Abstract
For a long time, Cannabis sativa has been used for therapeutic and industrial purposes. Due to its increasing demand in medicine, recreation, and industry, there is a dire need to apply new biotechnological tools to introduce new genotypes with desirable traits and enhanced secondary metabolite production. Micropropagation, conservation, cell suspension culture, hairy root culture, polyploidy manipulation, and Agrobacterium-mediated gene transformation have been studied and used in cannabis. However, some obstacles such as the low rate of transgenic plant regeneration and low efficiency of secondary metabolite production in hairy root culture and cell suspension culture have restricted the application of these approaches in cannabis. In the current review, in vitro culture and genetic engineering methods in cannabis along with other promising techniques such as morphogenic genes, new computational approaches, clustered regularly interspaced short palindromic repeats (CRISPR), CRISPR/Cas9-equipped Agrobacterium-mediated genome editing, and hairy root culture, that can help improve gene transformation and plant regeneration, as well as enhance secondary metabolite production, have been highlighted and discussed.
1. Introduction
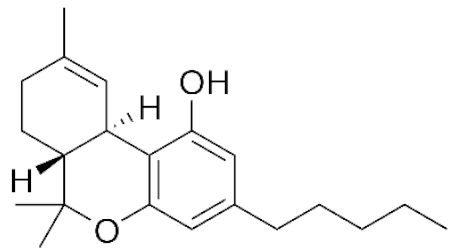
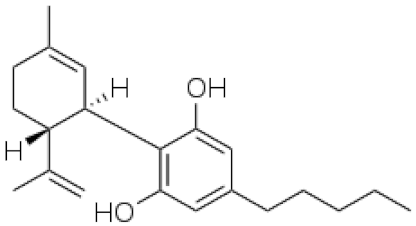
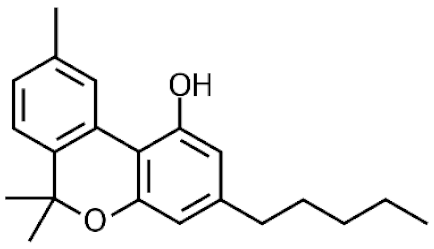
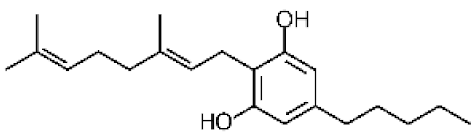
Cannabis sativa L. is a high-demand plant with a long history of medicinal, industrial, recreational, and agricultural uses [1,2]. Cannabis can be categorized based on taxonomic relationships or chemotype but is often divided into two main groups and regulated based on the level of psychoactive cannabinoids that are produced. In most countries, anything below 0.3% Δ9-tetrahydrocannabinol (THC) is classified as hemp and plants that produce 0.3% or greater are categorized as marijuana [3]. To date, more than 560 secondary metabolites are known in cannabis [1,2]. Although cannabinoids and terpenes are the predominant biomolecules in cannabis, phenolic compounds and flavonoids have also been detected. Currently, more than 115 cannabinoids, isoprenylated polyketides, have been identified in cannabis, which are mainly produced in glandular trichomes of female flowers. Cannabidiol (CBD), THC, and cannabichromene (CBC) can be considered as the major cannabinoids in the crop, but new genetics that express other cannabinoids such as cannabigerol (CBG) are now emerging [4].
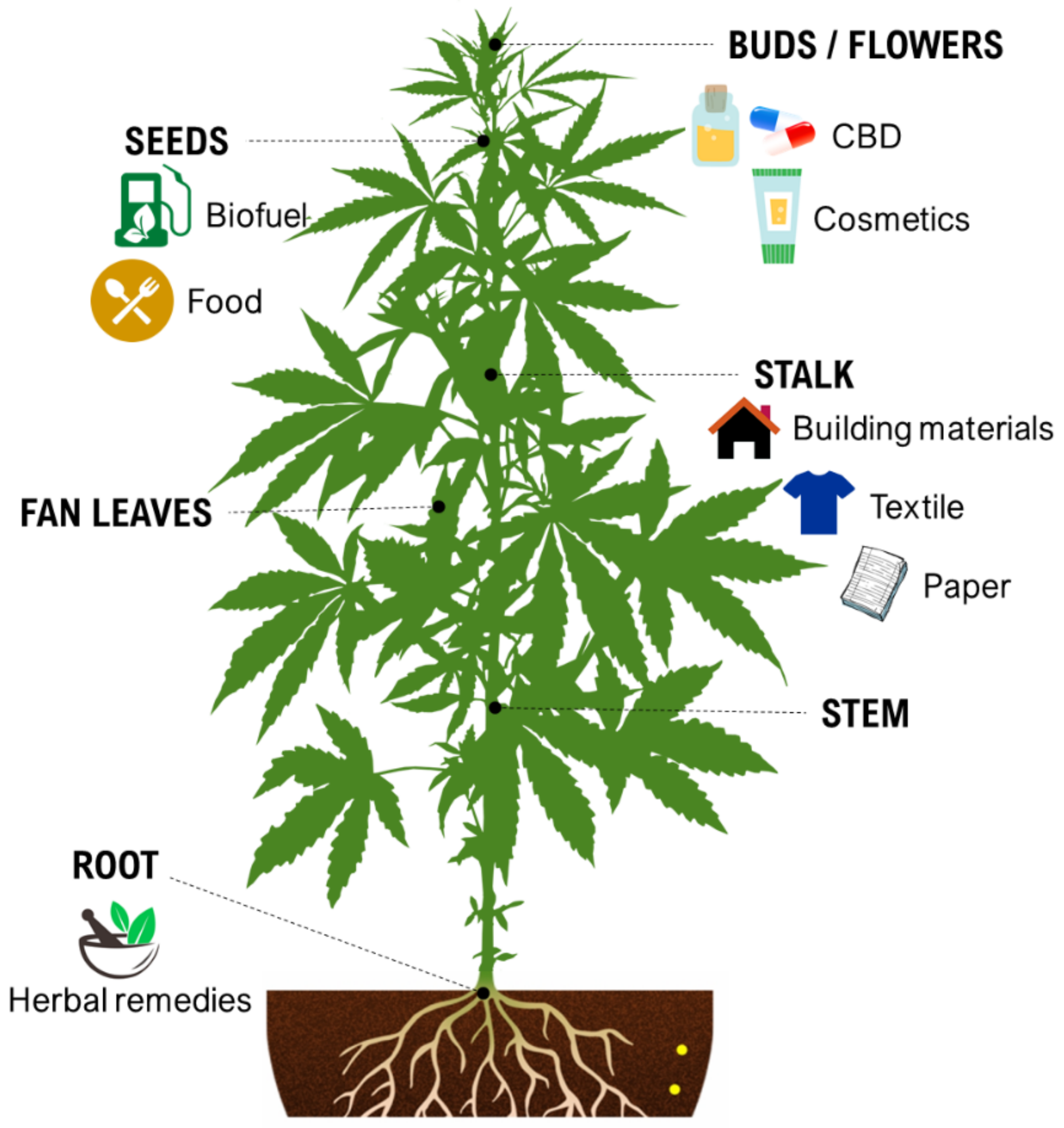
During the last decade, the industrial properties of cannabis (Figure 1) for applications in textiles, paper, building materials, cosmetics, and foods [5,6,7], as well as pharmacological properties (Table 1) such as the palliation of chronic pains associated with cancer, neutralizing the adverse impacts of chemotherapy with cytostatic drugs, eating disorders related to anorexia and AIDS, inflammatory diseases, epilepsy, and anti-spastic activity in Tourette’s syndrome or sclerosis multiplex cases have been broadly studied and supported [5,8].
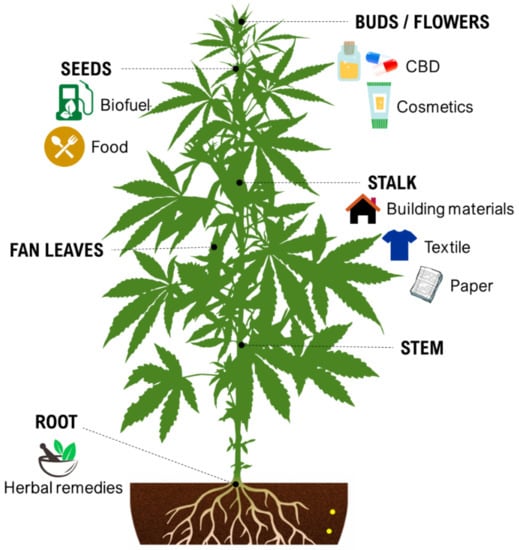
Figure 1.
Some industrial properties of Cannabis.

Table 1.
Some pharmacological properties of Cannabis.
While cannabinoids and cannabinoid-containing products are a new market, they are exponentially growing and a recent market report estimated that the global value of CBD alone will reach 16 billion by 2025 [9]. As the demand for these products increases, there is a pressing need to develop improved genetics and cultivation techniques [10,11].
Conventional plant breeding involves directed crosses of parent plants with desirable characteristics, population evaluation, selection, and fixing desired traits (selfing). In cannabis, these are difficult criteria to meet due to plant biology (e.g., dioecy) and regulations. Cannabis plants are predominantly dioecious but selfing can be achieved through the induction of male flowers on female plants to produce feminized seeds [40]. These limitations in cannabis make conventional breeding methods time-consuming, costly, and laborious. The composition and content of cannabis secondary metabolites, in particular cannabinoids and terpenes, is also greatly related to various factors such as genotypes, age of plants, sex, developmental phase, growth and environmental conditions, harvesting time, storage conditions, and methods of cultivation [3,5].
For most crops with this economic importance, biotechnological tools (i.e., genetic engineering methods including transcription activator-like effector nucleases (TALENs) [41], zinc-finger nucleases (ZFNs) [42], and clustered regularly interspaced short palindromic repeats (CRISPR) [43] are well developed and have been implemented into breeding programs for decades. However, due to the long history of the prohibition of recreational/drug type cannabis, along with the strict regulation and lower market value of hemp, these tools are rudimentary, and many common techniques used in other crops have yet to be applied to cannabis [44]. With recent shifts towards the legalization of cannabis for medicinal and recreational purposes in many jurisdictions and the establishment of a legal market for cannabinoids, cannabis production is becoming a large-scale enterprise similar to other major crops [3]. Along with the emergence of legal commercial producers, the need for modern technologies for genetic improvement is steadily increasing. While biotechnology of cannabis is still relatively new and unrefined, with the advent of affordable large-scale sequencing technologies (i.e., next-generation sequencing (NGS)) and the increasing body of candidate genes for traits of interest, we argue that it is time for a paradigm shift toward improving cannabis genetics through genetic engineering.
Recently, whole genomic and transcriptomic information of cannabis has been obtained using NGS methods [1]. Cannabis NGS information can be applied for robust molecular tools such as DNA barcoding to detect genetic diversity, sex determination, and chemotype inheritance [5]. Moreover, these data can be merged with metabolomics and proteomics to identify unknown secondary metabolites of cannabis [3]. More rapid and accurate transcriptome analysis to detect key enzymes and genes in the biosynthetic pathway of secondary metabolites, mapping of unknown and wild populations using restriction site-associated DNA sequencing (e.g., genotyping by sequencing (GBS)) [45], and interpretation of targeting-induced local lesions in genomes (TILLING) populations are applicable based on the NGS information in cannabis [3,5]. Above all, NGS-derived data facilitate the introduction of genetic engineering methods in cannabis [46].
In vitro tissue culture techniques (e.g., callus and cell culture, de novo regeneration, hairy root culture) are the basis of micropropagation and breeding in cannabis [10,47]. In vitro culture methods coupled with genetic engineering techniques (e.g., Agrobacterium-mediated gene transformation and genome editing) as well as polyploidy induction offer opportunities for producing new genotypes and manipulating secondary metabolite production in cannabis [46]. Although conventional genetic engineering tools (e.g., Agrobacterium-mediated gene transformation and A. rhizogenes-mediated hairy root cultures) can alter the production of some secondary metabolites, it seems that the CRISPR/Cas9 system has more potential than these tools to introduce new germplasms and enhance secondary metabolite production in cannabis in a faster manner [10,47]. Therefore, biotechnological methods can be employed in order to develop improved genetics to help satisfy the demands of producers and consumers.
In the current review, all applied in vitro propagation and genetic engineering methods in cannabis along with other possibly applicable techniques such as designing new culture media, machine learning algorithms, and morphogenic genes that can help cannabis propagation and improvements, as well as enhance secondary metabolite yield, have been highlighted and discussed. The principles, benefits, weaknesses, and concerns of different methods have also been presented.
2. In vitro Culture in Cannabis
In vitro culture is the basis of most biotechnological tools [10,47]. Many methods such as micropropagation, in situ and ex situ conservation, cell culture, Agrobacterium-mediated gene transformation, and polyploidy induction completely depend on in vitro culture techniques [48]. Moreover, plant cell and tissue culture is also a robust method for assessing the secondary metabolite production and endogenous phytohormone metabolism signaling in many plants [46]. Indeed, in vitro culture techniques are useful to propagate plants, but also to produce engineered biomolecules and initiate synthetic biology approaches [10,47].
Callus and cell suspension cultures were one of the main objectives of early in vitro culture in cannabis. The first attempts of callus cultures to produce cannabinoids were performed by Hemphill et al. [49], Loh et al. [50], and Braemer and Paris [51] and led to the conversion of olivetol and CBD to cannabielsoin. However, unstable and inadequate levels of cannabinoid production were achieved. Furthermore, cannabinoids could not be synthesized without adding exogenous cannabigerolic acid (CBGA) as a precursor to the callogenesis medium. Further studies [52,53] revealed that cannabinoids could not be produced even from an inflorescence-derived callus. In another study, Flores-Sanchez et al. [54] used various biotic (Pythium aphanidermatum and Botrytis cinerea) and abiotic (methyl jasmonate, salicylic acid, jasmonic acid, UV-B, AgNO3, NiSO4·6H2O, and CoCl2·6H2O) elicitors in cannabis cell suspension cultures; however, improved cannabinoid production was not obtained.
These results suggest that the biosynthesis of cannabinoids is completely linked to tissue and organ-specific development and complex gene regulatory networks that can only be efficiently produced by trichomes, which are most abundant in differentiated floral tissues. However, cell suspension cultures may still be promising for producing other secondary metabolites such as terpenes, polyphenols, lignans, and alkaloids [7,10]. For instance, Gabotti et al. [55] reported that the activity and expression of tyrosine aminotransferase (TAT) and phenylalanine ammonia-lyase (PAL) increased in cannabis cell suspension cultures using a methyl jasmonate elicitor in combination with tyrosine precursor. Some aromatic compounds such as 4-hydroxyphenylpyruvate (4-HPP) were also identified. This is relevant as highly biologically active flavonoids have been isolated from cannabis [55].
Hairy root culture is another application of in vitro methods that have been used for secondary metabolite production in many species and investigated in cannabis [10]. Affordable and high production of secondary metabolites, high genetic stability, and rapid accumulation and growth of biomass are only some of the merits of hairy root cultures [46]. A larger scale and more profitable process can also be achieved by the cultivation of hairy roots in bioreactors [56]. Sirikantaramas et al. [57] isolated Δ9-tetrahydrocannabinolic acid synthase (THCAS) from cannabis leaves and cloned its cDNA. Then, the cDNA was transformed in tobacco hairy roots using A. rhizogenes. Although THCA was produced through THCAS expression and by adding CBGA, the THCA production rate was low. Farag and Kayser [58] reported 1 µg THCA g−1 dry weight (DW), 1.7 µg CBDA g−1 DW, 1.6 µg CBGA g−1 DW, and 2 µg cannabinoids g−1 DW obtained from adventitious roots from callus cultures. Given that floral tissues from whole plants can produce over 20% THC w/dw, these levels are very low [58]. These results are not surprising given that cannabinoids are generally produced in trichomes, which are not found in root tissues and this approach is likely not suitable for cannabinoids production. Generally, many compounds require differentiated tissues for efficient production. Moher et al. [59] demonstrated that in vitro plants respond to photoperiod and that they develop “normal” looking flowers. While the cannabinoid content of these flowers has not been examined, it is likely that they produce much higher levels than would be observed in undifferentiated tissues, or roots. Therefore, this could be an alternative approach to producing cannabinoids in vitro but it has yet to be explored.
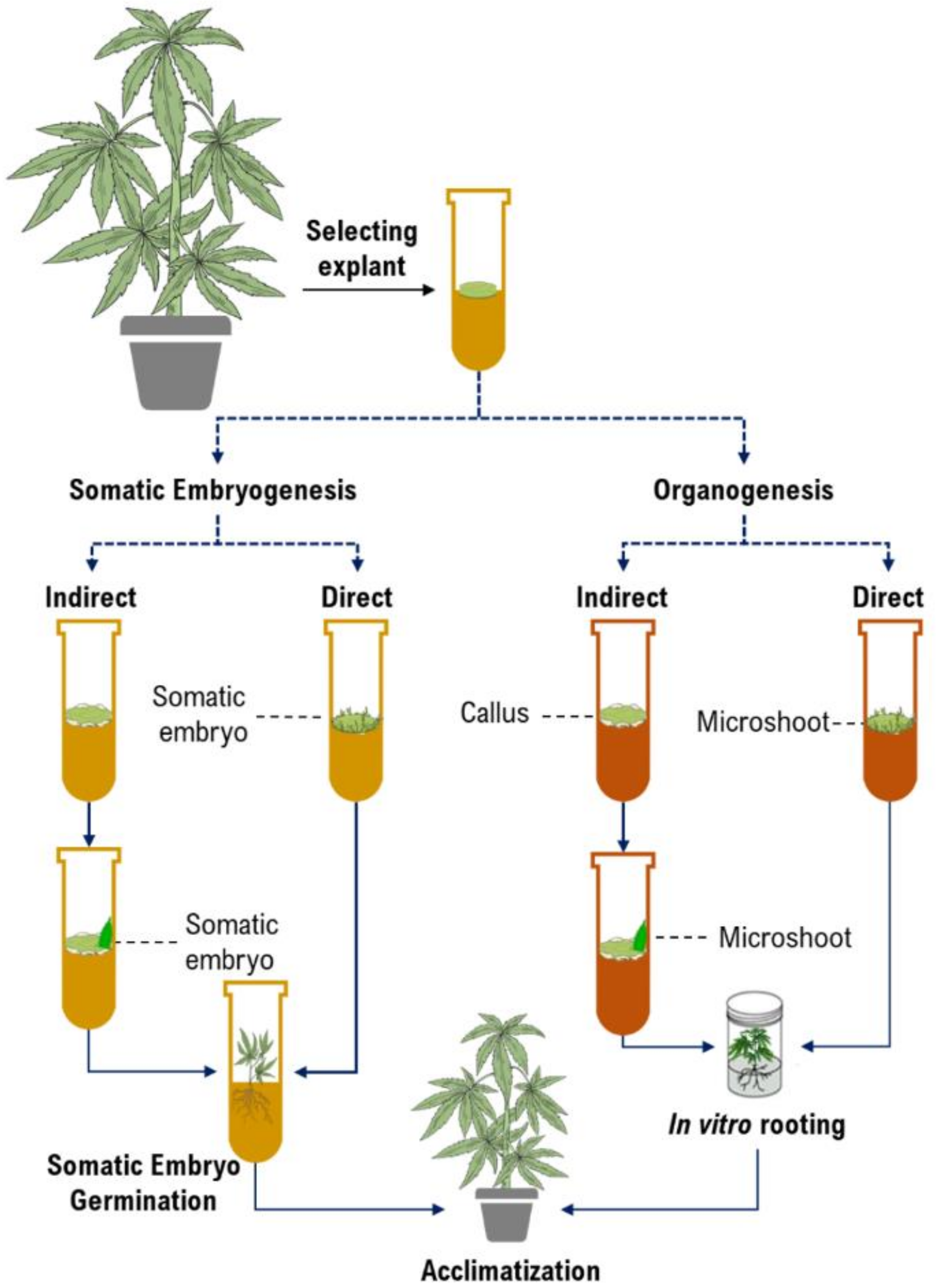
Micropropagation is the first and foremost application of in vitro culture in cannabis [10,47,60,61]. While micropropagation for applications in genetic preservation or propagation are generally achieved through shoot proliferation from existing meristems, many applications in biotechnology require the establishment of de novo regeneration in which plants are produced from non-meristematic tissues. Somatic embryogenesis and organogenesis through either direct or indirect regeneration are the most important platforms for developing regeneration protocols (Figure 2). Although somatic embryogenesis is considered the ideal approach since they regenerate from single cells and reduce chimerism in transformed plants [46], it has been rarely achieved in cannabis. Table 2 represents callogenesis and organogenesis studies in cannabis to date. As can be seen in Table 2, most studies have investigated the effects of plant growth regulators (PGRs) and type of explants and genotypes on micropropagation of cannabis. However, there are many factors (e.g., medium composition and incubation conditions, discussed in the following sections) that affect cannabis micropropagation. Therefore, it is necessary to study these factors for obtaining high-frequency protocols.
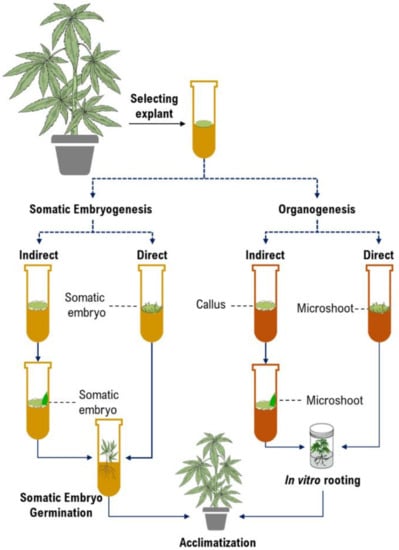
Figure 2.
The schematic diagram of plant tissue culture procedures.

Table 2.
In vitro regeneration studies in cannabis.
2.1. Strategies to Improve In Vitro Culture Procedures
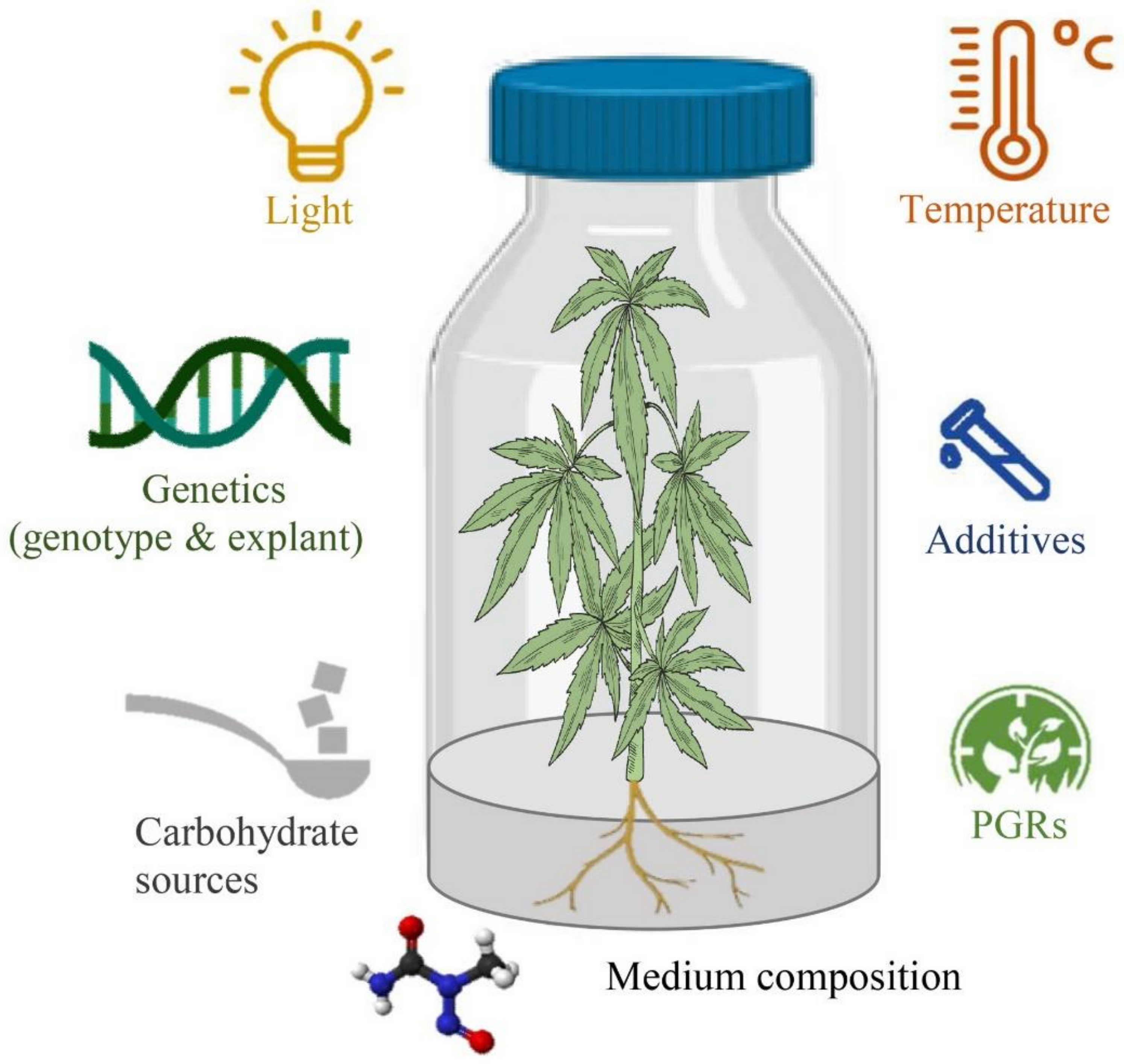
Despite advances in in vitro cell and tissue culture of cannabis in recent years, efficient cannabis regeneration remains one of the main obstacles to applying biotechnology for cannabis improvement and the species is generally considered to be relatively recalcitrant [94]. Genotypes, type and concentration of PGRs, size, age, and type of explant, gelling agent, carbohydrate sources, type and concentration of macro- and micro-nutrients, type and concentration of vitamins, type and concentration of additives (casein hydrolysate, nanoparticles, phloroglucinol, activated charcoal, etc.), pH of the medium, type and volume of the vessels, volume of the medium per culture vessels, and culture conditions (intensity and quality of the light, temperature, photoperiod, and light source) are the most important factors affecting in vitro culture systems [94,95] (Figure 3). However, most studies have investigated the effects of PGRs and the type of explants and genotypes on cannabis micropropagation and little information on many other factors is available. Therefore, studying other factors may result in high-frequency regeneration systems or even obtaining somatic embryogenesis and haploid production protocols. In this section, several promising strategies for improving in vitro culture protocols have been highlighted based on the mentioned factors, new computational methodologies such as machine learning algorithms, and new genetic engineering methods.
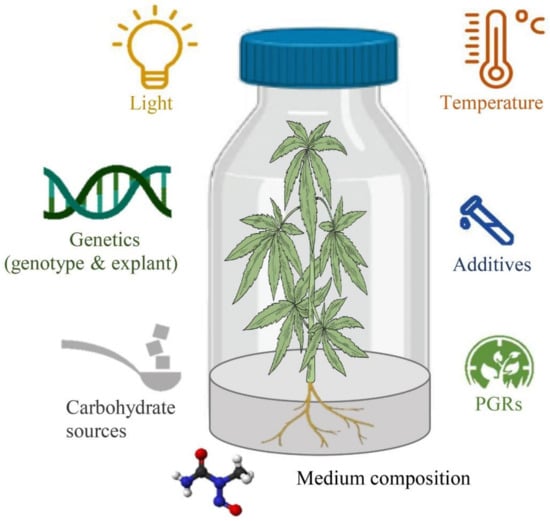
Figure 3.
The schematic diagram of factors affecting in vitro culture procedures.
Although a few studies [78,81] have tested newer PGRs and additives such as 6-benzylamino-9-(tetrahydroxypyranyl) purin (BAP9THP) and meta-topolin (mT) for cannabis micropropagation, there are still some promising PGRs and additives, such as polyamines, brassinosteroids, nano-particles, and nitric oxide (NO) that have not been used for developing cannabis micropropagation protocols. NO, a messenger molecule regulating plant development such as flowering, germination, fruit ripening, and organ senescence, has been recently characterized as a phytohormone [96,97]. It was shown that NO can be experimentally applied in the media as sodium nitroprusside (SNP), which eliminates the difficulty in the application of NO in its gaseous form [98]. Several studies showed that NO is one of the main signaling pathways in in vitro organogenesis and somatic embryogenesis [96,99]. Therefore, the application of SNP may pave the way for obtaining somatic embryogenesis or improving organogenesis protocols in cannabis. Recent studies showed that adding nanoparticles to the culture media improves callogenesis, organogenesis, somatic embryogenesis, and rhizogenesis by inhibiting the production of ROS and ethylene and altering gene expression and antioxidant enzyme activities [100,101,102,103,104,105]. Thus, the application of nanoparticles can be investigated as a promising approach to enhance the in vitro regeneration capacity of cannabis.
The source of carbohydrates is another factor affecting in vitro culture systems. The effect of sucrose, glucose, and fructose as the most important carbohydrates have been widely studied in in vitro morphogenic responses of different plants [106]. While sucrose has resulted in the maximum in vitro organogenesis and embryogenesis in some plants (e.g., Agave angustifolia [107], Sapindus trifoliatus [108], and Pinus koraiensis [109]), other plants (e.g., Vitis Vinifera [110], Brassica napus [111], and Chrysanthemum ×grandiflorum [112]) had better in vitro morphogenic responses to glucose and fructose [106]. Therefore, it is essential to study the effect of different carbohydrate sources on cannabis micropropagation.
The source, intensity, and quality of light play a pivotal role in in vitro organogenesis and embryogenesis [112,113]. The usefulness of light emitting diodes (LEDs) in different micropropagation procedures has been widely demonstrated [114,115]. LEDs provide an appropriate light spectrum and therefore can be considered promising light sources for improving micropropagation studies [113,116]. However, it has been shown that each step of in vitro culture needs a particular light spectrum [113]. For instance, there is an ongoing debate on using red or blue light. However, several studies showed that the red light is better than blue light for somatic embryogenesis [112,117].
Many cannabis micropropagation studies [65,66,67,69,71,72,73,74,75,76,77,78,79,80,81,83] used MS [118] as a basal medium while the composition of MS medium was initially defined for the analysis of tissue ashes of tobacco. Several factors related to the basal medium, such as macro- and micro-elements and vitamins, are known as major factors that are affected in vitro morphogenesis in different species or plant organs [119]. Recently, Page et al. [92] reported that plants cultivated on MS medium displayed a number of physiological defects and that DKW [120] basal salts were much better. Additionally, they reported that DKW basal salts also supported greater callus growth from leaf explants. Together, this suggests that MS salts are sub-optimal for shoot and callus growth in cannabis, but the authors also stated that plants cultured on DKW basal salts still displayed some symptoms and further improvement is likely possible and they did not report regeneration so it is unknown if DKW is suitable for that application.
The challenges in designing a de novo medium and optimizing these myriad factors for specific purposes are expensive and time consuming due to the large number of variables and their interactions with one another. Therefore, new approaches such as new computational methodologies (i.e., machine learning algorithms) are needed to design regeneration protocols. Artificial intelligence models and optimization algorithms provide a complementary outlook for calibrating in vitro protocols, as these algorithms find optimal solutions in terms of genotype, explant source, plant growth regulators, medium composition, and incubation conditions, without the requirement for large-scale, costly, time-consuming, and tedious experimental trials [95,121]. Recently, different machine learning algorithms have been successfully used for predicting and optimizing different in vitro culture processes such as shoot proliferation [122,123,124,125,126], callogenesis [127,128], somatic embryogenesis [129], secondary metabolite production [130,131,132], and gene transformation [133]. Hence, the combination of the experimental approach and machine learning algorithms can be considered a powerful and reliable method to develop a specific protocol for cannabis.
It has been shown that micropropagation after mechanical wounding induced by brushing tissue surfaces has been significantly increased [134]. Although there are no reports regarding the effect of wounding on cannabis micropropagation, from our observations callusing has generally initialed at wound sites and tissue wounding may be a promising approach to improve plant regeneration in cannabis. Three consecutive stages improve in vitro organogenesis by tissue wounding: (i) organogenesis is stimulated by some signals related to tissue damages, (ii) subsequently, endogenous phytohormones are accumulated, which results in (iii) cell fate transition [134].
Thin cell layer culture can be considered as another promising approach that can be used in cannabis micropropagation [135]. Although this method has been used in different recalcitrant plants such as Hedychium coronarium [136], Withania coagulans [135], and Agave fourcroydes [137], there is no report of the application of thin cell layer culture in cannabis. In this method, a thin layer of tissue as the explant is selected, which causes close contact between wounded cells and medium composition and finally leads to improvement of regeneration [135].
Bioreactors (e.g., continuous immersion and temporary immersion) can be considered as useful tools for cannabis micropropagation and for studying plant development [138]. The use of these devices can help overcome the recalcitrance of cannabis genotypes to proliferation, rooting, and acclimation. In addition, they can also be used to reduce the cost of large-scale propagation. The number of cannabis plants cultured in bioreactors is steadily increased, and frequently the physiological state of plant propagules improves with these systems of culture, which also facilitate photoautotrophic propagation.
The use of morphogenic genes is another strategy that may help alleviate the bottlenecks in cannabis regeneration. This strategy has been extensively discussed in section “4.2. Strategies to Improve Gene Transformation Efficiency”.
Protoplast culture can be considered a powerful method for many purposes such as plant regeneration, functional genetic analyses, genome editing, and studying cell processes (e.g., membrane function, cell structure, and hormonal signalization) [139,140]. The development of reproducible and stable protoplast isolation is one of the most important prerequisites for the success of protoplast-based technology. Although there are a few studies about protoplast isolation in cannabis (Table 3), there is no report regarding protoplast-mediated plant regeneration.

Table 3.
Protoplast isolation studies in Cannabis.
Beard et al. [141] showed protoplast isolation from the mesophyll of young, not fully expanded leaves of in vitro grown plantlets of C. sativa var. Cherry x Otto II: Sweetened. The authors reported that an enzymolysis solution composed of 0.3% w/v Macerozyme R-10, 20 mM MES (2-(N-morpholino) ethanesulfonic acid), 1.25% w/v Cellulase R-10, 0.4 M mannitol, 0.1% w/v bovine serum albumin, 10 mM calcium chloride, 20 mM potassium chloride, and 0.075% w/v Pectolyase Y23, adjusted to pH 5.7 and heated to 55 °C for 10 min, resulted in the maximum number of protoplasts (2.27 × 106 protoplasts per gram of leaf segments). Lazič [142] showed protoplast isolation from etiolated hypocotyls and the mesophyll of leaf cells of cannabis. The author also reported that an enzyme solution composed of 0.4% Macerozyme R–10 and 1.5% Cellulase Onozuka R-10 resulted in the maximum number of protoplasts from leaves whereas the highest number of protoplasts from etiolated hypocotyls was achieved from enzyme solution supplemented with 0.1% Macerozyme R-10 and 1% Cellulase Onozuka R-10. In another study, cannabis protoplasts were isolated using a digestion solution supplemented with 88 mM sucrose, 0.4 M mannitol, 1% (w/v) Cellulase Onozuka R-10, 0.1% (w/v) pectolyase Y-23, and 0.2% (w/v) Macerozyme R-10 at 30 °C for 4 h with gentle agitation [143].
Cannabis is a dioecious species, with separate male and female plants, and the most economically important product is unfertilized, seedless, female flowers [1]. Some of the challenges that result from these factors include producers not being able to have pollen-producing plants in their production facility, plants must be unfertilized for accurate phenotyping, which complicates breeding strategies, and it is difficult to self-pollenate plants to produce inbred lines for F1 hybrid seed production [48]. To address these challenges, in vitro techniques for the production of homozygous double haploids for F1 hybrid production can be considered as a robust solution [128]. In vitro haploid production consists of different methods such as wide hybridization-chromosome elimination, parthenogenesis, gynogenesis, and androgenesis [144]. Although there are no reports regarding haploid production in cannabis, it seems that haploid production protocols are needed for further genetic engineering studies. Recently, knockdown and/or knockout of the centromere-specific histone H3 (CENH3) gene, which connects spindle microtubules to chromosome centromere regions, provides a robust tool for producing haploid plants [145,146]. For instance, Wang et al. [145] and Kelliher et al. [146] successfully used the CRISPR/Cas9 system for the knockout of the CENH3 gene in maize genotypes to produce haploid inducer lines. It seems that such methodologies are very useful for haploid production in cannabis [147]; however, it is vital to develop stable gene transformation and plant regeneration systems before this can be done.
A combination of polyploidy induction and CRISPR/Cas9-equipped Agrobacterium rhizogenes-mediated hairy root culture can be considered as a robust strategy for increasing secondary metabolites production and changing the chemical profile [46]. This strategy has been recently applied for the knockout of the SmCPS1, an important gene in the tanshinone biosynthesis pathway [148] and SmRAS, a key gene in rosmarinic acid biosynthesis, [149] in Salvia miltiorrhiza, as well as DzFPS, a key gene in farnesyl pyrophosphate biosynthesis, in Dioscorea zingiberensis [150]. It seems that this strategy can be used to overcome the problems in hairy root culture of cannabis.
2.2. Somaclonal Variation
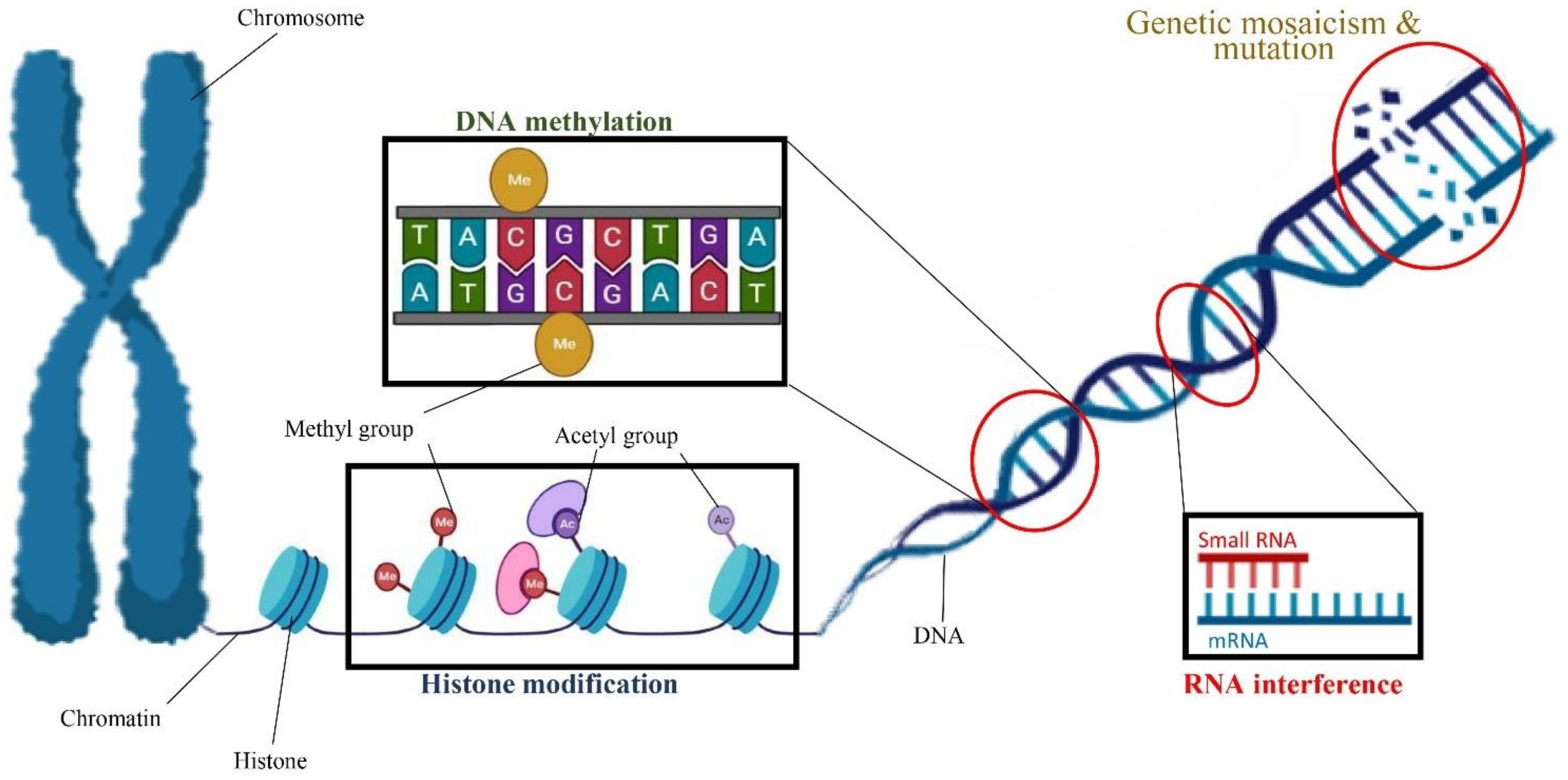
Most previous cannabis tissue culture studies have focused on optimizing culture conditions to increase the cannabis micropropagation rate. However, the optimal condition for in vitro propagation may not be optimal to preserve the genetic integrity of the regenerated genotype [151]. Indeed, in vitro conditions such as medium composition, PGRs, high humidity, the number of subcultures, length of the culture period, temperature, light quality, and light intensity can eventually result in several developmental and physiological aberrations of the micropropagated plants [60]. The term “somaclonal variation” refers to any phenotypic variation detected among micropropagated plants [151]. Somaclonal variation is created by either chromosome mosaics and spontaneous mutation or epigenetic regulations such as histone modification (e.g., histone methylation and histone acetylation), DNA methylation, and RNA interference [151,152] (Figure 4).
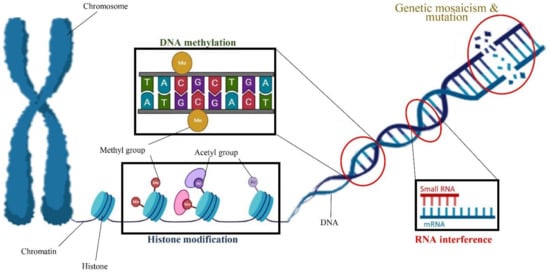
Figure 4.
A schematic view of factors involved in somaclonal variation including genetic mosaicism and mutation as well as epigenetic regulations such as DNA methylation, histone modification, and RNA interference.
Somaclonal variation can be considered as a double-edged sword that has its own merits and demerits based on the objective of the micropropagation experiment. If the objective of micropropagation is breeding, increasing diversity, and generating new variants, somaclonal variation can be considered a beneficial event. On the other hand, if the objective of micropropagation is the production of true-to-type clones, somaclonal variation can be considered as an obstacle.
Although previous cannabis tissue culture studies have shown that regenerated cannabis plants are phenotypically similar to the mother plants and genetically stable with a low mutation rate [78,81,153,154,155], they employed low-resolution molecular markers such as Inter Simple Sequence Repeats (ISSR), which leads to the detection of somaclonal variation, only, at specific genomic regions. Recently, Adamek et al. [156] employed deep whole-genome sequencing to determine the accumulation of somatic mutations within different parts of an individual Cannabis sativa cv. “Honey Banana” plant. They identified a significant number of intra-plant genetic diversity that could impact the long-term genetic fidelity of clonal lines and potentially contribute to the phenotypic variation. Application of the new approaches based on NGS technologies in combination with epigenetic studies is required for the future investigation of the mutation rate in micropropagated cannabis.
3. Ploidy Engineering in Cannabis
Polyploidy is common in many cultivated crop species including wheat, banana, potato, sugar cane, rye, alfalfa, apple, and strawberry [157]; however, the way in which each crop harnesses the benefits of polyploidy is unique. Polyploid can be used as a method of increasing heterosis in a population or can help to mask deleterious alleles [158]. The ploidy level of crops can also be manipulated to induce desired characteristics such as seedless fruits and is achieved by crossing two individuals with unique ploidy levels to produce progeny with an odd number of chromosomes [157]. This is a desired characteristic in cannabis production as seedless flowers produce a greater economic yield [48]. As the production of cannabis moves outdoors, seedless cultivars will likely become increasingly popular. Cannabis is naturally a diploid and multiple successful artificial inductions of polyploidy in cannabis have been reported [159,160,161]. Kurtz et al. [160] have used this technique to produce triploid plants, but field performance and lack of seed development have not yet been reported. These reports provide promising results for the potential for polyploidy to be used to improve cannabis cultivars. Polyploidy induction studies in cannabis have been summarized in Table 4.

Table 4.
Polyploidy induction studies in Cannabis.
3.1. Types of Polyploids
Polyploidy occurs when the normal somatic cells of an organism have more than two sets of homologous chromosomes [158]. An organism can also be a chimera where the individual is composed of cells with different numbers of chromosomes. If the DNA content within the chimera has various ploidy levels, then the organism is a mixoploid [157]. Autopolyploid is defined as polyploidization within a single species and can be produced either somatically or sexually [158].
Somatic polyploids are produced using anti-mitotic agents that are intended to alter the process of mitosis inducing irregular cell division [157]. Oryzalin has been proven to effectively disrupt the action of mitosis in plants through disruption of microtubule action [158]. In contrast, allopolyploid is described as a polyploidization as a result of a hybridization between two unique species, which has not yet been documented for cannabis [158,161].
3.2. Advantages to Polyploidy in Breeding Programs
The natural production of polyploids is considered to be one of the major mechanisms of speciation [158]. The formation of polyploids in nature creates increased heterosis, which could potentially be exploited by modern breeders.
A major benefit of polyploidy is the ability to produce seedless triploids [157]. The production of seedless plants requires crossing two individuals with different ploidy levels. This is usually done by crossing a tetraploid and diploid plant [158]. As both the diploid and tetraploid organisms contain even sets of chromosomes the pairs segregate normally [163]. The two gametes fuse in the mother and produce a triploid (2n = 3x) embryo [163]. The triploid embryo is viable and can undergo regular cell division. The seedless mechanism in triploids alters meiosis so that viable gametes are not produced. Due to the inability of the triploid plant to produce viable gametes, seed production is aborted [164]. Seedless cultivars of cannabis are particularly valuable as studies have shown that seed sets reduce the production of secondary metabolites [165]. This is of particular interest to commercial operations as production moves outdoor.
3.3. Disadvantages to Polyploid Breeding
While autopolyploids provide many benefits to breeders, there are also some fundamental problems with breeding at higher ploidy levels. One drawback to polyploid breeding is that heterozygotes and homozygotes do not separate into classic mendelian ratios [157]. This becomes a significant issue when selecting for disease resistance or selecting more than one recessive trait [158]. Another issue breeders face is that combining two recessive traits becomes more difficult where the chance of getting a double recessive genotype decreases from 1/16 to 1/1296 in tetraploids [157]. This suggests that if the goal of a polyploid breeding program is to combine two or more recessive alleles it would be beneficial to make these improvements at the diploid level before polyploidization. Finally, severe inbreeding depression in polyploid cannabis could render this mechanism useless [163].
3.4. Effects of Polyploidy
Studies observing the morphological traits of tetraploid hemp-type cannabis have shown differences in leaf width, stomate count, and stomate size compared to diploid plants (Figure 5) [159]. The tetraploid leaves were 47% larger than diploid leaves and the tetraploid flowers were more than twice the diameter in comparison to the control flowers [159]. Stomates in the tetraploid plants were twice the length of the diploid stomates but the stomate density was lower [159]. The above-ground shoot weight of the tetraploid plants was almost twice the mass of the diploids. At the cellular level, it is noted that tetraploid individuals have larger mesophyll cells and less intercellular space [162].

Figure 5.
Comparing the morphological traits of diploid and tetraploid cannabis (a) Diploid Cannabis leaf, (b) Tetraploid Cannabis leaf, which is noticeably wider than the diploid, (c) Bright light image of diploid Cannabis stomata, (d) Bright light image of tetraploid Cannabis stomata.
A recent study [161] reported a successful in vitro polyploidy induction in a drug type cannabis using oryzalin. In this study, growth media was supplemented with various concentrations of oryzalin ranging from 20–150 µM and clonal explants from a greenhouse were exposed to treatments for 24 h. The most successful treatments reported were 20 and 40 µM concentrations [161]. Two cultivars were used in this trial and results did vary between treatments. For one cultivar, the 20 µM treatment was sufficient to induce polyploidy; however, for the second cultivar the 20 µM concentration did not produce tetraploids and the 40 µM treatment was most successful. It was noted that following treatments it took several weeks for explants to show any signs of growth. Following treatments, explants were acclimatized and transferred to a greenhouse to observe growth. Tetraploid plants showed an increase in rooting time and a decrease in rooting success compared to diploids [161]. The polyploid plants had slight morphological differences compared to diploids. Tetraploid plants had wider leaves, larger stomates, and a lower density of stomates compared to diploids [161]. The effect of polyploidy on phytochemical composition was noted and CBDA was the only cannabinoid that increased in the polyploid population. In addition, the terpene content of the cannabis plants was also increased in the polyploid population [161].
3.5. Secondary Metabolites
Many species that produce secondary metabolites have witnessed an increased production of these compounds at higher ploidy levels [158]. An increase of secondary metabolite production yield has been reported in polyploid Vetiveria zizanioides L. Nash compared to its diploid counterpart [166]. This species produces aromatic compounds valuable to the fragrance industry and production of these compounds was increased by over 62% when polyploidy was induced [166]. As secondary metabolites produced by cannabis are becoming a legal commodity, the production of these compounds needs to be optimized. In recent literature, it has been reported that polyploid cannabis plants had lower THC production compared to diploid controls but also had increased CBD production [161]. In hemp, the polyploid individuals produced on average 50% less THC in the female flowers compared to the control, but CBD production in the female leaves was more than three times greater in the polyploid population [162]. This study utilized a hemp variety of cannabis, which is bred for low secondary metabolite production [162]. Duplication of some deleterious recessive alleles may be responsible for the decreased THC concentration observed in polyploid cannabis plants. A deleterious allele affecting an important enzyme involved in the metabolic pathway of cannabinoid synthesis could inhibit the entire process. Secondary metabolites such as cannabinoids are heavily dependent on the presence of chemical precursors and enzymes [158].
3.6. Limitations of Existing Polyploidy Literature and Future Potential
Based on the current literature there is very little reported work in the interest of polyploidization in cannabis, with only moderate morphological/chemical differences [161,162]. However, it should be noted that the existing literature does not evaluate many agronomically important traits and only evaluated the first generation of artificially induced autotetraploids. It is worthwhile to mention that, while the tetraploids hold twice as many chromosomes, they do not contain a greater number of unique alleles. Further studies including crosses of unique tetraploids are needed to fully understand the effects of tetraploidy that includes greater allelic diversity. It is possible that while the initial generation of tetraploids is similar to their diploid progenitors, subsequent generations may demonstrate unique phenotypes that are of use to modern breeding programs. Regardless, the use of tetraploids to produce seedless triploids has great potential for the cannabis industry. Moreover, ploidy engineering can be used in cannabis for terpene manipulation, CBD-to-THC ratio in hemp, biomass improvements, and novel cannabinoid production.
4. Genetic Engineering Approaches in Cannabis
Plant genetic engineering can be considered a basic approach to studying gene function and genetic improvement. Generally, plant cells can be either transiently or stably transformed [167]. Although there are a few studies [168,169] that used targeting-induced local lesions in genomes (TILLING) and virus-induced gene silencing (VIGS) approaches for studying the function of some genes in cannabis, there is still a dire need for developing a stable gene transformation system [93]. TILLING, as a powerful method for selecting mutations in specific genes, was used by Bielecka et al. [168] to find cannabis plants with mutations in CsFAD2 and CsFAD3 genes that result in the modification of the seed-oil composition. The requirement of large mutant populations and homozygous mutations are the flip side of the TILLING method. Recently, the VIGS system using Cotton leaf crumple virus (CLCrV) was successfully applied in cannabis to knockdown endogenous phytoene desaturase (PDS) and magnesium chelatase subunit I (ChlI) genes [169].
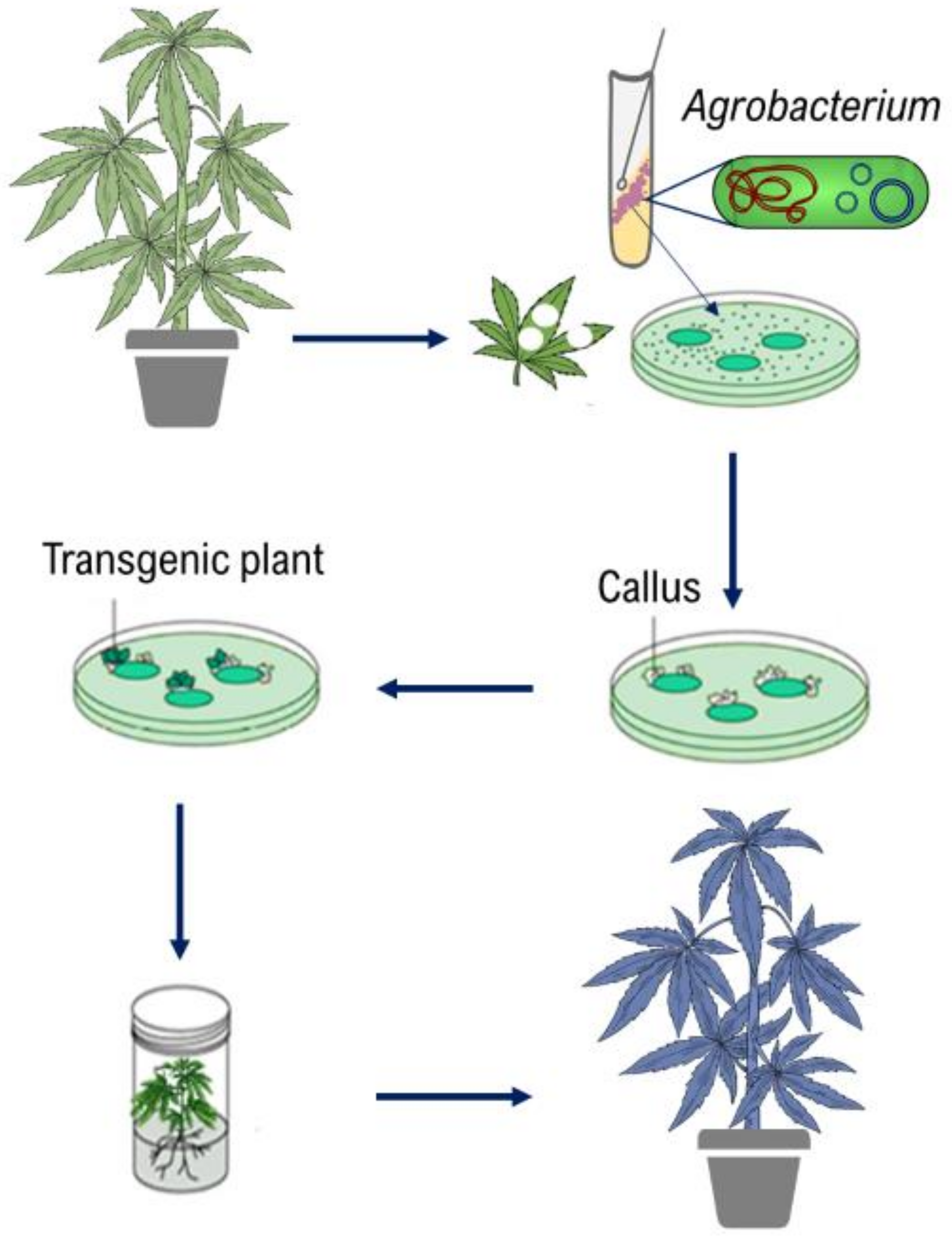
Genetic transformation allows foreign genes to be introduced into a crop and has been extensively used to introduce a variety of important traits (herbicide resistance, pro-vitamin A production, insect resistance, etc.) into major crops for decades [167]. CRISPR/Cas systems have also been recently applied for modifying major crops such as wheat and rice [170]. Developing a gene transformation and/or genome editing systems in cannabis are not only useful for modifying horticultural traits, growth morphology, and biotic and abiotic stress resistance but are also important for studying gene functions. Agrobacterium- and Biolistic-mediated gene transformation systems, de novo meristem induction, and virus-assisted gene editing are applicable to cannabis. Biolistic-mediated gene transformation, which uses particle bombardment to transfer the gene into the plant, and genome editing methods have not yet been reported in cannabis. On the other hand, several studies have investigated Agrobacterium-mediated gene transformation in cannabis. The Agrobacterium-mediated gene transformation system is directly dependent on plant tissue culture (Figure 6).

Figure 6.
The schematic diagram of Agrobacterium-mediated gene transformation.
Recently, Beard et al. [141] used the protoplast of C. sativa var. Cherry x Otto II: Sweetened for transient transformation with plasmid DNA containing a fluorescent marker gene. The authors reported that more than 31% of the cells were successfully transformed. Although gene transformation has been achieved in cannabis by different studies [141,171,172,173,174,175,176], there is only one report regarding transgenic plant regeneration [93].
In the following section, cannabis transformation studies, factors involved in gene transformation, and strategies for improving gene transformation have been discussed.
4.1. Agrobacterium-Mediated Gene Transformation
As soon as the susceptibility of cannabis genotypes to Agrobacterium was revealed [171], Agrobacterium-mediated gene transformation in cannabis became of great interest to many. However, several obstacles have been reported for establishing and developing gene transformation in cannabis such as low efficiency of gene transformation, low rates of regeneration, chimeric regeneration including both non-transgenic and transgenic cells and tissues, as well as inactivation of the transgene [10,47]. Therefore, it is crucial to study different factors involved in gene transformation such as Agrobacterium strains, treatments for explants infection, selection markers, eliminating chimerism, promoters, and translational enhancer. Agrobacterium-mediated gene transformation studies have been summarized in Table 5.

Table 5.
Agrobacterium-mediated gene transformation studies in Cannabis.
4.1.1. Agrobacterium Strains
Agrobacterium strain selection is one of the most important factors in gene transformation (Table 5). The first study of successful gene transformation with more than 50% transformation frequency in fiber-type cannabis (hemp) was performed by MacKinnon et al. [171]. However, they did not report which strain of Agrobacterium was used. Feeney and Punja [172] obtained an acceptable transformation efficiency (15.1 to 55.3%) by using A. tumefaciens EHA101. Wahby et al. [173] used three A. tumefaciens strains including LBA4404, C58, and IVIA 251, as well as eight A. rhizogenes strains including 476, 477, 478, A424, AR10GUS, A4, AR10, and R1601 for establishing hairy root cultures in different genotypes of cannabis. According to their results, genotypes had different responses to Agrobacterium strains in such a way that transformation efficiency ranged between 43% for AR10GUS to 98% for R1601 in A. rhizogenes strains, and between 33.7% for IVIA251 and 63% for C58 for A. tumefaciens strains. Generally, Wahby et al. [173] reported that the gene transformation frequency in cannabis is dependent not only on the strains of Agrobacterium but also on cannabis genotypes, consisting of their sensitivity to agro-infection and their potential to regenerate transgenic tissues.
Deguchi et al. [175] compared transformation efficiency among several hemp genotypes in including Ferimon, Fedora 17, USO31, Felina 32, Santhica 27, Futura 75, CRS-1, and CFX-2 using different A. tumefaciens strains including LBA4404, GV3101, and EHA105 and found high transformation efficiency (>50%) for some genotypes. Based on their results, the maximum GUS expression was observed in the CRS-1 genotype and A. tumefaciens GV3101 led to the highest transformation frequency. In another study, Sorokin et al. [176] investigated the potential of A. tumefaciens EHA105 in transforming different cannabis genotypes (Candida CD-1, Holy Grail x CD-1, Green Crack CBD, and Nightingale) and obtained a high transformation efficiency (45–70.6%). Different responses to Agrobacterium strains are not unique to cannabis and it has been previously documented that various Agrobacterium strains differ in their capacity to transform different recalcitrant plants such as maize [177]. Therefore, it is necessary to investigate more strains to obtain efficient strains for a high-frequency gene transformation protocol.
4.1.2. Infection of Explant
The physiological condition and source of explants play a pivotal role in Agrobacterium-mediated gene transformation. Different explants such as shoot tip and hypocotyl have been employed for gene transformation in cannabis (Table 5). Most studies used different parts of in vitro grown seedlings. MacKinnon et al. [171] succeeded in gene transformation using shoot tip explants that were selected from greenhouse-grown cannabis. Feeney and Punja [172] used callus cells derived from stem and leaf segments of cannabis for Agrobacterium-mediated gene transformation. In another study, Wahby et al. [173] used different parts of 5-day-old in vitro grown seedling of hemp including hypocotyls, cotyledons, cotyledonary node, and primary leaves for gene transformation and reported that the best gene transformation results were obtained from hypocotyl segments. Sorokin et al. [176] also used cotyledons and true leaves of 4-day-old in vitro grown seedling of hemp for gene transformation. Deguchi et al. [175] reported a successful gene transformation using male and female flowers, stem, leaf, and root tissues derived from 2-month-old in vitro grown seedling of hemp.
The co-cultivation period and concentration of Agrobacterium inoculum (optical density (OD)) have a significant impact on successful gene transformation. Feeney and Punja [172] suggested three-day co-cultivation and OD600nm 1.6–1.8 for gene transformation of callus cells. In another study, different explants were co-cultured for two days [173]. Sorokin et al. [176] suggested three days of co-cultivation and OD600nm 0.6 for gene transformation of different parts of in vitro grown seedling of hemp.
Agrobacterium infection efficiency in cannabis can be increased by adding chemical compounds, such as sodium citrate, acetosyringone, and mannose, to the co-cultivation medium. Feeney and Punja [172] reported increased Agrobacterium infection using 100 µM acetosyringone and 2% mannose for hemp gene transformation in the co-cultivation medium. Wahby et al. [173] studied the effect of different concentrations of acetosyringone (20, 100, and 200 µM), sucrose (0.5 and 2%), sodium citrate (20 mM), and 2-N-morpholineethanesulfonic acid (MES) (30 mM) on the gene transformation of cannabis and reported that different chemical compounds had little impact on Agrobacterium infection efficiency, and 20 µM acetosyringone resulted in the best results. On the other hand, Deguchi et al. [175] applied 200 μM acetosyringone, 2% glucose, and 10 mM MES for increasing strain virulence in the gene transformation of hemp. Sorokin et al. [176] also reported increasing the Agrobacterium infectability by using 100 µM acetosyringone for cannabis gene transformation in the co-cultivation medium. Recently, Karthik et al. [178] reported that SNP improved the efficiency of Agrobacterium-mediated gene transformation in soybean. Therefore, it is reasonable to investigate the effects of SNP on the gene transformation of cannabis.
4.1.3. Selection Markers
Although kanamycin has been the main selection agent of transgenic cannabis cells and tissues, other antibiotics, such as spectinomycin, rifampicin, and chloramphenicol, have also been successfully applied for selecting the transformed cells and tissues of cannabis [10,47]. However, it is necessary to study the effect of other antibiotics on cannabis gene transformation because the response of various tissues and genotypes to different antibiotics may vary. For instance, Sorokin et al. [176] and Feeney and Punja [174] used spectinomycin- and kanamycin-resistant genes as selectable markers in the Agrobacterium vectors. Wahby et al. [173] used Agrobacterium vectors carrying kanamycin-, carbenicillin-, and rifampicin-resistant genes for transforming cannabis. Sorokin et al. [176] also used kanamycin- and rifampicin-resistant genes in Agrobacterium vectors. Moreover, Deguchi et al. [175] considered the chloramphenicol-resistant gene as a selective marker in Agrobacterium vectors.
Twin T-DNA binary vectors have also been successfully used for generating marker-free transgenic plants. This would be a very useful and promising method for generating marker-free transgenic cannabis and to mitigate scientific and public concerns regarding dispersing herbicide- and antibiotic-resistant genes of GMO products into the environment.
4.1.4. Eliminating Chimerism
The regeneration of chimeric tissue with both non-transformed and transformed cells and tissues is one of the most crucial challenges in developing a stable gene transformation system in different plants [179]. Therefore, it is essential to use an approach to eliminate the chimeric cells and regenerate only transgenic cells. Feeney and Punja [172] studied the gene transformation frequency and chimerism using the phosphomannose isomerase (PMI) selection strategy, which is based on the existence of sugar (mannose) in the medium. They compared two transformation procedures including 1% mannose and 300 mg/L Timentin (treatment 1) and 2% mannose and 150 mg/L Timentin (treatment 2) and reported that treatment 1 was not capable of distinguishing non-transgenic cells from transgenic cells, and, therefore, they suggested treatment 2 for gene transformation in cannabis. Wahby et al. [173] compared the transformation performance and chimerism of two procedures of gene transformation: complex media MI1 (100 µM acetosyringone, 0.5% sucrose, and 30 mM MES) and MI2 (200 µM acetosyringone, 2% sucrose, 20 mM sodium citrate) and reported that these media could not completely detect transgenic tissues from non-transgenic tissues. Chimerism in transformation systems is not unique to cannabis and is a challenge in many species and is highly dependent on the regeneration system. Moving forward, developing an efficient somatic embryogenesis-based regeneration system will be important to mitigate this issue.
4.1.5. Promoters and Translational Enhancer
Sorokin et al. [176] and Feeney and Punja [174] used the binary vector pNOV3635 and pCAMBIA1301, respectively, carrying a coding region for PMI under control of the nopaline synthase terminator (NOS) and the ubiquitin promoter derived from Arabidopsis thaliana (Ubq3), as well as a spectinomycin and kanamycin selectable marker genes. They also used chlorophenol-red PMI assay, PCR, and southern blot for confirming gene transformation.
When the β-glucuronidase (GUS) reporter gene was employed using the cauliflower mosaic virus 35S RNA (CaMV35S) promoter, GUS activities were observed [173]. In another study, Deguchi et al. [175] used a pEarleyGate 101 vector harboring the eGFP gene and uidA gene under the control of the CaMV 35 S promoter and OCS terminator for GFP fluorescence and GUS staining assays. Moreover, Sorokin et al. [176] reported that when the GUS gene in the pCAMBIA1301 vector was under the control of a CaMV 35S:: Intron, the highest GUS activity in the transgenic cannabis was achieved. While future studies will undoubtedly establish more efficient, or tissue/age-specific promotors, existing promoters are generally effective in cannabis.
4.2. Strategies to Improve Gene Transformation Efficiency
Despite advances in the gene transformation of cannabis over the past few years, efficient transgenic regeneration remains an obstacle. The ability to express and introduce transgene and to regenerate de novo shoots or embryos are two important obstacles to producing transgenic cannabis. Recent studies have confirmed that cannabis cells can be efficiently transformed [10,47,176]; however, there is only one report of transgenic cannabis regeneration [93]. The application of genes related to regulating plant growth and development such as WUSCHEL (WUS) [180], BABY BOOM (BBM) [181], and Growth-Regulating Factors (GRFs) alone or in combination with GRF-Interacting Factor (GIF) [182] is reported as a promising approach to increase plant regeneration efficiency [170]. In this way, ectopic overexpression of genes involved in meristem maintenance, somatic embryogenesis, or phytohormone metabolism can be used to overcome plant regeneration obstacles in recalcitrant plants [183]. Numerous genes involved in meristem maintenance, somatic embryogenesis, and phytohormone metabolism have been identified in different plants [180,183,184]. Accordingly, these studies led to novel approaches for in vitro plant regeneration and Agrobacterium-mediated gene transformation research on recalcitrant species. The downside of this approach is that morphogenic genes have adverse pleiotropic impacts and should be removed from transformed or edited plants [180,182,183,184]. The next sections presented recent progress in using morphogenic genes and highlighted possible approaches to overcome negative pleiotropic effects.
4.2.1. Morphogenic Genes, Key Factors in Plant Regeneration
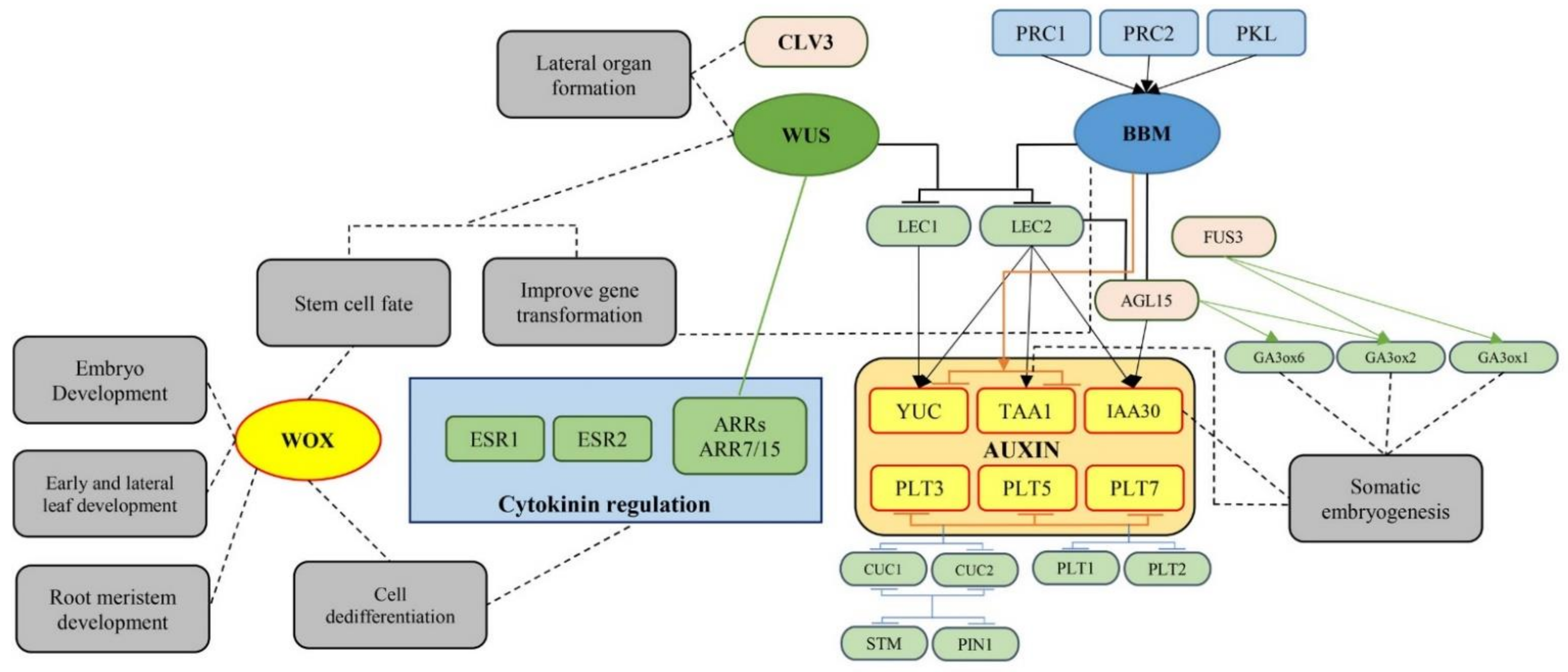
Morphogenic genes that induce regeneration, when overexpressed, are categorized into two classes according to their growth reactions. The first group is composed of genes (e.g., SERK1, AGL15, WUS, and STM) that improve a pre-existing response of embryogenesis under in vitro conditions [180]. The second group includes genes (e.g., BBM, EMK, RKD4, LEC1, LEC2, L1L, HAP3A, FUS3, WUS, WOX5, KN1, CUC1, CUC2, ESR1, and ESR2) that stimulate ectopic embryogenesis or meristem induction under in vitro conditions where such events are usually not seen [180,183,185,186]. The roles of morphogenic genes in plant regeneration have been summarized in Figure 7.

Figure 7.
A schematic view of the morphogenic genes demonstrating their roles in plant growth and development as well as in vitro plant regeneration (AGL15: AGAMOUS-LIKE15; ARR: ARABIDOPSIS RESPONSE REGULATOR; BBM: BABY BOOM; CLV3: CLAVATA3; CUC: CUP-SHAPED COTYLEDON; ESR: ENHANCER OF SHOOT REGENERATION; FUS3: FUSCA3; GA3ox: Gibberellin 3-beta-dioxygenase; IAA30: Indole acetic acid inducible 30; LEC: LEAFY COTYLEDON; PIN1: PIN-FORMED 1; PKL: PICKLE; PLT: PLETHORA; PRC: Polycomb repressive complex; STM: SHOOT MERISTEMLESS; TAA: TRYPTOPHAN AMINOTRANSFERASE ARABIDOPSIS; WOX: WUSCHEL-related homeobox; WUS: WUSCHEL; YUC: YUCCA).
In the first class, the overexpression of genes leads to an increase in embryogenesis under in vitro conditions in which the formation of somatic embryos is already observed. For instance, overexpression of SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 (SERK1) using CaMV 35S promoter resulted in ~4-fold and 2-fold increases in somatic embryogenesis of Arabidopsis thaliana [187] and Coffea canephora [188], respectively. Similarly, further studies in Arabidopsis [189], cotton [190], and soybean [191] have shown an increase in somatic embryogenesis with overexpression of the AGAMOUS-LIKE15 (AGL15) gene, a member of the SERK1 protein complex [192]. It is also well documented that SERK1 and SERK3 are the co-receptors of the Brassinosteroid Insensitive 1 (BRI1) protein. Brassinosteroid is a known PGR that plays a key role in plant embryogenesis. The members of SERK proteins seem to act as mediators across the plasma membrane for brassinosteroid signaling [193].
The use of meristem formation-related genes such as SHOOT MERISTEMLESS (STM) [194] or A. thaliana WUSCHEL (AtWUS), a key gene for regulating meristem cell fate [195], have also resulted in improving embryogenic responses. Arroyo-Herrera et al. [196] reported that somatic embryogenesis in Coffea canephora was increased up to 3–5 fold using an estradiol-inducible AtWUS construct. In line with this result, Bouchabké-Coussa et al. [197] reported that using the 35S::AtWUS cassette led to a 3-fold increase in somatic embryogenesis of Gossypium hirsutum after Agrobacterium-mediated gene transformation. In another study, microspore-derived embryogenesis of Brassica napus was increased using the 35S::BnSTM construct [198].
In the second class, gene overexpression leads to spontaneous regeneration or direct ectopic regeneration of meristems or embryo-like structures in the lack of inductive in vitro conditions. The early studies have shown that overexpression of the AtWUS gene [199] and B. napus BABY BOOM (BnBBM) gene, a member of the AP2/ERF transcription factors (TFs) [200], leads to embryonic morphogenesis. Consequently, Boutilier et al. [200] reported that the constitutive expression of the BnBBM gene in A. thaliana led to ectopic somatic embryogenesis and these ectopic embryos could generate plantlets in the lack of PGRs. This approach has been successfully used to regenerate the transgenic T0 plantlets in different plants using orthologs of the BnBBM gene such as transforming 35S::AtBBM into Nicotiana tabacum [201], soybean 35S::GmBBM into A. thaliana [202], oil palm 35S::EgBBM into A. thaliana [203], and 35S::TcBBM into Theobroma cacao [204]. In another study, Tsuwamoto et al. [205] reported that ectopic overexpression of A. thaliana EMBRYOMAKER (AtEMK) led to embryo-like structures at 23% of cotyledon tips; however, these structures could not regenerate the plantlets. They concluded that AtEMK, a member of the AP2/ERF TFs and related to BBM, should be expressed under a regulated network for avoiding pleiotropic effects and achieving normal plantlets. RKD4 as a key gene in the RWP-RK TF family is another gene that has an important role during early embryo development in A. thaliana and plays a pivotal role in the first asymmetrical zygotic division [206,207]. For instance, ectopic somatic embryogenesis of Phalaenopsis, a plant typically reluctant to direct somatic embryogenesis, was obtained using the chemical induction of transgenic RKD4 [208].
The overexpression of genes related to embryo maturation (e.g., FUSCA3 (FUS3), LEAFY COTYLEDON1 (LEC1), and LEC2) causes similar morphogenic reactions. Lotan et al. [209] characterized the first of these genes namely LEC1, in which a cassette of 35S::AtLEC1 was transformed to A. thaliana. Although the embryo-like structures were produced, functional ectopic embryogenesis was not observed [209]. In another study, Zhu et al. [210] used a 35S::CsL1L cassette for overexpression of Citrus sinensis LEC1 after gene transformation in epicotyls of sweet orange and reported that embryo-like structures were obtained after two months. They concluded that overexpression of L1L in Citrus is sufficient for recovering functional somatic embryos. However, Uddenberg et al. [211] indicated that although the overexpression of the LEC1/L1L (PaHAP3A) did not lead to ectopic embryogenesis in vegetative tissues, ectopic embryogenesis was obtained during zygotic embryo maturation. These findings showed that certain types of cells and tissues may be more receptive to morphogenic genes for enhancing ectopic embryogenesis or meristem maintenance [211]. Later studies have shown that the overexpression of LEC2 leads to more somatic embryogenesis in comparison with LEC1, FUS3, or L1L [212,213,214].
However, several studies [190,197,198,200] showed that pre-existing embryogenic responses are increased through overexpression of morphogenic genes, and additional studies revealed that ectopic overexpression of morphogenic genes resulted in improving somatic embryogenesis where such events are usually not seen [183]. Zuo et al. [199] identified the first “meristem” gene in Arabidopsis (AtWUS) which enhances somatic embryogenesis. They suggested that the vegetative-to-embryonic transition can be stimulated by the WUS gene. Moreover, Gallois et al. [215] demonstrated that the overexpression of WUS and STM genes generates the bulk of cells contiguous to the WUS foci showing primary meristems. In another study, Gallois et al. [216] indicated that unique phenotypes obtained from WUS overexpression were related to different factors such as co-expression of other morphogenic TFs, phytohormone regime, and type of WUS activation (such as GAL4-VP16 activation method and HSP::CRE-mediated excision). Rashid et al. [217] also reported that a member of the WUS/WOX gene family namely AtWOX5 led to the direct organogenic response in Nicotiana tabacum. Luo et al. [218] showed that shoot organogenesis in N. tabacum was increased 3-fold by overexpression of the maize STM ortholog KNOTTED1 (KN1) through 35S::ZmKN1 cassette. Similar results were reported by Nishimura et al. [219].
The CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 genes are known to have a key role in the shoot meristem formation. Daimon et al. [220] indicated that the overexpression of AtCUC1 and AtCUC2 under a CaMV 35S promoter resulted in a significant increase (about 6.5–9-fold) in shoot organogenesis of A. thaliana transgenic calli. However, no micro-shoots were recovered in the lack of hormones, showing the hormone-dependent function of CUC1 and CUC2 genes. Genes related to the phytohormone signal transduction (both downstream targets and receptors), play a pivotal role in improving plant regeneration; genes like ENHANCER OF SHOOT REGENERATION 1 (ESR1) and ESR2 that are involved in the cytokinin response pathway and MONOPTEROS (MP) that is involved in the auxin response pathway. Several studies have shown that the overexpression of these genes led to an increase in the shoot meristem formation [221,222,223]. Moreover, the phenotypic response of morphogenic genes can be impacted by levels of phytohormones. For instance, Wójcikowska et al. [224] reported that the high and low concentrations of auxin led to callogenesis and somatic embryogenesis, respectively, in the transformed A. thaliana overexpressing LEC2 under a dexamethasone (DEX)-inducible system.
4.2.2. Strategies to Overcome Pleiotropic Effects
Based on the aforementioned literature and other references therein, it is clear that the use of morphogenic genes may help to increase regeneration efficiency in plants. However, strong and constitutive overexpression of these genes can lead to unwanted pleiotropic effects such as infertility of regenerated plants [170]. A promising approach to mitigate this is to implement an inducible expression system for these genes in a stable transformation system. This means, it is necessary to apply an additional step that includes optimization of the morphogenic gene expression level with restricting expression after transformation. Such an approach causes enhanced gene transformation and improved fertile and healthy T0 plantlets regeneration. Generally, there are five strategies to cope with these challenges including (i) removing the morphogenic gene when no longer needed, (ii) using the inducible expression of the gene for improving morphogenic growth response followed by excision of the inducing ligand for silencing the expression, (iii) using Agrobacterium-mediated gene transformation in such a way that promotes the transient expression of the morphogenic genes, (iv) using promoter that can turn off the morphogenic regulatory gene when no longer required, and (v) using GRF-GIF Chimeras [180,182,183,185,186].
The excision-based method has provided a reliable strategy for applying morphogenic genes to regenerate fertile and healthy transgenic plantlets. The first successful application of this method through BBM and Flippase (FLP)-recombinase was reported in Populus tomentosa [225]. A T-DNA construct including a single pair of FLP Recombination Target Sites (FRT) encompassing both a CaMV 35S promoter stimulating B. campestris BBM (BcBBM) expression and a heat-shock inducible promoter stimulating expression of FLP recombinase was designed. Deng et al. [225] reported that after transforming this T-DNA construct, more than 28% of calli produced normal plantlets on phytohormone-free media and also showed that 42 °C heat shock treatment for 2 h resulted in the removal of both the BBM and FLP recombinase cassettes. In another study, Lowe et al. [226] reported that high transformation efficiency in inbred maize was obtained by low expression of ZmWUS2 (using the Agrobacterium NOPALINE SYNTHASE, or NOS, promoter) and overexpression of ZmBBM (using the Zea mays UBIQUITIN promoter). The explants were inoculated on dry filter paper to induce a desiccation-stimulated maize promoter derived from an ABA-responsive gene (RAB17) carrying CRE recombinase, which then removed these 3 expression cassettes. After the removal of the BBM, WUS2, and CRE transgenes, only the T-DNA construct with the genes of interest were remained. This method has been successfully used in other recalcitrant plants such as sorghum, rice, corn, and sugarcane [227].
Inducible expression to control morphogenic gene expression is another robust alternative approach. Heidmann et al. [228] reported that gene transformation in Capsicum annum, a recalcitrant plant species, was achieved using the 35S::BnBBM~GR vector for gene transformation and cultivation of explants in the medium consisting of DEX and TDZ. In a similar study, Lutz et al. [229] reported that transgenic fertile and healthy A. thaliana plantlets were achieved using the DEX-inducible AtBBM~GR cassette.
The overexpression of PLANT GROWTH ACTIVATION genes, such as PGA37 in A. thaliana led to the transition of the vegetative phase to the embryogenic phase by using the estradiol-inducible system [230]. The PGA37 gene, based on the DNA-binding domain similarities, encodes the MYB118. Wang et al. [230] reported that root segments developed somatic embryos by PGA37 expression under inducible control, which were related to overexpression of LEC1. The green-yellowish embryonic calli were produced through the expression of PGA37 using the estradiol-inducible system in the medium containing auxin after 7–10 days of inoculation and somatic embryos were obtained after 3–5 weeks. When estradiol was removed from the medium, which causes downregulating PGA37 expression, healthy, fertile plantlets were obtained from the somatic embryos. Moreover, Wang et al. [230] showed that estradiol-induced expression of MYB115, a closely related homolog, resulted in somatic embryogenesis from root segments. A similar strategy using DEX-induced expression of TcLEC2 was successfully used for the regeneration of transgenic somatic embryos in Theobroma cacao [231].
Using Agrobacterium-mediated gene transformation in such a way that promotes the transient expression of the morphogenic genes is another alternative strategy [232,233,234,235]. Negative selectable markers located outside the T-DNA (beyond the left border) have been used to remove plant cells containing these sequences [233]. Other studies have located a positive marker gene outside the T-DNA, which could be transiently expressed for generating marker-free transgenic plants [236]. A mixture of T-DNAs could be received by placing an Agrobacterium-derived isopentyl transferase (IPT) gene outside the T-DNA, with the greater part of the T-strands containing the trait and a minority of T-strands not terminated properly outside the T-DNA, including the flanking IPT gene. Therefore, the transient expression of the IPT gene could stimulate the signaling of cytokinin and subsequently improve shoot proliferation, which results in the recovery of transgenic plants without a selectable marker. This strategy was successfully used in maize genotypes by positioning WUS2 and BBM beyond the left border for transient somatic embryogenesis and subsequent transgenic plantlet regeneration [183].
Recently, a new strategy has been developed using the maize phospholipid transfer protein (PLTP) promoter driving BBM and the maize auxin inducible (AXIG1) promoter for improving gene transformation [237]. The application of these two promoters in the expression cassettes led to somatic embryogenesis within a week, and germination of these somatic embryos within 3–4 weeks. Similarly, the use of a combination of GRF and GIF has been proposed as a new method to tackle negative pleiotropic effects [170,182,238]. Debernardi et al. [170] demonstrated that fertile transgenic wheat, rice, and citrus without obvious developmental defects can be achieved by the expression of a fusion protein combining wheat GRF4 and GIF1. Moreover, they reported that GRF4–GIF1 induced efficient plant regeneration in the absence of exogenous PGRs which helps transgenic plant selection without selectable markers. They also combined CRISPR–Cas9 with GRF4– GIF1 and regenerated edited transgenic plants. Therefore, the combination of CRISPR–Cas9 and GRF4– GIF1 can be considered a powerful and promising strategy for the regeneration of healthy and fertile transgenic plants [170]. This strategy has been recently used in cannabis. Zhang et al. [93] reported that using GRF3– GIF1 in the CRISPR vector resulted in a 1.7-fold increase in edited plant regeneration.
As a future perspective, morphogenic genes can be considered as targets of gene transformation in order to overcome current obstacles in cannabis tissue culture and successful regeneration of transformed plants. Furthermore, useful techniques enabled us to control the side effects of ectopic expression of these genes through transient and inducible gene expression in the host.
4.3. Strategies to Prevent Transgene Escape
The frequency of the alleles/genes in a population can be changed due to outcrossing of gametes or gene flow [239]. Thus, transgenes can move from a transgenic plant to their non-transgenic counterparts or wild relatives, a process called transgene escape. It is not uncommon for transgenic plants to mate with their wild relatives. Spontaneous hybridization will occur among transgenic and non-transgenic plants unless the proper distances are maintained [239]. This is an area much of concern, specifically in outcrossing plants, in plant biotechnology. Outcrossing poses negative impacts in terms of contamination in non-transgenic crops but this problem depends on whether a new allele causes an increase in transgene escape or not [240]. While dealing with trans-gene flow one should consider the situations according to transgenic crops [240]. In general, the possible containment and mitigation strategies are physical containment [241], biological/molecular containments (e.g., sterility [242], clistogamy [243], apomixes [244], maternal transformation [245], incompatible genome [246], gene splitting [247], expression in virus [248], genetic use restriction technology (GURT) [239]), and transgenic mitigation [249]. The appropriate approaches should be considered after transgenic cannabis production to prevent transgene escape.
4.4. CRISPR/Cas-Mediated Genome Editing
CRISPR/Cas-mediated genome editing has exceptionally improved plant biotechnology [43,250]. This system is robust and offers relatively high target programmability and specificity that can allow accurate genetic modification. The CRISPR/Cas system provides a unique opportunity to improve cannabis varieties with desired traits in a sustainable fashion. Recently, Zhang et al. [93] employed the CRISPR/Cas9 system to knock out the phytoene desaturase gene and they reported that four edited cannabis plantlets with albino phenotype have been successfully generated. However, the numerous new biotechnological methods such as base editing and prime editing based on CRISPR/Cas platforms would expand cannabis synthetic biology and the toolbox of fundamental research. A successful CRISPR/Cas system experiment requires designing target-specific guide RNAs (gRNAs) and an efficient regeneration protocol for developing transgenic/edited plantlets [251]. The gRNA is categorized as a chimeric RNA including a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA). The crRNA consists of a guide (spacer) sequence that accurately navigates the Cas9 protein to the targeted gene. Then, the targeted DNA is cleaved by the RNA-guided DNA endonuclease Cas9. Another key part of CRISPR/Cas-mediated genome editing is the protospacer adjacent motif (PAM), which is a conserved and CRISPR-dependent DNA sequence motif adjacent to the target site (protospacer) and is utilized by the endogenous CRISPR in archaea and bacteria to discriminate invading- and self-DNAs. Generally, the application of an effective and precise CRISPR/Cas system is strongly dependent on the selection of the best guide sequence (gRNA target site) [43,250,251].
In the previous sections, promising approaches for developing tissue culture protocols for genetic material delivery (e.g., Agrobacterium-mediated method) and regeneration methods have been discussed. In this section, we discuss the principles of designing gRNA in order to produce precise target mutation(s) and prevent off-target mutations as one of the most important prerequisites of genome editing technology. We also present the available bioinformatics tools for designing gRNAs that can be used in cannabis.
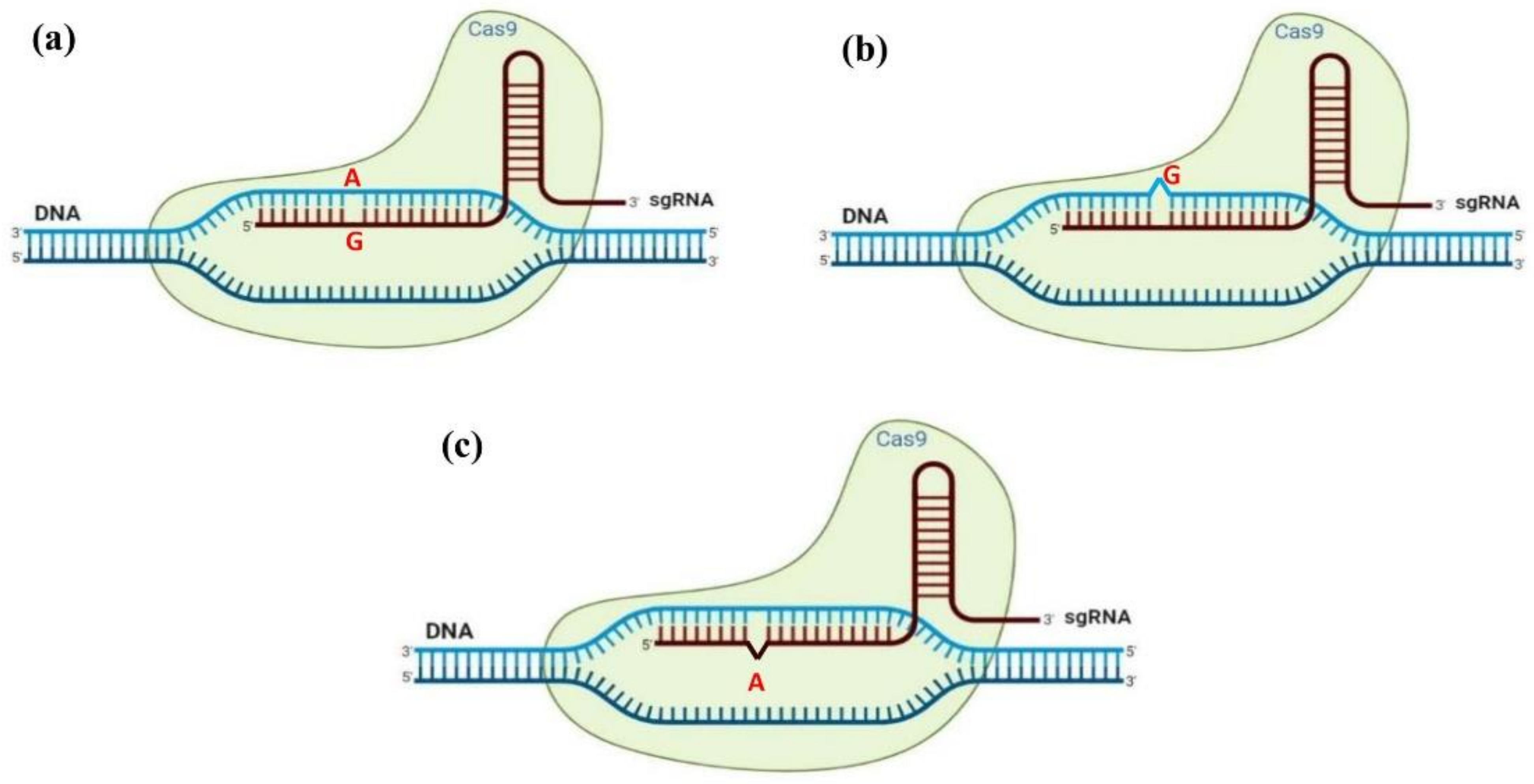
The prediction of the presence of off-target sites (i.e., unintended mutations) is one of the most important steps in designing gRNA for CRISPR/Cas-mediated genome editing. The design of the candidate gRNAs starts with the screening of the organism’s whole-genome sequence. Therefore, the availability and accessibility of a high-quality reference genome is required, which is the case in cannabis [252]. The genome of cannabis has been de novo assembled nearly a decade ago [253]; however, there are still some major challenges to using the cannabis reference genome. Currently, 12 different genomes (assembled and annotated) are available for cannabis [252]. Having multiple reference genomes can be considered a bonus, but, contradictorily, there are significant differences in reported genome size, chromosome order, and gene annotations among cannabis genome assemblies [1,2,252]. Therefore, designing a precise and accurate gRNA is an important starting point for high-quality CRISPR/Cas-mediated genome editing in cannabis with the minimum off-target activities. As can be seen in Figure 8, there are three types of off-targets induced by the CRISPR/Cas system including (a) off-target sites with a base mismatch, (b) off-target sites with extra base (DNA bulge or deletion), and (c) off-target site with missing base (RNA bulge or insertion) [254]. Cases (b) and (c) are considered as the indel (insertion or deletion) off-target events [254]. It is necessary to consider these off-targets during genome engineering projects.

Figure 8.
Three types of off-targets induced by CRISPR-mediated genome editing; (a) off-target sites with base mismatch, (b) off-target sites with extra base (DNA bulge or deletion), and (c) off-target site with missing base (RNA bulge or insertion).
Over the past few years, several bioinformatics tools have been developed to design gRNAs and predict, in silico, the off-targets. These tools have significantly facilitated the successful application of CRISPR/Cas-mediated genome editing technology [43,250]. A list of bioinformatics tools that contain cannabis genome information is presented in Table 6.

Table 6.
The main characteristics of some tools for the design of gRNA for genome editing in cannabis.
Generally, these tools exploit a similar algorithm to design gRNA [255]. However, some characteristics related to the gRNA spacer region (e.g., number and type of mismatch, GC-content, length, and indels) and the selection of specific Cas-nucleases vary between these tools [256]. In general, these tools provide multiple sequences for designing gRNA, which are appropriate for editing similar genome conservative motives of various evolutionarily related genotypes [256]. These tools require the sequence of the targeted gene and the Cas-nucleases type. Then they search the genome and provide information related to candidate gRNAs such as the potential protospacers, both non-ranked and ranked with respect to their fitness, and off-target sites [255]. Generally, considering variations in the nucleotide sequences and the increase in individual gene copy number, it is necessary to design multiple gRNAs for each target for an efficient genome editing.
CRISPR RGEN is a web-based tool that also includes BE-Designer [257], Cas-OFFinder [258], Cas-Designer [259], Microhomology-Predictor [260], and some other tools that can be employed for post-editing analysis. Implementation of multiple tools increases the flexibility of CRISPR RGEN Tools that can be illustrated by the following examples: The Cas-Designer can detect potential protospacers in the analyzed sequence. It also provides a broad range of nucleases containing mutant forms with various PAM-sequences: SpCas9 (NRG for the off-target sites and NGG for the target sites), SaCas9 (NNGRRT), StCas9 (NNAGAAW), VRER SpCas9 (NGCG), wtSpCas9 (NNGTGA), NmCas9 (NNNNGMTT), VQR SpCas9 (NGA), CjCas9 (NNNNRYAC), CjCas9 (NNNVRYAC), BhCas12b (TTN), FnCpf1 (KYTV), FnCpf1 (TTN), AsCpf1 (TTTV), AsCpf1 (TTTN), etc. In the protospacer searches, one may specify if it is essential to find the probability of single nucleotide indels. In this case, the tool notifies about enhancing the time of the search. However, if this option is not applied, the results of the search returns immediately. Other parameters are chosen by default. Cannabis genome should be selected from the list to run the search. Further changes to other options of the Cas-Designer can be applied when it is locally installed on a computer [255]. The Microhomology-Predictor facilitates the forecast of the type of microhomology-mediated end-joining (MMEJ) and nonhomologous end-joining (NHEJ) by analyzing the microhomology of nucleotide sequences in the sites of double-strand breaks [260]. The Cas-OFFinder tool can be used to screen possible off-target sites in the genome. Furthermore, this tool can be independently applied for previously selected gRNAs to find off-target sites. For analysis, it is essential to insert the selected gRNAs sequences, specify the number of indels (both in RNA and DNA) and the adequate number of mismatched nucleotides, similarly Cas-Designer [258]. The BE-Designer tool can be applied for designing gRNAs to edit individual nitrogenous bases. Although Cas-Designer works similarly to BE-Designer, there are some differences. For instance, the selection of the corresponding nucleases and the PAM-regions is slightly limited in the BE-Designer, and the “window” size should be chosen for searching the targeted substitution of nucleotides (A→G or C→T) and the editing sites. The results would be provided in a table, in which the substituted nucleotides are shown with a specific color [255].
The CRISPOR [261] is another user-friendly tool with a detailed user manual. Selecting optimal protospacers in this tool can be performed in three steps. In the first step, the sequence of the targeted gene should be provided by the user. In the second step, the genome (e.g., cannabis) should be selected. In the third step, the Cas9 nuclease must be selected out of 33 options. The results of the search are provided in both table and graphic formats. The analyzed sequence is represented, and the potential edited sites are highlighted with various colors, relating to low, medium, and high specificity. Moreover, the table of the results includes information regarding the “Cloning/PCR primers”, protospacers sequences, their positions with indicated DNA chains, expression of a specific gRNA, and cloning. Two algorithms are used to assess the efficiency of the proposed gRNAs and, also, specificity is used to rank the proposed gRNAs. In addition to predicting the types of microhomology, it foresees the off-target sites with the number of mismatches [262].
The CRISPR/Cas9 Target online predictor (CCTop) tool [263] is applied to design and screen gRNAs for on-target and off-target sites. It can use a wide range of Cas-9 nucleases from various bacterial strains (e.g., Campylobacter jejuni, Streptococcus pyogenes, Treponema denticola, Staphylococcus aureus, Streptococcus thermophilus, and Neisseria meningitidis), their mutant forms, as well as Cpf1 from Francisella novicida and Lachnospiraceae/Acadaminoccus. The most intriguing feature of this tool is its ability to add nucleotides in the form of one or two guanines at the 5′-end of the spacer region to improve in vitro transcription. Additionally, two favorable nucleotides adjacent to the PAM region can be considered in order to prevent them from being with the gRNA. This tool exploits the CRISPRater prognosis algorithm to rank predicted gRNAs. gRNAs in intergenic, introns, and exons spacers are presented in various colors. This tool shows information in both table and graphic formats [255].
The CHOPCHOP tool [264] can be used to design and screen gRNAs with the capability to work in complex with the Cpf1 nuclease and Cas9-nuclease and its nickase forms. The search for gRNA can be run either in all 5′- or 3′-non-coding regions, splicing sites, exons, and in the promoter sequence or in a specified coding region, the length of which would be chosen individually. Moreover, the restriction sites in the edited region can be screened by the corresponding enzymes with the manufacturer’s name. After finishing the search, an adjustable color graphic and an interactive table with ranked gRNA target sequences, consisting of DNA chains, the GC-composition, and other information are provided [255].
The Breaking Cas tool [265] can be applied for designing gRNAs and screening the off-target sites. It is essential to select the form of nuclease from the three types of Cpf1 and the four types of Cas9 with mismatch number and the length of pre-set spacers. It is also possible to select a “Custom” nuclease, which is followed by providing the corresponding lengths of the spacer sequence of gRNA, PAM-sequences, and an acceptable number of mismatches.
All in all, different types of CRISPR-mediated genome editing systems, including targeted mutagenesis to knock out a target gene [266], base editing systems for transition substitution using a combination of Cas9 nickase and either adenosine or cytidine deaminase [267], transversion substitution using Cas9 nickase, cytidine deaminase and uracil DNA glycosylase [268], as well as prime editing systems using DNA or RNA as donors [269], have been recently developed and applied in different species [270]. Very recently the application of the CRISPR system [93] and prediction systems through big data analysis and machine learning [147] have been reported in cannabis. Focusing on cannabis genome engineering, the design of genome sequences is as essential as the applied genome-editing technology. When the target gene is epigenetically regulated, not only genome sequences but also histone modifications, DNA methylation, and chromatin structure, may need to be modified to generate the desired cannabis plant. Furthermore, systems for highly efficient sequence-specific nuclease delivery and subsequent genome-edited cell and plant selection must also be developed to accelerate the breeding of designer cannabis plants (Figure 9).

Figure 9.
A schematic view of Cannabis genome engineering.
5. Conclusions and Future Objectives
Efficient and reliable in vitro culture procedures can be considered as an important prerequisite for successful gene transformation, genome editing, micropropagation, and conservation of cannabis. Micropropagation is a powerful tool that can develop and propagate new varieties. Polyploidy induction is also a method that can influence secondary metabolites. Although current shoot proliferation-based protocols are relatively developed for mass propagation of cannabis, it is necessary to develop somatic embryogenesis, organogenesis, and haploid production protocols especially for gene transformation and genome editing studies. Agrobacterium-mediated gene transformation has been described in cannabis. Recently, transgenic plant regeneration has been successfully obtained. It seems that using morphogenic genes can help overcome challenges in transgenic plant regeneration. The production of secondary metabolites is another important aspect of in vitro culture of cannabis. Although several studies have attempted to manipulate secondary metabolite production in cell suspension culture and hairy root culture of cannabis, small amounts of cannabinoids have been produced. Therefore, it is necessary to apply new methods such as CRISPR/Cas-equipped A. rhizogenes-mediated hairy root culture for increasing or changing secondary metabolite profiles. Machine learning algorithms can also be considered as robust computational biology for a comprehensive study of secondary metabolites and, also, modeling and optimizing hairy root culture or cell suspension culture for improving its yield. Generally, cannabis plants still provide the most efficient natural sources of secondary metabolites, and, in particular, cannabinoids and modern biotechnologies will play an important role in further genetic improvement.
Author Contributions
All authors have equal contribution in this review paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NSERC Discovery Grant, grant number RGPIN-2016-06252.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kovalchuk, I.; Pellino, M.; Rigault, P.; van Velzen, R.; Ebersbach, J.; Ashnest, J.; Mau, M.; Schranz, M.; Alcorn, J.; Laprairie, R. The Genomics of Cannabis and Its Close Relatives. Annu. Rev. plant Biol 2020, 71, 713–739. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Pepe, M.; Alizadeh, M.; Rakei, A.; Baiton, A.; Jones, A.M.P. Recent advances in cannabis biotechnology. Ind. Crop. Prod. 2020, 158, 113026. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Bernard-Perron, D. Cannabinomics: Application of Metabolomics in Cannabis (Cannabis sativa L.) Research and Development. Front. plant Sci. 2020, 11, 554. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Peris, A.; Vergara, K.; Matus, J.T. Comprehending and improving cannabis specialized metabolism in the systems biology era. plant Sci. 2020, 298, 110571. [Google Scholar] [CrossRef] [PubMed]
- Schultz, C.J.; Lim, W.L.; Khor, S.F.; Neumann, K.A.; Schulz, J.M.; Ansari, O.; Skewes, M.A.; Burton, R.A. Consumer and health-related traits of seed from selected commercial and breeding lines of industrial hemp, Cannabis sativa L. J. Agric. Food Res. 2020, 2, 100025. [Google Scholar] [CrossRef]
- Salami, S.A.; Martinelli, F.; Giovino, A.; Bachari, A.; Arad, N.; Mantri, N. It Is Our Turn to Get Cannabis High: Put Cannabinoids in Food and Health Baskets. Molecules 2020, 25, 4036. [Google Scholar] [CrossRef]
- Schilling, S.; Dowling, C.A.; Shi, J.; Ryan, L.; Hunt, D.; Oreilly, E.; Perry, A.S.; Kinnane, O.; McCabe, P.F.; Melzer, R. The Cream of the Crop: Biology, Breeding and Applications of Cannabis sativa. Authorea Prepr. 2020. [Google Scholar] [CrossRef]
- Devsi, A.; Kiyota, B.; Ouellette, T.; Hegle, A.P.; Rivera-Acevedo, R.E.; Wong, J.; Dong, Y.; Pugsley, M.K.; Fung, T. A pharmacological characterization of Cannabis sativa chemovar extracts. J. Cannabis Res. 2020, 2, 17. [Google Scholar] [CrossRef]
- Valleriani, J. Identity narratives in the face of market competition: The emerging legal medical cannabis market in Canada. Drugs Educ. Prev. Policy 2020, 27, 37–48. [Google Scholar] [CrossRef]
- Wróbel, T.; Dreger, M.; Wielgus, K.; Słomski, R. The application of plant in vitro cultures in cannabinoid production. Biotechnol. Lett. 2018, 40, 445–454. [Google Scholar] [CrossRef]
- Farag, S.; Kayser, O. Cultivation and breeding of Cannabis sativa L. for preparation of standardized extracts for medicinal purposes. In Medicinal and Aromatic Plants of the World; Máthé, Á., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 1, pp. 165–186. [Google Scholar]
- Maayah, Z.H.; Takahara, S.; Ferdaoussi, M.; Dyck, J.R.B. The molecular mechanisms that underpin the biological benefits of full-spectrum cannabis extract in the treatment of neuropathic pain and inflammation. Biochim. Biophys. Acta (Bba) Mol. Basis Dis. 2020, 1866, 165771. [Google Scholar] [CrossRef] [PubMed]
- Workman, C.D.; Kindred, J.H.; Boles Ponto, L.L.; Kamholz, J.; Rudroff, T. The effects of chronic Δ-9-tetrahydrocannabinol (THC) and cannabidiol (CBD) use on cerebral glucose metabolism in multiple sclerosis: A pilot study. Appl. Physiol. Nutr. Metab. 2019, 45, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Cassano, T.; Villani, R.; Pace, L.; Carbone, A.; Bukke, V.N.; Orkisz, S.; Avolio, C.; Serviddio, G. From Cannabis sativa to Cannabidiol: Promising Therapeutic Candidate for the Treatment of Neurodegenerative Diseases. Front. plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Alves, P.; Amaral, C.; Teixeira, N.; Correia-da-Silva, G. Cannabis sativa: Much more beyond Δ9-tetrahydrocannabinol. Pharmacol. Res. 2020, 157, 104822. [Google Scholar] [CrossRef]
- Baron, E.P. Medicinal Properties of Cannabinoids, Terpenes, and Flavonoids in Cannabis, and Benefits in Migraine, Headache, and Pain: An Update on Current Evidence and Cannabis Science. J. Head Face Pain 2018, 58, 1139–1186. [Google Scholar] [CrossRef]
- Nuutinen, T. Medicinal properties of terpenes found in Cannabis sativa and Humulus lupulus. Eur. J. Med. Chem. 2018, 157, 198–228. [Google Scholar] [CrossRef]
- Hwang, E.; Ngo, H.T.T.; Park, B.; Seo, S.-A.; Yang, J.-E.; Yi, T.-H. Myrcene, an Aromatic Volatile Compound, Ameliorates Human Skin Extrinsic Aging via Regulation of MMPs Production. Am. J. Chin. Med. 2017, 45, 1113–1124. [Google Scholar] [CrossRef]
- Aly, E.; Khajah, M.A.; Masocha, W. β-Caryophyllene, a CB2-Receptor-Selective Phytocannabinoid, Suppresses Mechanical Allodynia in a Mouse Model of Antiretroviral-Induced Neuropathic Pain. Molecules 2020, 25, 106. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, O.; Ozdemir, I.; Tanyildizi, S.; Yildiz, S.; Oguzturk, H. Antioxidative effects of curcumin, β-myrcene and 1,8-cineole against 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced oxidative stress in rats liver. Toxicol. Ind. Health 2011, 27, 447–453. [Google Scholar] [CrossRef]
- Shah, S.; Dhanani, T.; Sharma, S.; Singh, R.; Kumar, S.; Kumar, B.; Srivastava, S.; Ghosh, S.; Kumar, R.; Juliet, S. Development and Validation of a Reversed Phase High Performance Liquid Chromatography-Photodiode Array Detection Method for Simultaneous Identification and Quantification of Coumarin, Precocene-I, β-Caryophyllene Oxide, α-Humulene, and β-Caryophyllene in Ageratum Conyzoides Extracts and Essential Oils from Plants. J. AOAC Int. 2020, 103, 857–864. [Google Scholar] [CrossRef]
- Kołodziejczyk, A.; Pyrcz, P.; Błaziak, K.; Pobudkowska, A.; Sarang, K.; Szmigielski, R. Physicochemical Properties of Terebic Acid, MBTCA, Diaterpenylic Acid Acetate, and Pinanediol as Relevant α-Pinene Oxidation Products. ACS Omega 2020, 5, 7919–7927. [Google Scholar] [CrossRef]
- Guzmán-Gutiérrez, S.L.; Bonilla-Jaime, H.; Gómez-Cansino, R.; Reyes-Chilpa, R. Linalool and β-pinene exert their antidepressant-like activity through the monoaminergic pathway. Life Sci. 2015, 128, 24–29. [Google Scholar] [CrossRef]
- Jana, S.; Patra, K.; Sarkar, S.; Jana, J.; Mukherjee, G.; Bhattacharjee, S.; Mandal, D.P. Antitumorigenic Potential of Linalool Is Accompanied by Modulation of Oxidative Stress: An In Vivo Study in Sarcoma-180 Solid Tumor Model. Nutr. Cancer 2014, 66, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Srividya, N.; Lange, I.; Lange, B.M. Determinants of Enantiospecificity in Limonene Synthases. Biochemistry 2020, 59, 1661–1664. [Google Scholar] [CrossRef]
- Faria, G.M.; Soares, I.D.P.; D’Alincourt Salazar, M.; Amorim, M.R.; Pessoa, B.L.; da Fonseca, C.O.; Quirico-Santos, T. Intranasal perillyl alcohol therapy improves survival of patients with recurrent glioblastoma harboring mutant variant for MTHFR rs1801133 polymorphism. BMC Cancer 2020, 20, 294. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, Y.; Huang, W.; He, L.; Lin, Z.; Zhou, J.; He, Q. Toxic effects of terpinolene on Microcystis aeruginosa: Physiological, metabolism, gene transcription, and growth effects. Sci. Total Environ. 2020, 719, 137376. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.A.M.d.; Monteiro, O.S.; Coutinho, D.F.; Rodrigues, A.A.C.; Silva, J.K.R.d.; Maia, J. Seasonal and Circadian Study of a Thymol/3-Terpinene/p-Cymene Type Oil of Ocimum gratissimum L. and Its Antioxidant and Antifungal Effects. J. Braz. Chem. Soc. 2019, 30, 930–938. [Google Scholar] [CrossRef]
- de Oliveira, M.G.B.; Marques, R.B.; de Santana, M.F.; Santos, A.B.D.; Brito, F.A.; Barreto, E.O.; De Sousa, D.P.; Almeida, F.R.C.; Badauê-Passos, D., Jr.; Antoniolli, Â.R.; et al. α-Terpineol Reduces Mechanical Hypernociception and Inflammatory Response. Basic Clin. Pharmacol. Toxicol. 2012, 111, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Vieira, G.; Cavalli, J.; Gonçalves, E.C.D.; Braga, S.F.P.; Ferreira, R.S.; Santos, A.R.S.; Cola, M.; Raposo, N.R.B.; Capasso, R.; Dutra, R.C. Antidepressant-Like Effect of Terpineol in an Inflammatory Model of Depression: Involvement of the Cannabinoid System and D2 Dopamine Receptor. Biomolecules 2020, 10, 792. [Google Scholar] [CrossRef] [PubMed]
- Lira, M.H.P.d.; Andrade Júnior, F.P.d.; Moraes, G.F.Q.; Macena, G.d.S.; Pereira, F.d.O.; Lima, I.O. Antimicrobial activity of geraniol: An integrative review. J. Essent. Oil Res. 2020, 32, 187–197. [Google Scholar] [CrossRef]
- Barros Silva Soares de Souza, E.P.; Trindade, G.d.G.G.; Lins Dantas Gomes, M.V.; Santos Silva, L.A.; Grespan, R.; Quintans Junior, L.J.; Cavalcanti de Albuquerque Júnior, R.L.; Shanmugan, S.; Antunes de Souza Araújo, A. Anti-inflammatory effect of nano-encapsulated nerolidol on zymosan-induced arthritis in mice. Food Chem. Toxicol. 2020, 135, 110958. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liu, Q.; Lv, H.; Wang, F.; Liu, R.; Zeng, N. Effect of pulegone on the NLPR3 inflammasome during inflammatory activation of THP-1 cells. Exp. Ther. Med. 2020, 19, 1304–1312. [Google Scholar] [CrossRef]
- Xu, C.; Sheng, S.; Dou, H.; Chen, J.; Zhou, K.; Lin, Y.; Yang, H. α-Bisabolol suppresses the inflammatory response and ECM catabolism in advanced glycation end products-treated chondrocytes and attenuates murine osteoarthritis. Int. Immunopharmacol. 2020, 84, 106530. [Google Scholar] [CrossRef] [PubMed]
- Gogineni, V.; Nael, M.A.; León, F.; Núñez, M.J.; Cutler, S.J. Computationally aided stereochemical assignment of undescribed bisabolenes from Calea urticifolia. Phytochemistry 2019, 157, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Liu, Y.; Jiang, L.; Wang, J. Multi-Targeting by β-Elemene and Its Anticancer Properties: A Good Choice for Oncotherapy and Radiochemotherapy Sensitization. Nutr. Cancer 2020, 72, 554–567. [Google Scholar] [CrossRef]
- Müller, A.D.; Kutscher, E.; Artemyev, A.N.; Demekhin, P.V. Photoelectron circular dichroism in the multiphoton ionization by short laser pulses. III. Photoionization of fenchone in different regimes. J. Chem. Phys. 2020, 152, 044302. [Google Scholar] [CrossRef]
- de Christo Scherer, M.M.; Marques, F.M.; Figueira, M.M.; Peisino, M.C.O.; Schmitt, E.F.P.; Kondratyuk, T.P.; Endringer, D.C.; Scherer, R.; Fronza, M. Wound healing activity of terpinolene and α-phellandrene by attenuating inflammation and oxidative stress in vitro. J. Tissue Viability 2019, 28, 94–99. [Google Scholar] [CrossRef]
- Kotawong, K.; Chaijaroenkul, W.; Roytrakul, S.; Phaonakrop, N.; Na-Bangchang, K. Proteomics Analysis for Identification of Potential Cell Signaling Pathways and Protein Targets of Actions of Atractylodin and β-Eudesmol Against Cholangiocarcinoma. Asian Pac. J. Cancer Prev. 2020, 21, 621–628. [Google Scholar] [CrossRef]
- Lubell, J.D.; Brand, M.H. Foliar sprays of silver thiosulfate produce male flowers on female hemp plants. HortTechnology 2018, 28, 743–747. [Google Scholar] [CrossRef]
- Barnett, P. Transcription Activator Like Effector Nucleases (TALENs): A New, Important, and Versatile Gene Editing Technique with a Growing Literature. Sci. Technol. Libr. 2018, 37, 100–112. [Google Scholar] [CrossRef]
- Novak, S. Plant biotechnology applications of zinc finger technology. In Transgenic Plants. Methods in Molecular Biology; Kumar, S., Barone, P., Smith, M., Eds.; Humana Press: New York, NY, USA, 2019; Volume 1864, pp. 295–310. [Google Scholar]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Barcaccia, G.; Palumbo, F.; Scariolo, F.; Vannozzi, A.; Borin, M.; Bona, S. Potentials and Challenges of Genomics for Breeding Cannabis Cultivars. Front. plant Sci. 2020, 11, 1472. [Google Scholar] [CrossRef] [PubMed]
- Torkamaneh, D.; Boyle, B.; Belzile, F. Efficient genome-wide genotyping strategies and data integration in crop plants. Theor. Appl. Genet. 2018, 131, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Niazian, M. Application of genetics and biotechnology for improving medicinal plants. Planta 2019, 249, 953–973. [Google Scholar] [CrossRef] [PubMed]
- Schachtsiek, J.; Warzecha, H.; Kayser, O.; Stehle, F. Current perspectives on biotechnological cannabinoid production in plants. Planta Med. 2018, 84, 214–220. [Google Scholar] [CrossRef]
- Salentijn, E.M.J.; Petit, J.; Trindade, L.M. The Complex Interactions Between Flowering Behavior and Fiber Quality in Hemp. Front. plant Sci. 2019, 10, 614. [Google Scholar] [CrossRef]
- Hemphill, J.K.; JK, H.; JC, T.; PG, M. Studies on growth and cannabinoid composition of callus derived from different strains of Cannabis sativa. Lloydia 1978, 41, 453–462. [Google Scholar]
- Loh, W.H.T.; Hartsel, S.C.; Robertson, L.W. Tissue Culture of Cannabis sativa L. and in vitro Biotransformation of Phenolics. Z. Pflanzenphysiol. 1983, 111, 395–400. [Google Scholar] [CrossRef]
- Braemer, R.; Paris, M. Biotransformation of cannabinoids by a cell suspension culture of Cannabis sativa L. plant Cell Rep. 1987, 6, 150–152. [Google Scholar] [CrossRef]
- Sirikantaramas, S.; Taura, F.; Tanaka, Y.; Ishikawa, Y.; Morimoto, S.; Shoyama, Y. Tetrahydrocannabinolic Acid Synthase, the Enzyme Controlling Marijuana Psychoactivity, is Secreted into the Storage Cavity of the Glandular Trichomes. plant Cell Physiol. 2005, 46, 1578–1582. [Google Scholar] [CrossRef]
- Staginnus, C.; Zörntlein, S.; de Meijer, E. A PCR marker Linked to a THCA synthase Polymorphism is a Reliable Tool to Discriminate Potentially THC-Rich Plants of Cannabis sativa L. J. Forensic Sci. 2014, 59, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Flores-Sanchez, I.J.; Peč, J.; Fei, J.; Choi, Y.H.; Dušek, J.; Verpoorte, R. Elicitation studies in cell suspension cultures of Cannabis sativa L. J. Biotechnol. 2009, 143, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Gabotti, D.; Locatelli, F.; Cusano, E.; Baldoni, E.; Genga, A.; Pucci, L.; Consonni, R.; Mattana, M. Cell Suspensions of Cannabis sativa (var. Futura): Effect of Elicitation on Metabolite Content and Antioxidant Activity. Molecules 2019, 24, 4056. [Google Scholar] [CrossRef]
- Baek, S.; Ho, T.-T.; Lee, H.; Jung, G.; Kim, Y.E.; Jeong, C.-S.; Park, S.-Y. Enhanced biosynthesis of triterpenoids in Centella asiatica hairy root culture by precursor feeding and elicitation. plant Biotechnol. Rep. 2020, 14, 45–53. [Google Scholar] [CrossRef]
- Sirikantaramas, S.; Morimoto, S.; Shoyama, Y.; Ishikawa, Y.; Wada, Y.; Shoyama, Y.; Taura, F. The gene controlling marijuana psychoactivity molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J. Biol. Chem. 2004, 279, 39767–39774. [Google Scholar] [CrossRef]
- Farag, S.; Kayser, O. Cannabinoids production by hairy root cultures of Cannabis sativa L. Am. J. plant Sci. 2015, 6, 1874. [Google Scholar] [CrossRef]
- Moher, M.; Jones, M.; Zheng, Y. Photoperiodic Response of in vitro Cannabis sativa Plants. HortScience 2021, 56, 108–113. [Google Scholar] [CrossRef]
- Monthony, A.S.; Page, S.R.G.; Hesami, M.; Jones, A.M.P. The Past, Present and Future of Cannabis sativa Tissue Culture. Plants 2021, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, D.; Kulkarni, M.; El-Mezawy, A.; Mobini, S.; Elhiti, M.; Gjuric, R.; Ray, A.; Polowick, P.; Slaski, J.J.; Jones, M.P.; et al. Medical Cannabis and Industrial Hemp Tissue Culture: Present Status and Future Potential. Front. Plant Sci. 2021, 12, 627240. [Google Scholar] [CrossRef]
- Veliky, I.; Genest, K. Growth and metabolites of Cannabis sativa cell suspension cultures. Lloydia 1972, 35, 450–456. [Google Scholar] [PubMed]
- Itokawa, H.; Takeya, K.; Akasu, M. Studies on the constituents isolated from the callus of Cannabis sativa. Shoyakugaku Zasshi 1975, 29, 106–112. [Google Scholar]
- Hartsel, S.; Loh, W.-T.; Robertson, L. Biotransformation of cannabidiol to cannabielsoin by suspension cultures of Cannabis sativa and Saccharum officinarum. Planta Med. 1983, 48, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Richez-Dumanois, C.; Braut-Boucher, F.; Cosson, L.; Paris, M. Multiplication végétative in vitro du chanvre (Cannabis sativa L.). Application à la conserva-tion des clones sélectionnés. Agronomie 1986, 6, 487–495. [Google Scholar] [CrossRef]
- Mandolino, G.; Ranalli, P. Advances in biotechnological approaches for hemp breeding and industry. In Advances in Hemp Research; Ranalli, P., Ed.; Haworth Press: Binghamton, NY, USA, 1999; pp. 185–212. [Google Scholar]
- Slusarkiewicz-Jarzina, A.; Ponitka, A.; Kaczmarek, Z. Influence of cultivar, explant source and plant growth regulator on callus induction and plant regeneration of Cannabis sativa L. Acta Biol. Crac. Ser. Bot. 2005, 47, 145–151. [Google Scholar]
- Plawuszewski, M.; Lassocinski, W.; Wielgus, K. Regeneration of Polish cultivars of monoecious hemp (Cannabis sativa L.) grown in vitro. In Renewable Resources and Plant Biotechnology; Kozłowski, R., Zaikov, G., Pudel, F., Eds.; Nova Publishers: Poznan, Poland, 2006; pp. 149–154. [Google Scholar]
- Bing, X.; Ning, L.; Jinfeng, T.; Nan, G. Rapid Tissue Culture Method of Cannabis sativa for Industrial Uses. CN Patent 1887043, 2007. [Google Scholar]
- Wielgus, K.; Luwanska, A.; Lassocinski, W.; Kaczmarek, Z. Estimation of Cannabis sativa L. tissue culture conditions essential for callus induction and plant regeneration. J. Nat. Fibers 2008, 5, 199–207. [Google Scholar] [CrossRef]
- Wang, R.; He, L.; Xia, B.; Tong, J.; Li, N.; Peng, F. A micropropagation system for cloning of hemp (Cannabis sativa L.) by shoot tip culture. Pak. J. Bot. 2009, 41, 603–608. [Google Scholar]
- Lata, H.; Chandra, S.; Khan, I.; ElSohly, M.A. Thidiazuron-induced high-frequency direct shoot organogenesis of Cannabis sativa L. Vitr. Cell. Dev. Biol. Plant 2009, 45, 12–19. [Google Scholar] [CrossRef]
- Lata, H.; Chandra, S.; Khan, I.A.; ElSohly, M.A. Propagation through alginate encapsulation of axillary buds of Cannabis sativa L.—an important medicinal plant. Physiol. Mol. Biol. Plants 2009, 15, 79–86. [Google Scholar] [CrossRef]
- Lata, H.; Chandra, S.; Khan, I.A.; ElSohly, M.A. High Frequency Plant Regeneration from Leaf Derived Callus of High Δ9-Tetrahydrocannabinol Yielding Cannabis sativa L. Planta Med. 2010, 76, 1629–1633. [Google Scholar] [CrossRef]
- Movahedi, M.; Ghasemi-Omran, V.; Torabi, S. The effect of different concentrations of TDZ and BA on in vitro regeneration of Iranian cannabis (Cannabis sativa) using cotyledon and epicotyl explants. J. plant Mol. Breed. 2015, 3, 20–27. [Google Scholar] [CrossRef]
- Movahedi, M.; Ghasemiomran, V.; Torabi, S. In vitro callus induction and regeneration of medicinal plant Cannabis sativa L. Iran. J. Med. Aromat. Plants 2016, 32, 758–768. [Google Scholar] [CrossRef]
- Movahedi, M.; Ghasemiomran, V.; Torabi, S. Effect of explants type and plant growth regulators on in vitro callus induction and shoot regeneration of Cannabis sativa L. Iran. J. Med. Aromat. Plants 2016, 32, 83–96. [Google Scholar] [CrossRef]
- Lata, H.; Chandra, S.; Techen, N.; Khan, I.A.; ElSohly, M.A. In vitro mass propagation of Cannabis sativa L.: A protocol refinement using novel aromatic cytokinin meta-topolin and the assessment of eco-physiological, biochemical and genetic fidelity of micropropagated plants. J. Appl. Res. Med. Aromat. Plants 2016, 3, 18–26. [Google Scholar] [CrossRef]
- Chaohua, C.; Gonggu, Z.; Lining, Z.; Chunsheng, G.; Qing, T.; Jianhua, C.; Xinbo, G.; Dingxiang, P.; Jianguang, S. A rapid shoot regeneration protocol from the cotyledons of hemp (Cannabis sativa L.). Ind. Crop. Prod. 2016, 83, 61–65. [Google Scholar] [CrossRef]
- Piunno, K.F.; Golenia, G.; Boudko, E.A.; Downey, C.; Jones, A.M.P. Regeneration of shoots from immature and mature inflorescences of Cannabis sativa. Can. J. plant Sci. 2019, 99, 556–559. [Google Scholar] [CrossRef]
- Smýkalová, I.; Vrbová, M.; Cvečková, M.; Plačková, L.; Žukauskaitė, A.; Zatloukal, M.; Hrdlička, J.; Plíhalová, L.; Doležal, K.; Griga, M. The effects of novel synthetic cytokinin derivatives and endogenous cytokinins on the in vitro growth responses of hemp (Cannabis sativa L.) explants. plant Cell Tissue Organ. Cult. 2019, 139, 381–394. [Google Scholar] [CrossRef]
- Monthony, A.S.; Kyne, S.T.; Grainger, C.M.; Jones, A.M.P. Recalcitrance of Cannabis sativa to de novo regeneration; a multi-genotype replication study. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wróbel, T.; Dreger, M.; Wielgus, K.; Słomski, R. Modified Nodal Cuttings and Shoot Tips Protocol for Rapid Regeneration of Cannabis sativa L. J. Nat. Fibers 2020, 8, 1–10. [Google Scholar] [CrossRef]
- Galán-Ávila, A.; García-Fortea, E.; Prohens, J.; Herraiz, F.J. Development of a Direct in vitro Plant Regeneration Protocol From Cannabis sativa L. Seedling Explants: Developmental Morphology of Shoot Regeneration and Ploidy Level of Regenerated Plants. Front. plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Monthony, A.S.; Bagheri, S.; Zheng, Y.; Jones, A.M.P. Flower Power: Floral reversion as a viable alternative to nodal micropropagation in Cannabis sativa. bioRxiv 2020. [Google Scholar] [CrossRef]
- Codesido, V.; Meyer, S.; Casano, S. Influence of media composition and genotype for successful Cannabis sativa L. in vitro introduction. Acta Hortic. 2020, 1285, 75–80. [Google Scholar] [CrossRef]
- Mubi, Š.M.; Svetik, S.; FlajŠMan, M.; Murovec, J.; Mestinšek Mubi, Š.; Svetik, S.; Flajšman, M.; Murovec, J. In vitro tissue culture and genetic analysis of two high-CBD medical cannabis (Cannabis sativa L.) breeding lines. GENETIKA 2020, 52, 925–941. [Google Scholar] [CrossRef]
- Ioannidis, K.; Dadiotis, E.; Mitsis, V.; Melliou, E.; Magiatis, P. Biotechnological Approaches on Two High CBD and CBG Cannabis sativa L. (Cannabaceae) Varieties: In vitro Regeneration and Phytochemical Consistency Evaluation of Micropropagated Plants Using Quantitative 1H-NMR. Molecules 2020, 25, 5928. [Google Scholar] [CrossRef]
- Jessica, D.L.-B.; Lauren, E.K.; Mark, H.B. An In vitro–ex vitro Micropropagation System for Hemp. Horttechnol. Hortte 2021, 31, 199–207. [Google Scholar] [CrossRef]
- Murphy, R.; Adelberg, J. Physical factors increased quantity and quality of micropropagated shoots of Cannabis sativa L. in a repeated harvest system with ex vitro rooting. Vitr. Cell. Dev. Biol. Plant 2021, 57, 1–9. [Google Scholar] [CrossRef]
- Zarei, A.; Behdarvandi, B.; Tavakouli Dinani, E.; Maccarone, J. Cannabis sativa L. photoautotrophic micropropagation: A powerful tool for industrial scale in vitro propagation. Vitr. Cell. Dev. Biol. Plant 2021, 57, 1–10. [Google Scholar] [CrossRef]
- Page, S.R.G.; Monthony, A.S.; Jones, A.M.P. DKW basal salts improve micropropagation and callogenesis compared to MS basal salts in multiple commercial cultivars of Cannabis sativa. Botany 2021, 99, 269–279. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, G.; Cheng, C.; Lei, L.; Sun, J.; Xu, Y.; Deng, C.; Dai, Z.; Yang, Z.; Chen, X.; et al. Establishment of an Agrobacterium-mediated genetic transformation and CRISPR/Cas9-mediated targeted mutagenesis in Hemp (Cannabis Sativa L.). plant Biotechnol. J. 2021, 19, 1–25. [Google Scholar] [CrossRef]
- Boonsnongcheep, P.; Pongkitwitoon, B. Factors affecting micropropagation of Cannabis sativa L.: A review. Pharm. Sci. Asia 2020, 47, 21–29. [Google Scholar] [CrossRef]
- Hesami, M.; Jones, A.M.P. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl. Microbiol. Biotechnol. 2020, 104, 9449–9485. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M. Introducing a hybrid artificial intelligence method for high-throughput modeling and optimizing plant tissue culture processes: The establishment of a new embryogenesis medium for chrysanthemum, as a case study. Appl. Microbiol. Biotechnol. 2020, 104, 10249–10263. [Google Scholar] [CrossRef]
- Mira, M.M.; Wally, O.S.; Elhiti, M.; El-Shanshory, A.; Reddy, D.S.; Hill, R.D.; Stasolla, C. Jasmonic acid is a downstream component in the modulation of somatic embryogenesis by Arabidopsis Class 2 phytoglobin. J. Exp. Bot. 2016, 67, 2231–2246. [Google Scholar] [CrossRef] [PubMed]
- Sarropoulou, V.; Maloupa, E. Effect of the NO donor “sodium nitroprusside”(SNP), the ethylene inhibitor “cobalt chloride”(CoCl 2) and the antioxidant vitamin E “α-tocopherol” on in vitro shoot proliferation of Sideritis raeseri Boiss. & Heldr. subsp. raeseri. plant Cell Tissue Organ. Cult. 2017, 128, 619–629. [Google Scholar] [CrossRef]
- Ötvös, K.; Pasternak, T.P.; Miskolczi, P.; Domoki, M.; Dorjgotov, D.; Sz˝ cs, A.; Bottka, S.; Dudits, D.; Fehér, A. Nitric oxide is required for, and promotes auxin-mediated activation of, cell division and embryogenic cell formation but does not influence cell cycle progression in alfalfa cell cultures. Plant J. 2005, 43, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.; Ilyas, S.; Naz, S. Nanotechnology and Plant Tissue Culture. In Nanoagronomy; Javad, S., Ed.; Springer: Cham, Switzerland, 2020; pp. 23–35. [Google Scholar]
- Ahmad, M.A.; Javed, R.; Adeel, M.; Rizwan, M.; Ao, Q.; Yang, Y. Engineered ZnO and CuO Nanoparticles Ameliorate Morphological and Biochemical Response in Tissue Culture Regenerants of Candyleaf (Stevia rebaudiana). Molecules 2020, 25, 1356. [Google Scholar] [CrossRef]
- Zaeem, A.; Drouet, S.; Anjum, S.; Khurshid, R.; Younas, M.; Blondeau, J.P.; Tungmunnithum, D.; Giglioli-Guivarc’h, N.; Hano, C.; Abbasi, B.H. Effects of Biogenic Zinc Oxide Nanoparticles on Growth and Oxidative Stress Response in Flax Seedlings vs. in vitro Cultures: A Comparative Analysis. Biomolecules 2020, 10, 918. [Google Scholar] [CrossRef]
- Nalci, O.B.; Nadaroglu, H.; Pour, A.H.; Gungor, A.A.; Haliloglu, K. Effects of ZnO, CuO and γ-Fe3O4 nanoparticles on mature embryo culture of wheat (Triticum aestivum L.). plant Cell Tissue Organ. Cult. 2019, 136, 269–277. [Google Scholar] [CrossRef]
- Hesami, M.; Daneshvar, M.H.; Yoosefzadeh-Najafabadi, M. An efficient in vitro shoot regeneration through direct organogenesis from seedling-derived petiole and leaf segments and acclimatization of Ficus religiosa. J. For. Res. 2019, 30, 807–815. [Google Scholar] [CrossRef]
- Hesami, M.; Tohidfar, M.; Alizadeh, M.; Daneshvar, M.H. Effects of sodium nitroprusside on callus browning of Ficus religiosa: An important medicinal plant. J. For. Res. 2020, 31, 789–796. [Google Scholar] [CrossRef]
- Yaseen, M.; Ahmad, T.; Sablok, G.; Standardi, A.; Hafiz, I.A. Review: Role of carbon sources for in vitro plant growth and development. Mol. Biol. Rep. 2013, 40, 2837–2849. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Díaz, J.I.; Arzate-Fernández, A.M.; Piña-Escutia, J.L.; Vázquez-García, L.M. Media culture factors affecting somatic embryogenesis in Agave angustifolia Haw. Ind. Crop. Prod. 2017, 108, 81–85. [Google Scholar] [CrossRef]
- Asthana, P.; Rai, M.K.; Jaiswal, U. Somatic embryogenesis from sepal explants in Sapindus trifoliatus, a plant valuable in herbal soap industry. Ind. Crop. Prod. 2017, 100, 228–235. [Google Scholar] [CrossRef]
- Peng, C.; Gao, F.; Wang, H.; Shen, H.; Yang, L. Optimization of maturation process for somatic embryo production and cryopreservation of embryogenic tissue in Pinus koraiensis. plant Cell Tissue Organ. Cult. 2020. [Google Scholar] [CrossRef]
- Yancheva, S.D.; Roichev, V. Carbohydrate Source can Influence the Efficiency of Somatic Embryogenesis in Seedless Grapes (Vitis Vinifera L.). Biotechnol. Biotechnol. Equip. 2005, 19, 62–66. [Google Scholar] [CrossRef]
- Yadollahi, A.; Abdollahi, M.R.; Moieni, A.; Danaee, M. Effects of carbon source, polyethylene glycol and abscisic acid on secondary embryo induction and maturation in rapeseed (Brassica napus L.) microspore-derived embryos. Acta Physiol. plant 2011, 33, 1905–1912. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M.; Yoosefzadeh-Najafabadi, M. Application of Adaptive Neuro-Fuzzy Inference System-Non-dominated Sorting Genetic Algorithm-II (ANFIS-NSGAII) for Modeling and Optimizing Somatic Embryogenesis of Chrysanthemum. Front. plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.R.; Singh, A.S.; Piunno, K.; Saxena, P.K.; Jones, A.M.P. Application of 3D printing to prototype and develop novel plant tissue culture systems. plant Methods 2017, 13, 6. [Google Scholar] [CrossRef]
- Chen, J.-R.; Wu, L.; Hu, B.-W.; Yi, X.; Liu, R.; Deng, Z.-N.; Xiong, X.-Y. The Influence of Plant Growth Regulators and Light Quality on Somatic Embryogenesis in China Rose (Rosa chinensis Jacq.). J. plant Growth Regul. 2014, 33, 295–304. [Google Scholar] [CrossRef]
- Li, H.; Xu, Z.; Tang, C. Effect of light-emitting diodes on growth and morphogenesis of upland cotton (Gossypium hirsutum L.) plantlets in vitro. plant Cell Tissue Organ. Cult. 2010, 103, 155–163. [Google Scholar] [CrossRef]
- Miler, N.; Kulus, D.; Woźny, A.; Rymarz, D.; Hajzer, M.; Wierzbowski, K.; Nelke, R.; Szeffs, L. Application of wide-spectrum light-emitting diodes in micropropagation of popular ornamental plant species: A study on plant quality and cost reduction. Vitr. Cell. Dev. Biol. Plant 2019, 55, 99–108. [Google Scholar] [CrossRef]
- D’Onofrio, C.; Morini, S.; Bellocchi, G. Effect of light quality on somatic embryogenesis of quince leaves. plant Cell Tissue Organ. Cult. 1998, 53, 91–98. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Hesami, M.; Jones, A.M.P. Modeling and optimizing callus growth and development in Cannabis sativa using random forest and support vector machine in combination with a genetic algorithm. Appl. Microbiol. Biotechnol. 2021, 105, in press. [Google Scholar]
- Driver, J.A.; Kuniyuki, A.H. In vitro propagation of Paradox walnut rootstock. HortScience 1984, 19, 507–509. [Google Scholar]
- Hesami, M.; Naderi, R.; Tohidfar, M. Modeling and Optimizing in vitro Sterilization of Chrysanthemum via Multilayer Perceptron-Non-dominated Sorting Genetic Algorithm-II (MLP-NSGAII). Front. plant Sci. 2019, 10, 282. [Google Scholar] [CrossRef]
- Akin, M.; Eyduran, S.P.; Eyduran, E.; Reed, B.M. Analysis of macro nutrient related growth responses using multivariate adaptive regression splines. plant Cell Tissue Organ. Cult. 2020, 140, 661–670. [Google Scholar] [CrossRef]
- García-Pérez, P.; Lozano-Milo, E.; Landín, M.; Gallego, P.P. Machine Learning Technology Reveals the Concealed Interactions of Phytohormones on Medicinal Plant In Vitro Organogenesis. Biomolecules 2020, 10, 746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Deng, D.; Dai, W.; Li, J.; Jin, X. Optimization of culture conditions for differentiation of melon based on artificial neural network and genetic algorithm. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Naderi, R.; Tohidfar, M. Modeling and Optimizing Medium Composition for Shoot Regeneration of Chrysanthemum via Radial Basis Function-Non-dominated Sorting Genetic Algorithm-II (RBF-NSGAII). Sci. Rep. 2019, 9, 18237. [Google Scholar] [CrossRef]
- Hesami, M.; Condori-Apfata, J.A.; Valderrama Valencia, M.; Mohammadi, M. Application of Artificial Neural Network for Modeling and Studying in vitro Genotype-Independent Shoot Regeneration in Wheat. Appl. Sci. 2020, 10, 5370. [Google Scholar] [CrossRef]
- Munasinghe, S.P.; Somaratne, S.; Weerakoon, S.R.; Ranasinghe, C. Prediction of chemical composition for callus production in Gyrinops walla Gaetner through machine learning. Inf. Process. Agric. 2020, 7, 1–12. [Google Scholar] [CrossRef]
- Niazian, M.; Shariatpanahi, M.E.; Abdipour, M.; Oroojloo, M. Modeling callus induction and regeneration in an anther culture of tomato (Lycopersicon esculentum L.) using image processing and artificial neural network method. Protoplasma 2019, 256, 1317–1332. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M.; Yoosefzadeh-Najafabadi, M. Development of support vector machine-based model and comparative analysis with artificial neural network for modeling the plant tissue culture procedures: Effect of plant growth regulators on somatic embryogenesis of chrysanthemum, as a case study. plant Methods 2020, 16, 112. [Google Scholar] [CrossRef] [PubMed]
- Salehi, M.; Farhadi, S.; Moieni, A.; Safaie, N.; Hesami, M. A hybrid model based on general regression neural network and fruit fly optimization algorithm for forecasting and optimizing paclitaxel biosynthesis in Corylus avellana cell culture. plant Methods 2021, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, S.; Salehi, M.; Moieni, A.; Safaie, N.; Sabet, M.S. Modeling of paclitaxel biosynthesis elicitation in Corylus avellana cell culture using adaptive neuro-fuzzy inference system-genetic algorithm (ANFIS-GA) and multiple regression methods. PLoS ONE 2020, 15, e0237478. [Google Scholar] [CrossRef]
- Salehi, M.; Farhadi, S.; Moieni, A.; Safaie, N.; Ahmadi, H. Mathematical Modeling of Growth and Paclitaxel Biosynthesis in Corylus avellana Cell Culture Responding to Fungal Elicitors Using Multilayer Perceptron-Genetic Algorithm. Front. plant Sci 2020, 11, 1148. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Alizadeh, M.; Naderi, R.; Tohidfar, M. Forecasting and optimizing Agrobacterium-mediated genetic transformation via ensemble model- fruit fly optimization algorithm: A data mining approach using chrysanthemum databases. PLoS ONE 2020, 15, e0239901. [Google Scholar] [CrossRef] [PubMed]
- Bidabadi, S.S.; Jain, S.M. Cellular, Molecular, and Physiological Aspects of in vitro Plant Regeneration. Plants 2020, 9, 702. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Rai, K.K.; Rai, S.K.; Rai, S.P. An improved thin cell layer culture system for efficient clonal propagation and in vitro withanolide production in a medicinal plant Withania coagulans Dunal. Ind. Crop. Prod. 2018, 119, 172–182. [Google Scholar] [CrossRef]
- Tu, H.-Y.; Zhang, A.-L.; Xiao, W.; Lin, Y.-R.; Shi, J.-H.; Wu, Y.-W.; Wu, S.-T.; Zhong, C.-H.; Mo, S.-X. Induction and identification of tetraploid Hedychium coronarium through thin cell layer culture. plant CellTissue Organ. Cult. 2018, 135, 395–406. [Google Scholar] [CrossRef]
- Monja-Mio, K.M.; Robert, M.L. Direct somatic embryogenesis of Agave fourcroydes Lem. through thin cell layer culture. Vitr. Cell. Dev. Biol. Plant 2013, 49, 541–549. [Google Scholar] [CrossRef]
- Vidal, N.; Sánchez, C. Use of bioreactor systems in the propagation of forest trees. Eng. Life Sci. 2019, 19, 896–915. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Su, W.-P.; Zheng, H.; Yu, B.-C.; Ma, Z.-F.; Zhang, P.; Guo, W.-W. Plant regeneration via protoplast electrofusion in cassava. J. Integr. Agric. 2020, 19, 632–642. [Google Scholar] [CrossRef]
- Jones, A.M.P.; Shukla, M.R.; Biswas, G.C.G.; Saxena, P.K. Protoplast-to-plant regeneration of American elm (Ulmus americana). Protoplasma 2015, 252, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Beard, K.M.; Boling, A.W.H.; Bargmann, B.O.R. Protoplast isolation, transient transformation, and flow-cytometric analysis of reporter-gene activation in Cannabis sativa L. Ind. Crop. Prod. 2021, 164, 113360. [Google Scholar] [CrossRef]
- Lazič, S. Izolacija Protoplastov Navadne Konoplje (Cannabis sativa L.). Ph.D. Thesis, University of Ljubljana, Ljubljana, Slovenia, 2020. [Google Scholar]
- Morimoto, S.; Tanaka, Y.; Sasaki, K.; Tanaka, H.; Fukamizu, T.; Shoyama, Y.; Shoyama, Y.; Taura, F. Identification and characterization of cannabinoids that induce cell death through mitochondrial permeability transition in Cannabis leaf cells. J. Biol. Chem. 2007, 282, 20739–20751. [Google Scholar] [CrossRef]
- Niazian, M.; Shariatpanahi, M.E. In vitro-based doubled haploid production: Recent improvements. Euphytica 2020, 216, 69. [Google Scholar] [CrossRef]
- Wang, B.; Zhu, L.; Zhao, B.; Zhao, Y.; Xie, Y.; Zheng, Z.; Li, Y.; Sun, J.; Wang, H. Development of a Haploid-Inducer Mediated Genome Editing System for Accelerating Maize Breeding. Mol. plant 2019, 12, 597–602. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef]
- Hesami, M.; Yoosefzadeh Najafabadi, M.; Adamek, K.; Torkamaneh, D.; Jones, A.M. Synergizing off-target predictions for in silico insights of CENH3 knockout in cannabis through CRISPR/CAS. Molecules 2021, 26, 2053. [Google Scholar] [CrossRef]
- Li, B.; Cui, G.; Shen, G.; Zhan, Z.; Huang, L.; Chen, J.; Qi, X. Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza. Sci. Rep. 2017, 7, 43320. [Google Scholar] [CrossRef]
- Zhou, Z.; Tan, H.; Li, Q.; Chen, J.; Gao, S.; Wang, Y.; Chen, W.; Zhang, L. CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry 2018, 148, 63–70. [Google Scholar] [CrossRef]
- Feng, S.; Song, W.; Fu, R.; Zhang, H.; Xu, A.; Li, J. Application of the CRISPR/Cas9 system in Dioscorea zingiberensis. plant Cell Tissue Organ Cult. 2018, 135, 133–141. [Google Scholar] [CrossRef]
- Azizi, P.; Hanafi, M.M.; Sahebi, M.; Harikrishna, J.A.; Taheri, S.; Yassoralipour, A.; Nasehi, A. Epigenetic changes and their relationship to somaclonal variation: A need to monitor the micropropagation of plantation crops. Funct. plant Biol. 2020, 47, 508–523. [Google Scholar] [CrossRef]
- Rodriguez-Enriquez, J.; Dickinson, H.G.; Grant-Downton, R.T. MicroRNA misregulation: An overlooked factor generating somaclonal variation. Trends plant Sci. 2011, 16, 242–248. [Google Scholar] [CrossRef]
- Lata, H.; Chandra, S.; Techen, N.; Khan, I.A.; ElSohly, M.A. Assessment of the Genetic Stability of Micropropagated Plants of Cannabis sativa by ISSR Markers. Planta Med. 2010, 76, 97–100. [Google Scholar] [CrossRef]
- Chandra, S.; Lata, H.; Mehmedic, Z.; Khan, I.A.; ElSohly, M.A. Assessment of Cannabinoids Content in Micropropagated Plants of Cannabis sativa and Their Comparison with Conventionally Propagated Plants and Mother Plant during Developmental Stages of Growth. Planta Med. 2010, 76, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Lata, H.; Chandra, S.; Techen, N.; Khan, I.A.; ElSohly, M.A. Molecular analysis of genetic fidelity in Cannabis sativa L. plants grown from synthetic (encapsulated) seeds following in vitro storage. Biotechnol. Lett. 2011, 33, 2503–2508. [Google Scholar] [CrossRef] [PubMed]
- Adamek, K.; Jones, A.M.P.; Torkamaneh, D. Accumulation of somatic mutations leads to genetic mosaicism in Cannabis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Niazian, M.; Nalousi, A.M. Artificial polyploidy induction for improvement of ornamental and medicinal plants. plant Cell Tissue Organ. Cult. 2020, 142, 11–23. [Google Scholar] [CrossRef]
- Iannicelli, J.; Guariniello, J.; Tossi, V.E.; Regalado, J.J.; Di Ciaccio, L.; van Baren, C.M.; Pitta Álvarez, S.I.; Escandón, A.S. The “polyploid effect” in the breeding of aromatic and medicinal species. Sci. Hortic. 2020, 260, 108854. [Google Scholar] [CrossRef]
- Bagheri, M.; Mansouri, H. Effect of Induced Polyploidy on Some Biochemical Parameters in Cannabis sativa L. Appl. Biochem. Biotechnol. 2015, 175, 2366–2375. [Google Scholar] [CrossRef]
- Kurtz, L.E.; Brand, M.H.; Lubell-Brand, J.D. Production of Tetraploid and Triploid Hemp. HortScience 2020, 55, 1703–1707. [Google Scholar] [CrossRef]
- Parsons, J.L.; Martin, S.L.; James, T.; Golenia, G.; Boudko, E.A.; Hepworth, S.R. Polyploidization for the genetic improvement of Cannabis sativa. Front. plant Sci. 2019, 10, 476. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, H.; Bagheri, M. Induction of Polyploidy and Its Effect on Cannabis sativa L. In Cannabis sativa L.-Botany and Biotechnology; Chandra, S., Lata, H., ElSohly, M., Eds.; Springer: Cham, Switzerland, 2017; pp. 365–383. [Google Scholar]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Corneillie, S.; De Storme, N.; Van Acker, R.; Fangel, J.U.; De Bruyne, M.; De Rycke, R.; Geelen, D.; Willats, W.G.; Vanholme, B.; Boerjan, W. Polyploidy affects plant growth and alters cell wall composition. plant Physiol. 2019, 179, 74–87. [Google Scholar] [CrossRef] [PubMed]
- McLaren, J.; Swift, W.; Dillon, P.; Allsop, S. Cannabis potency and contamination: A review of the literature. Addiction 2008, 103, 1100–1109. [Google Scholar] [CrossRef]
- Lavania, U.C. Enhanced productivity of the essential oil in the artificial autopolyploid of vetiver (Vetiveria zizanioides L. Nash). Euphytica 1988, 38, 271–276. [Google Scholar] [CrossRef]
- Tabatabaei, I.; Dal Bosco, C.; Bednarska, M.; Ruf, S.; Meurer, J.; Bock, R. A highly efficient sulfadiazine selection system for the generation of transgenic plants and algae. plant Biotechnol. J. 2019, 17, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Bielecka, M.; Kaminski, F.; Adams, I.; Poulson, H.; Sloan, R.; Li, Y.; Larson, T.R.; Winzer, T.; Graham, I.A. Targeted mutation of Δ12 and Δ15 desaturase genes in hemp produce major alterations in seed fatty acid composition including a high oleic hemp oil. plant Biotechnol. J. 2014, 12, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Schachtsiek, J.; Hussain, T.; Azzouhri, K.; Kayser, O.; Stehle, F. Virus-induced gene silencing (VIGS) in Cannabis sativa L. plant Methods 2019, 15, 157. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef]
- MacKinnon, L.; McDougall, G.; Aziz, N.; Millam, S. Progress towards transformation of fibre hemp. Annu. Rep. Scott. Crop. Res. Inst. 2000, 18, 84–86. [Google Scholar]
- Feeney, M.; Punja, Z.K. Tissue culture and Agrobacterium-mediated transformation of hemp (Cannabis sativa L.). Vitr. Cell. Dev. Biol. Plant 2003, 39, 578–585. [Google Scholar] [CrossRef]
- Wahby, I.; Caba, J.M.; Ligero, F. Agrobacterium infection of hemp (Cannabis sativa L.): Establishment of hairy root cultures. J. plant Interact. 2013, 8, 312–320. [Google Scholar] [CrossRef]
- Feeney, M.; Punja, Z.K. Hemp (Cannabis sativa L.). In Agrobacterium Protocols. Methods in Molecular Biology; Wang, K., Ed.; Springer: New York, NY, USA, 2015; Volume 1224, pp. 319–329. [Google Scholar]
- Deguchi, M.; Bogush, D.; Weeden, H.; Spuhler, Z.; Potlakayala, S.; Kondo, T.; Zhang, Z.J.; Rudrabhatla, S. Establishment and optimization of a hemp (Cannabis sativa L.) agroinfiltration system for gene expression and silencing studies. Sci. Rep. 2020, 10, 3504. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.; Yadav, N.S.; Gaudet, D.; Kovalchuk, I. Transient expression of the β-glucuronidase gene in Cannabis sativa varieties. plant Signal. Behav. 2020, 15, 1780037. [Google Scholar] [CrossRef]
- Anand, A.; Wu, E.; Li, Z.; TeRonde, S.; Arling, M.; Lenderts, B.; Mutti, J.S.; Gordon-Kamm, W.; Jones, T.J.; Chilcoat, N.D. High efficiency Agrobacterium-mediated site-specific gene integration in maize utilizing the FLP-FRT recombination system. plant Biotechnol. J. 2019, 17, 1636–1645. [Google Scholar] [CrossRef]
- Karthik, S.; Pavan, G.; Manickavasagam, M. Nitric oxide donor regulates Agrobacterium-mediated genetic transformation efficiency in soybean [Glycine max (L.) Merrill]. plant Cell Tissue Organ. Cult. 2020, 141, 655–660. [Google Scholar] [CrossRef]
- Das Bhowmik, S.S.; Cheng, A.Y.; Long, H.; Tan, G.Z.H.; Hoang, T.M.L.; Karbaschi, M.R.; Williams, B.; Higgins, T.J.V.; Mundree, S.G. Robust Genetic Transformation System to Obtain Non-chimeric Transgenic Chickpea. Front. plant Sci. 2019, 10, 524. [Google Scholar] [CrossRef]
- Jha, P.; Ochatt, S.J.; Kumar, V. WUSCHEL: A master regulator in plant growth signaling. plant Cell Rep. 2020, 39, 431–444. [Google Scholar] [CrossRef]
- Jha, P.; Kumar, V. BABY BOOM (BBM): A candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 2018, 40, 1467–1475. [Google Scholar] [CrossRef]
- Luo, G.; Palmgren, M. GRF-GIF Chimeras Boost Plant Regeneration. Trends plant Sci. 2020. [Google Scholar] [CrossRef]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, T. Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Paul, P.; Joshi, S.; Perry, S.E. Genetic activity during early plant embryogenesis. Biochem. J. 2020, 477, 3743–3767. [Google Scholar] [CrossRef] [PubMed]
- Hoerster, G.; Wang, N.; Ryan, L.; Wu, E.; Anand, A.; McBride, K.; Lowe, K.; Jones, T.; Gordon-Kamm, B. Use of non-integrating Zm-Wus2 vectors to enhance maize transformation. Vitr. Cell. Dev. Biol. Plant 2020, 56, 265–279. [Google Scholar] [CrossRef]
- Szczygieł-Sommer, A.; Gaj, M.D. The miR396–GRF regulatory module controls the embryogenic response in Arabidopsis via an auxin-related pathway. Int. J. Mol. Sci. 2019, 20, 5221. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Vielle-Calzada, J.-P.; Hartog, M.V.; Schmidt, E.D.; Boutilier, K.; Grossniklaus, U.; de Vries, S.C. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. plant Physiol. 2001, 127, 803–816. [Google Scholar] [CrossRef]
- Pérez-Pascual, D.; Jiménez-Guillen, D.; Villanueva-Alonzo, H.; Souza-Perera, R.; Godoy-Hernández, G.; Zúñiga-Aguilar, J.J. Ectopic expression of the Coffea canephora SERK1 homolog-induced differential transcription of genes involved in auxin metabolism and in the developmental control of embryogenesis. Physiol. plant 2018, 163, 530–551. [Google Scholar] [CrossRef] [PubMed]
- Harding, E.W.; Tang, W.; Nichols, K.W.; Fernandez, D.E.; Perry, S.E. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. plant Physiol. 2003, 133, 653–663. [Google Scholar] [CrossRef]
- Yang, Z.; Li, C.; Wang, Y.; Zhang, C.; Wu, Z.; Zhang, X.; Liu, C.; Li, F. GhAGL15s, preferentially expressed during somatic embryogenesis, promote embryogenic callus formation in cotton (Gossypium hirsutum L.). Mol. Genet. Genom. 2014, 289, 873–883. [Google Scholar] [CrossRef]
- Thakare, D.; Tang, W.; Hill, K.; Perry, S.E. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. plant Physiol. 2008, 146, 1663–1672. [Google Scholar] [CrossRef]
- Karlova, R.; Boeren, S.; Russinova, E.; Aker, J.; Vervoort, J.; de Vries, S. The Arabidopsis somatic embryogenesis receptor-like kinase1 protein complex includes brassinosteroid-insensitive1. plant Cell 2006, 18, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, U.; Lau, K.; Hothorn, M. The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. plant Biol. 2017, 68, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.; Berger, J.; Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Herrera, A.; Ku Gonzalez, A.; Canche Moo, R.; Quiroz-Figueroa, F.R.; Loyola-Vargas, V.M.; Rodriguez-Zapata, L.C.; Burgeff D’Hondt, C.; Suárez-Solís, V.M.; Castaño, E. Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. plant Cell Tissue Organ. Cult. 2008, 94, 171–180. [Google Scholar] [CrossRef]
- Bouchabké-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Elhiti, M.; Tahir, M.; Gulden, R.H.; Khamiss, K.; Stasolla, C. Modulation of embryo-forming capacity in culture through the expression of Brassica genes involved in the regulation of the shoot apical meristem. J. Exp. Bot. 2010, 61, 4069–4085. [Google Scholar] [CrossRef]
- Zuo, J.; Niu, Q.W.; Frugis, G.; Chua, N.H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; van Lammeren, A.A.; Miki, B.L. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef]
- Srinivasan, C.; Liu, Z.; Heidmann, I.; Supena, E.D.J.; Fukuoka, H.; Joosen, R.; Lambalk, J.; Angenent, G.; Scorza, R.; Custers, J.B.M.; et al. Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta 2006, 225, 341. [Google Scholar] [CrossRef] [PubMed]
- El Ouakfaoui, S.; Schnell, J.; Abdeen, A.; Colville, A.; Labbé, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of somatic embryogenesis and embryo development by AP2 transcription factors. plant Mol. Biol. 2010, 74, 313–326. [Google Scholar] [CrossRef]
- Morcillo, F.; Gallard, A.; Pillot, M.; Jouannic, S.; Aberlenc-Bertossi, F.; Collin, M.; Verdeil, J.L.; Tregear, J.W. EgAP2-1, an AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm. Planta 2007, 226, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Florez, S.L.; Erwin, R.L.; Maximova, S.N.; Guiltinan, M.J.; Curtis, W.R. Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC plant Biol. 2015, 15, 121. [Google Scholar] [CrossRef] [PubMed]
- Tsuwamoto, R.; Yokoi, S.; Takahata, Y. Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. plant Mol. Biol. 2010, 73, 481–492. [Google Scholar] [CrossRef]
- Waki, T.; Hiki, T.; Watanabe, R.; Hashimoto, T.; Nakajima, K. The Arabidopsis RWP-RK Protein RKD4 Triggers Gene Expression and Pattern Formation in Early Embryogenesis. Curr. Biol. 2011, 21, 1277–1281. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Palmer, T.M.; Lukowitz, W. The RWP-RK Factor GROUNDED Promotes Embryonic Polarity by Facilitating YODA MAP Kinase Signaling. Curr. Biol. 2011, 21, 1268–1276. [Google Scholar] [CrossRef] [PubMed]
- Mursyanti, E.; Purwantoro, A.; Moeljopawiro, S.; Semiarti, E. Induction of somatic embryogenesis through overexpression of ATRKD4 genes in Phalaenopsis “Sogo Vivien”. Indones. J. Biotechnol. 2015, 20, 42–53. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.-a.; Yee, K.M.; West, M.A.L.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 Is Sufficient to Induce Embryo Development in Vegetative Cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef]
- Zhu, S.-P.; Wang, J.; Ye, J.-L.; Zhu, A.-D.; Guo, W.-W.; Deng, X.-X. Isolation and characterization of LEAFY COTYLEDON 1-LIKE gene related to embryogenic competence in Citrus sinensis. plant Cell Tissue Organ. Cult. 2014, 119, 1–13. [Google Scholar] [CrossRef]
- Uddenberg, D.; Abrahamsson, M.; von Arnold, S. Overexpression of PaHAP3A stimulates differentiation of ectopic embryos from maturing somatic embryos of Norway spruce. Tree Genet. Genomes 2016, 12, 18. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Tsuchiya, Y.; Lumba, S.; Okamoto, M.; McCourt, P. The Transcription Factor FUSCA3 Controls Developmental Timing in Arabidopsis through the Hormones Gibberellin and Abscisic Acid. Dev. Cell 2004, 7, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef] [PubMed]
- Santos Mendoza, M.; Dubreucq, B.; Miquel, M.; Caboche, M.; Lepiniec, L. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 2005, 579, 4666–4670. [Google Scholar] [CrossRef] [PubMed]
- Gallois, J.-L.; Woodward, C.; Reddy, G.V.; Sablowski, R. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 2002, 129, 3207–3217. [Google Scholar] [CrossRef] [PubMed]
- Gallois, J.-L.; Nora, F.R.; Mizukami, Y.; Sablowski, R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004, 18, 375–380. [Google Scholar] [CrossRef]
- Rashid, S.Z.; Yamaji, N.; Kyo, M. Shoot formation from root tip region: A developmental alteration by WUS in transgenic tobacco. plant Cell Rep. 2007, 26, 1449–1455. [Google Scholar] [CrossRef]
- Luo, K.; Zheng, X.; Chen, Y.; Xiao, Y.; Zhao, D.; McAvoy, R.; Pei, Y.; Li, Y. The maize Knotted1 gene is an effective positive selectable marker gene for Agrobacterium-mediated tobacco transformation. plant Cell Rep. 2006, 25, 403–409. [Google Scholar] [CrossRef]
- Nishimura, A.; Tamaoki, M.; Sakamoto, T.; Matsuoka, M. Over-expression of tobacco knotted 1-type class1 homeobox genes alters various leaf morphology. plant Cell Physiol. 2000, 41, 583–590. [Google Scholar] [CrossRef]
- Daimon, Y.; Takabe, K.; Tasaka, M. The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. plant Cell Physiol. 2003, 44, 113–121. [Google Scholar] [CrossRef]
- Banno, H.; Ikeda, Y.; Niu, Q.-W.; Chua, N.-H. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. plant Cell 2001, 13, 2609–2618. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Banno, H.; Niu, Q.-W.; Howell, S.H.; Chua, N.-H. The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. plant Cell Physiol. 2006, 47, 1443–1456. [Google Scholar] [CrossRef] [PubMed]
- Ckurshumova, W.; Smirnova, T.; Marcos, D.; Zayed, Y.; Berleth, T. Irrepressible MONOPTEROS/ARF 5 promotes de novo shoot formation. New Phytol. 2014, 204, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Jaskóła, K.; Gąsiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440. [Google Scholar] [CrossRef]
- Deng, W.; Luo, K.; Li, Z.; Yang, Y. A novel method for induction of plant regeneration via somatic embryogenesis. plant Sci. 2009, 177, 43–48. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef]
- Mookkan, M.; Nelson-Vasilchik, K.; Hague, J.; Zhang, Z.J.; Kausch, A.P. Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. plant Cell Rep. 2017, 36, 1477–1491. [Google Scholar] [CrossRef] [PubMed]
- Heidmann, I.; de Lange, B.; Lambalk, J.; Angenent, G.C.; Boutilier, K. Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. plant Cell Rep. 2011, 30, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Lutz, K.A.; Martin, C.; Khairzada, S.; Maliga, P. Steroid-inducible BABY BOOM system for development of fertile Arabidopsis thaliana plants after prolonged tissue culture. plant Cell Rep. 2015, 34, 1849–1856. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Q.-W.; Teng, C.; Li, C.; Mu, J.; Chua, N.-H.; Zuo, J. Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 2009, 19, 224–235. [Google Scholar] [CrossRef]
- Shires, M.E.; Florez, S.L.; Lai, T.S.; Curtis, W.R. Inducible somatic embryogenesis in Theobroma cacao achieved using the DEX-activatable transcription factor-glucocorticoid receptor fusion. Biotechnol. Lett. 2017, 39, 1747–1755. [Google Scholar] [CrossRef]
- Kononov, M.E.; Bassuner, B.; Gelvin, S.B. Integration of T-DNA binary vector ‘backbone’sequences into the tobacco genome: Evidence for multiple complex patterns of integration. plant J. 1997, 11, 945–957. [Google Scholar] [CrossRef]
- Hanson, B.; Engler, D.; Moy, Y.; Newman, B.; Ralston, E.; Gutterson, N. A simple method to enrich an Agrobacterium-transformed population for plants containing only T-DNA sequences. plant J. 1999, 19, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-R.; Lee, J.; Jun, S.-H.; Park, S.; Kang, H.-G.; Kwon, S.; An, G. Transgene structures in T-DNA-inserted rice plants. plant Mol. Biol. 2003, 52, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Podevin, N.; De Buck, S.; De Wilde, C.; Depicker, A. Insights into recognition of the T-DNA border repeats as termination sites for T-strand synthesis by Agrobacterium tumefaciens. Transgenic Res. 2006, 15, 557. [Google Scholar] [CrossRef] [PubMed]
- Richael, C.M.; Kalyaeva, M.; Chretien, R.C.; Yan, H.; Adimulam, S.; Stivison, A.; Weeks, J.T.; Rommens, C.M. Cytokinin vectors mediate marker-free and backbone-free plant transformation. Transgenic Res. 2008, 17, 905–917. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; La Rota, M.; Hoerster, G.; Hastings, C.; Wang, N.; Chamberlin, M.; Wu, E.; Jones, T.; Gordon-Kamm, W. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. Vitr. Cell. Dev. Biol. Plant 2018, 54, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Martin-Ortigosa, S.; Finer, J.; Orchard, N.; Gunadi, A.; Batts, L.A.; Thakare, D.; Rush, B.; Schmitz, O.; Stuiver, M.; et al. Overexpression of the Transcription Factor GROWTH-REGULATING FACTOR5 Improves Transformation of Dicot and Monocot Species. Front. plant Sci. 2020, 11, 572319. [Google Scholar] [CrossRef]
- Hills, M.J.; Hall, L.; Arnison, P.G.; Good, A.G. Genetic use restriction technologies (GURTs): Strategies to impede transgene movement. Trends plant Sci. 2007, 12, 177–183. [Google Scholar] [CrossRef]
- Daniell, H. Molecular strategies for gene containment in transgenic crops. Nat. Biotechnol. 2002, 20, 581–586. [Google Scholar] [CrossRef]
- Linder, C.R.; Taha, I.; Rieseberg, L.H.; Seiler, G.J.; Snow, A.A. Long-term introgression of crop genes into wild sunflower populations. Theor. Appl. Genet. 1998, 96, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Schernthaner, J.P.; Fabijanski, S.F.; Arnison, P.G.; Racicot, M.; Robert, L.S. Control of seed germination in transgenic plants based on the segregation of a two-component genetic system. Proc. Natl. Acad. Sci. USA 2003, 100, 6855. [Google Scholar] [CrossRef]
- Hüsken, A.; Prescher, S.; Schiemann, J. Evaluating biological containment strategies for pollen-mediated gene flow. Environ. Biosaf. Res. 2010, 9, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Bicknell, R.A.; Koltunow, A.M. Understanding Apomixis: Recent Advances and Remaining Conundrums. plant Cell 2004, 16, S228. [Google Scholar] [CrossRef] [PubMed]
- Haider, N.; Allainguillaume, J.; Wilkinson, M.J. Spontaneous capture of oilseed rape (Brassica napus) chloroplasts by wild B. rapa: Implications for the use of chloroplast transformation for biocontainment. Curr. Genet. 2009, 55, 139–150. [Google Scholar] [CrossRef]
- Lu, B.-R. Editorial. Transgene containment by molecular means—is it possible and cost effective. Environ. Biosaf. Res. 2003, 2, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, X.; Tang, Q.; Wang, Z. Theoretical Basis of Gene Splitting Technique and Its Application in the Control of Transgene Flow. Agric. Biotechnol. 2015, 4, 1–4. [Google Scholar]
- Kelloniemi, J.; Mäkinen, K.; Valkonen, J.P.T. Three heterologous proteins simultaneously expressed from a chimeric potyvirus: Infectivity, stability and the correlation of genome and virion lengths. Virus Res. 2008, 135, 282–291. [Google Scholar] [CrossRef]
- Saurabh, S.; Vidyarthi, A.S.; Prasad, D. RNA interference: Concept to reality in crop improvement. Planta 2014, 239, 543–564. [Google Scholar] [CrossRef]
- Lv, J.; Yu, K.; Wei, J.; Gui, H.; Liu, C.; Liang, D.; Wang, Y.; Zhou, H.; Carlin, R.; Rich, R.; et al. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Jacinto, F.V.; Link, W.; Ferreira, B.I. CRISPR/Cas9-mediated genome editing: From basic research to translational medicine. J. Cell. Mol. Med. 2020, 24, 3766–3778. [Google Scholar] [CrossRef]
- Hurgobin, B.; Tamiru-Oli, M.; Welling, M.T.; Doblin, M.S.; Bacic, A.; Whelan, J.; Lewsey, M.G. Recent advances in Cannabis sativa genomics research. New Phytol. 2020. [Google Scholar] [CrossRef]
- van Bakel, H.; Stout, J.M.; Cote, A.G.; Tallon, C.M.; Sharpe, A.G.; Hughes, T.R.; Page, J.E. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011, 12, R102. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Cradick, T.J.; Brown, M.T.; Deshmukh, H.; Ranjan, P.; Sarode, N.; Wile, B.M.; Vertino, P.M.; Stewart, F.J.; Bao, G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014, 42, 7473–7485. [Google Scholar] [CrossRef] [PubMed]
- Gerashchenkov, G.A.; Rozhnova, N.A.; Kuluev, B.R.; Kiryanova, O.Y.; Gumerova, G.R.; Knyazev, A.V.; Vershinina, Z.R.; Mikhailova, E.V.; Chemeris, D.A.; Matniyazov, R.T.; et al. Design of Guide RNA for CRISPR/Cas Plant Genome Editing. Mol. Biol. 2020, 54, 24–42. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Cheng, L.; Luo, Y. An overview and metanalysis of machine and deep learning-based CRISPR gRNA design tools. RNA Biol. 2020, 17, 13–22. [Google Scholar] [CrossRef]
- Hwang, G.-H.; Park, J.; Lim, K.; Kim, S.; Yu, J.; Yu, E.; Kim, S.-T.; Eils, R.; Kim, J.-S.; Bae, S. Web-based design and analysis tools for CRISPR base editing. BMC Bioinform. 2018, 19, 542. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Park, J.; Kim, J.-S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef]
- Park, J.; Bae, S.; Kim, J.-S. Cas-Designer: A web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 2015, 31, 4014–4016. [Google Scholar] [CrossRef]
- Bae, S.; Kweon, J.; Kim, H.S.; Kim, J.-S. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 2014, 11, 705–706. [Google Scholar] [CrossRef]
- Haeussler, M.; Schönig, K.; Eckert, H.; Eschstruth, A.; Mianné, J.; Renaud, J.-B.; Schneider-Maunoury, S.; Shkumatava, A.; Teboul, L.; Kent, J.; et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016, 17, 148. [Google Scholar] [CrossRef]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, M.; Thumberger, T.; del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef] [PubMed]
- Labun, K.; Montague, T.G.; Gagnon, J.A.; Thyme, S.B.; Valen, E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016, 44, W272–W276. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C.; Franch, M.; Tabas-Madrid, D.; San-León, D.; Montoliu, L.; Cubas, P.; Pazos, F. Breaking-Cas—interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016, 44, W267–W271. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted Mutagenesis, Precise Gene Editing, and Site-Specific Gene Insertion in Maize Using Cas9 and Guide RNA. plant Physiol. 2015, 169, 931. [Google Scholar] [CrossRef]
- Li, C.; Zhang, R.; Meng, X.; Chen, S.; Zong, Y.; Lu, C.; Qiu, J.-L.; Chen, Y.-H.; Li, J.; Gao, C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020, 38, 875–882. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Zhu, J.-K. Expanding the base editing scope in rice by using Cas9 variants. plant Biotechnol. J. 2019, 17, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Sukegawa, S.; Saika, H.; Toki, S. Plant genome editing: Ever more precise and wide reaching. plant J. 2021, 106, 1–11. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).