyme Class | Enzyme Group | Enzyme Source | Type of Biodegraded Plastic | Reference |
|---|---|---|---|---|
| Hydrolases that split ester bonds | Esterase | Streptomyces sp. SM14 | PET | [33] |
| Esterase | Bacillus subtilis | PU | [29] | |
| Esterase | Alicycliphilus sp. | PU | [30] | |
| Aromatic polyesterase | Ideonella sakaiensis 201-F6 | PET | [2] | |
| Esterase E3576 | Commercially available by Proteus | PU | [31] | |
| Lipase | Alcaligenes faecalis | PCL | [34] | |
| Hydrolases that act on carbon–nitrogen bonds | amidase E4143 | Commercially available by Proteus | PU | [31] |
| 6-aminohexanoate-cyclic-dimer hydrolase, 6-aminohexanoate -dimer hydrolase and endo-type6-aminohexanoate-oligomer hydrolase | Flavobacterium sp. KI72 | 6-aminohexano-ate, an intermediate product of nylon | [35] | |
| Oxydase | PVA dehydrogenase | Sphingomonas sp. strain 113P3 | PVA | [36] |
| Alkane hydroxylase | Pseudomonas sp. E4 | PE | [37] | |
| Laccases | Rhodococcus ruber | PE | [38] | |
| Monoxygenases | Rhodococcus sp. TMP2 | PE | [39] |
| Plastic Degradation Type | Polymer | Microorganism | Isolation Source | Temperature for Polymer Degradation | Effectiveness of Degradation | Reference |
|---|---|---|---|---|---|---|
| Non-biodegradable | Polyethylene | Brevibaccillus borstelensis strain 707 | Soil | 50 °C | 11% after 30 days | [50] |
| Bacillus sp. BCBT21 | Composting agricultural residual | 55 °C | 44% decrease of average MW of the polymer for 30 d | [51] | ||
| Polyethylene terephthalate | Thermobifida fusca | 55 °C | ≈50% decrease of the average MW of polymer for 3 weeks | [28] | ||
| Clostridium thermocellum | 60 °C | 60% after 14 days | [52] | |||
| Nylon | Anoxybacillus rupiensis | Hydrocarbon contaminated soil | 65 °C | Optical density ≈1.8 after 7 days growth on nylon 6 | [53] | |
| Geobacillus thermocatenulatus | Soil | 60 °C | Decrease in nylon 12 and nylon 66 MW from ≈ 40,000 to ≈15,000 over 20 d | [32] | ||
| Biodegradable | Polycaprolactone | Consortium—Streptomyces thermonitrificans PDS-1 + Bacillus licheniformis HA1 | Compost | 50 °C | 70% (compost as a substrate) for 48 h | [54] |
| Streptomyces thermoviolaceus subsp. thermoviolaceus | Soil | 45 °C | 100% (0.1% substrate) for 6 h | [55] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atanasova, N.; Stoitsova, S.; Paunova-Krasteva, T.; Kambourova, M. Plastic Degradation by Extremophilic Bacteria. Int. J. Mol. Sci. 2021, 22, 5610. https://doi.org/10.3390/ijms22115610
Atanasova N, Stoitsova S, Paunova-Krasteva T, Kambourova M. Plastic Degradation by Extremophilic Bacteria. International Journal of Molecular Sciences. 2021; 22(11):5610. https://doi.org/10.3390/ijms22115610
Chicago/Turabian StyleAtanasova, Nikolina, Stoyanka Stoitsova, Tsvetelina Paunova-Krasteva, and Margarita Kambourova. 2021. "Plastic Degradation by Extremophilic Bacteria" International Journal of Molecular Sciences 22, no. 11: 5610. https://doi.org/10.3390/ijms22115610
APA StyleAtanasova, N., Stoitsova, S., Paunova-Krasteva, T., & Kambourova, M. (2021). Plastic Degradation by Extremophilic Bacteria. International Journal of Molecular Sciences, 22(11), 5610. https://doi.org/10.3390/ijms22115610







