Structural Basis for the Diminished Ligand Binding and Catalytic Ability of Human Fetal-Specific CYP3A7
Abstract
1. Introduction
2. Results and Discussion
2.1. Modification of Surface Residues in CYP3A7
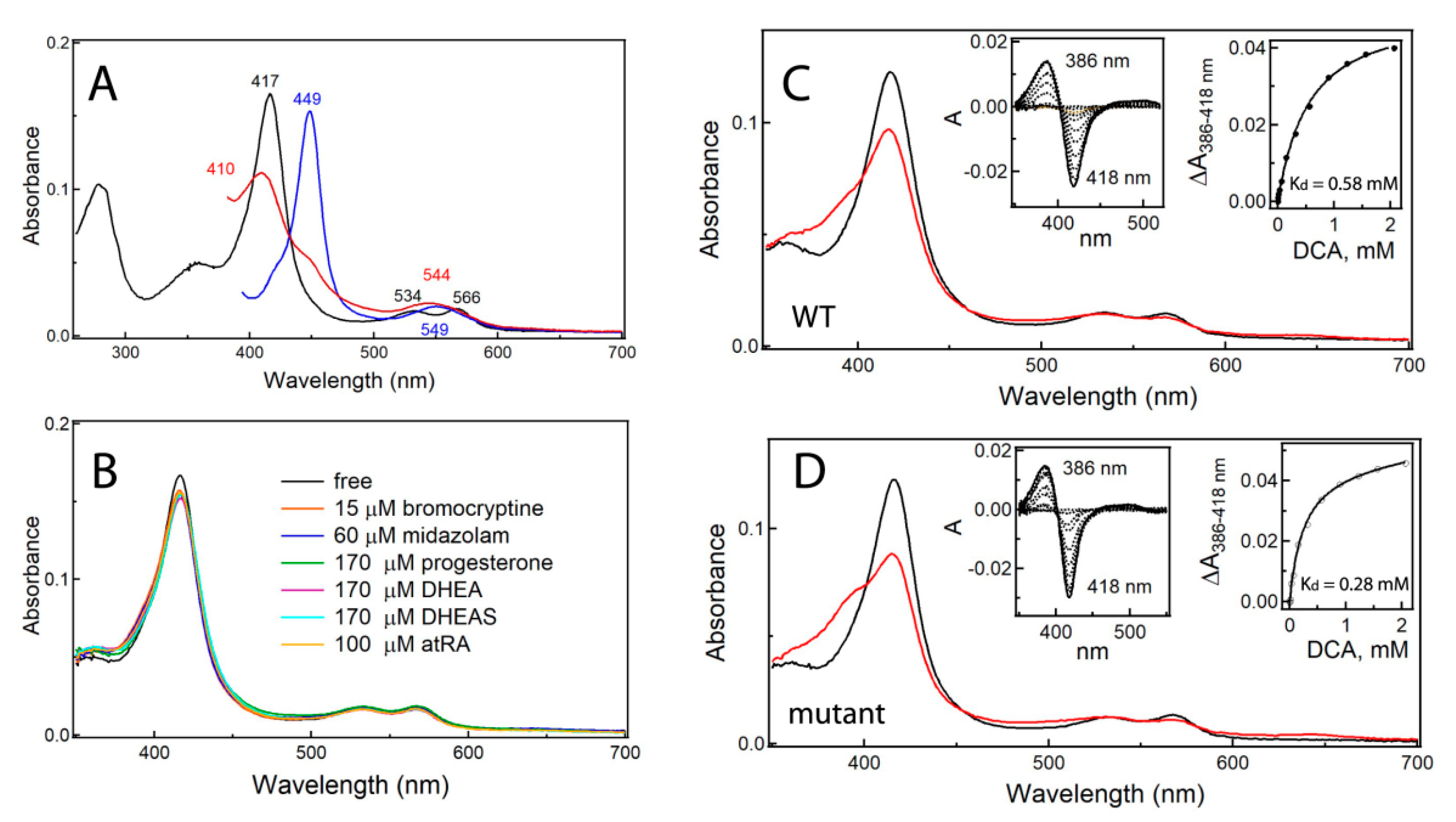
2.2. Impact of Surface Mutations on CYP3A7 Properties
2.3. Crystal Structure of CYP3A7
2.4. Unique Features of CYP3A7
2.4.1. Divergence in the F-G Fragment
2.4.2. Formation of the F304-Centered Hydrophobic Cluster
2.4.3. Tighter Packing of the I-E-D-Helix Bundle
2.4.4. Other Structural Distinctions
2.5. Functional Implications
2.6. CYP3A7 Polymorphism
2.7. Concluding Summary
3. Materials and Methods
3.1. Cloning, Expression, and Purification of CYP3A7
3.2. Spectral Measurements
3.3. Activity Assays
3.4. Gel Filtration
3.5. Crystallization and Structure Determination
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CYP | Cytochrome P450 |
| CPR | Cytochrome P450 reductase |
| BFC | 7-benzyloxy-4-(trifluoromethyl)coumarin |
| atRA | All-trans retinoic acid |
| DHEA | Dihydroepiandrosterone |
| DHEAS | Dihydroepiandrosterone sulfate |
| DTT | Dithiothreitol |
| WT | Wild type |
References
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Kuehl, P.; Zhang, J.; Lin, Y.; Lamba, J.; Assem, M.; Schuetz, J.; Watkins, P.B.; Daly, A.; Wrighton, S.A.; Hall, S.D.; et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 2001, 27, 383–391. [Google Scholar] [CrossRef]
- Lamba, J.K.; Lin, Y.S.; Schuetz, E.G.; Thummel, K.E. Genetic contribution to variable human CYP3A-mediated metabolism. Adv. Drug Deliv. Rev. 2002, 54, 1271–1294. [Google Scholar] [CrossRef]
- Gellner, K.; Eiselt, R.; Hustert, E.; Arnold, H.; Koch, I.; Haberl, M.; Deglmann, C.J.; Burk, O.; Buntefuss, D.; Escher, S.; et al. Genomic organization of the human CYP3A locus: Identification of a new, inducible CYP3A gene. Pharmacogenetics 2001, 11, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.C. New perspectives on the impact of cytochrome P450 3A expression for pediatric pharmacology. Drug Discov. Today 2006, 11, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, J.D.; Kauma, S.; Guzelian, P.S. Identification of the fetal liver cytochrome CYP3A7 in human endometrium and placenta. J. Clin. Investig. 1993, 92, 1018–1024. [Google Scholar] [CrossRef]
- Li, H.; Lampe, J.N. Neonatal cytochrome P450 CYP3A7: A comprehensive review of its role in development, disease, and xenobiotic metabolism. Arch. Biochem. Biophys. 2019, 673, 108078. [Google Scholar] [CrossRef]
- Miller, K.K.; Cai, J.; Ripp, S.L.; Pierce, W.M., Jr.; Rushmore, T.H.; Prough, R.A. Stereo- and regioselectivity account for the diversity of dehydroepiandrosterone (DHEA) metabolites produced by liver microsomal cytochromes P450. Drug Metab. Dispos. 2004, 32, 305–313. [Google Scholar] [CrossRef]
- Kitada, M.; Kamataki, T.; Itahashi, K.; Rikihisa, T.; Kanakubo, Y. P-450 HFLa, a form of cytochrome P-450 purified from human fetal livers, is the 16 alpha-hydroxylase of dehydroepiandrosterone 3-sulfate. J. Biol. Chem. 1987, 262, 13534–13537. [Google Scholar] [CrossRef]
- Marill, J.; Cresteil, T.; Lanotte, M.; Chabot, G.G. Identification of human cytochrome P450s involved in the formation of all-trans-retinoic acid principal metabolites. Mol. Pharmacol. 2000, 58, 1341–1348. [Google Scholar] [CrossRef]
- Lacroix, D.; Sonnier, M.; Moncion, A.; Cheron, G.; Cresteil, T. Expression of CYP3A in the human liver--evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur. J. Biochem. 1997, 247, 625–634. [Google Scholar] [CrossRef]
- Williams, J.A.; Ring, B.J.; Cantrell, V.E.; Jones, D.R.; Eckstein, J.; Ruterbories, K.; Hamman, M.A.; Hall, S.D.; Wrighton, S.A. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab. Dispos. 2002, 30, 883–891. [Google Scholar] [CrossRef]
- Gillam, E.M.; Wunsch, R.M.; Ueng, Y.F.; Shimada, T.; Reilly, P.E.; Kamataki, T.; Guengerich, F.P. Expression of cytochrome P450 3A7 in Escherichia coli: Effects of 5’ modification and catalytic characterization of recombinant enzyme expressed in bicistronic format with NADPH-cytochrome P450 reductase. Arch. Biochem. Biophys. 1997, 346, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Zhang, J.; Zhu, P.P.; Tan, X.W.; Lin, Q.H.; Wang, W.X.; Yin, S.S.; Gao, L.Z.; Su, M.M.; Liu, C.X.; et al. Stereoselective Oxidation Kinetics of Deoxycholate in Recombinant and Microsomal CYP3A Enzymes: Deoxycholate 19-Hydroxylation Is an In Vitro Marker of CYP3A7 Activity. Drug Metab. Dispos. 2019, 47, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Godamudunage, M.P.; Grech, A.M.; Scott, E.E. Comparison of antifungal azole interactions with adult cytochrome P450 3A4 versus neonatal cytochrome P450 3A7. Drug Metab. Dispos. 2018, 46, 1329–1337. [Google Scholar] [CrossRef]
- Kandel, S.E.; Han, L.W.; Mao, Q.; Lampe, J.N. Digging Deeper into CYP3A Testosterone Metabolism: Kinetic, Regioselectivity, and Stereoselectivity Differences between CYP3A4/5 and CYP3A7. Drug Metab. Dispos. 2017, 45, 1266–1275. [Google Scholar] [CrossRef]
- Goldschmidt, L.; Cooper, D.R.; Derewenda, Z.S.; Eisenberg, D. Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 2007, 16, 1569–1576. [Google Scholar] [CrossRef]
- Kaur, P.; Chamberlin, A.R.; Poulos, T.L.; Sevrioukova, I.F. Structure-based inhibitor design for evaluation of a CYP3A4 pharmacophore model. J. Med. Chem. 2016, 59, 4210–4220. [Google Scholar] [CrossRef]
- Samuels, E.R.; Sevrioukova, I.F. Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength. Int. J. Mol. Sci. 2021, 22, 852. [Google Scholar] [CrossRef]
- Strohmaier, S.J.; De Voss, J.J.; Jurva, U.; Andersson, S.; Gillam, E.M.J. Oxygen Surrogate Systems for Supporting Human Drug-Metabolizing Cytochrome P450 Enzymes. Drug Metab. Dispos. 2020, 48, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Samuels, E.R.; Sevrioukova, I. Structure-activity relationships of rationally designed ritonavir analogs: Impact of side-group stereochemistry, head-group spacing, and backbone composition on the interaction with CYP3A4. Biochemistry 2019, 58, 2077–2087. [Google Scholar] [CrossRef]
- Yano, J.K.; Wester, M.R.; Schoch, G.A.; Griffin, K.J.; Stout, C.D.; Johnson, E.F. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05—A resolution. J. Biol. Chem. 2004, 279, 38091–38094. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.H.; Savas, U.; Johnson, E.F. The X-Ray Crystal Structure of the Human Mono-Oxygenase Cytochrome P450 3A5-Ritonavir Complex Reveals Active Site Differences between P450s 3A4 and 3A5. Mol. Pharmacol. 2018, 93, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Sevrioukova, I.F.; Poulos, T.L. Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir. Proc. Natl. Acad. Sci. USA 2010, 107, 18422–18427. [Google Scholar] [CrossRef]
- Ekroos, M.; Sjogren, T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. USA 2006, 103, 13682–13687. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.A.; Cosme, J.; Vinkovic, D.M.; Ward, A.; Angove, H.C.; Day, P.J.; Vonrhein, C.; Tickle, I.J.; Jhoti, H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science 2004, 305, 683–686. [Google Scholar] [CrossRef]
- Denisov, I.G.; Grinkova, Y.V.; Nandigrami, P.; Shekhar, M.; Tajkhorshid, E.; Sligar, S.G. Allosteric Interactions in Human Cytochrome P450 CYP3A4: The Role of Phenylalanine 213. Biochemistry 2019, 58, 1411–1422. [Google Scholar] [CrossRef]
- Davydov, D.R.; Halpert, J.R. Allosteric P450 mechanisms: Multiple binding sites, multiple conformers or both? Expert Opin. Drug Metab. Toxicol. 2008, 4, 1523–1535. [Google Scholar] [CrossRef] [PubMed]
- Polic, V.; Auclair, K. Allosteric activation of cytochrome P450 3A4 via progesterone bioconjugation. Bioconjug. Chem. 2017, 28, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Svobodova Varekova, R.; Berka, K.; Pravda, L.; Navratilova, V.; Banas, P.; Ionescu, C.M.; Otyepka, M.; Koca, J. MOLE 2.0: Advanced approach for analysis of biomacromolecular channels. J. Cheminform. 2013, 5, 39. [Google Scholar] [CrossRef]
- Hsu, M.H.; Johnson, E.F. Active-site differences between substrate-free and ritonavir-bound cytochrome P450 (CYP) 3A5 reveal plasticity differences between CYP3A5 and CYP3A4. J. Biol. Chem. 2019, 294, 8015–8022. [Google Scholar] [CrossRef]
- Ohmori, S.; Nakasa, H.; Asanome, K.; Kurose, Y.; Ishii, I.; Hosokawa, M.; Kitada, M. Differential catalytic properties in metabolism of endogenous and exogenous substrates among CYP3A enzymes expressed in COS-7 cells. Biochim. Biophys. Acta 1998, 1380, 297–304. [Google Scholar] [CrossRef]
- Sevrioukova, I.F.; Poulos, T.L. Structural basis for regiospecific midazolam oxidation by human cytochrome P450 3A4. Proc. Natl. Acad. Sci. USA 2017, 114, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Paquin, A.; Oufqir, Y.; Sevrioukova, I.F.; Reyes-Moreno, C.; Berube, G. Innovative C2-symmetric testosterone and androstenedione dimers: Design, synthesis, biological evaluation on prostate cancer cell lines and binding study to recombinant CYP3A4. Eur. J. Med. Chem. 2021, 220, 113496. [Google Scholar] [CrossRef]
- Smit, P.; van Schaik, R.H.; van der Werf, M.; van den Beld, A.W.; Koper, J.W.; Lindemans, J.; Pols, H.A.; Brinkmann, A.O.; de Jong, F.H.; Lamberts, S.W. A common polymorphism in the CYP3A7 gene is associated with a nearly 50% reduction in serum dehydroepiandrosterone sulfate levels. J. Clin. Endocrinol. Metab. 2005, 90, 5313–5316. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burk, O.; Tegude, H.; Koch, I.; Hustert, E.; Wolbold, R.; Glaeser, H.; Klein, K.; Fromm, M.F.; Nuessler, A.K.; Neuhaus, P.; et al. Molecular mechanisms of polymorphic CYP3A7 expression in adult human liver and intestine. J. Biol. Chem. 2002, 277, 24280–24288. [Google Scholar] [CrossRef]
- Johnson, N.; De Ieso, P.; Migliorini, G.; Orr, N.; Broderick, P.; Catovsky, D.; Matakidou, A.; Eisen, T.; Goldsmith, C.; Dudbridge, F.; et al. Cytochrome P450 Allele CYP3A7*1C Associates with Adverse Outcomes in Chronic Lymphocytic Leukemia, Breast, and Lung Cancer. Cancer Res. 2016, 76, 1485–1493. [Google Scholar] [CrossRef]
- Thompson, E.E.; Kuttab-Boulos, H.; Yang, L.; Roe, B.A.; Di Rienzo, A. Sequence diversity and haplotype structure at the human CYP3A cluster. Pharm. J. 2006, 6, 105–114. [Google Scholar] [CrossRef]
- Rodriguez-Antona, C.; Jande, M.; Rane, A.; Ingelman-Sundberg, M. Identification and phenotype characterization of two CYP3A haplotypes causing different enzymatic capacity in fetal livers. Clin. Pharmacol. Ther. 2005, 77, 259–270. [Google Scholar] [CrossRef]
- Sevrioukova, I.F.; Li, H.; Zhang, H.; Peterson, J.A.; Poulos, T.L. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. USA 1999, 96, 1863–1868. [Google Scholar] [CrossRef]
- Estrada, D.F.; Laurence, J.S.; Scott, E.E. Cytochrome P450 17A1 Interactions with the FMN Domain of Its Reductase as Characterized by NMR. J. Biol. Chem. 2016, 291, 3990–4003. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R. The carbon monoxide-binding pigment of liver microsomes. Solubilization, purification, and properties. J. Biol. Chem. 1964, 239, 2379–2385. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40 Pt 4, 658–674. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 2010, 66 Pt 4, 486–501. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D 2010, 66 Pt 2, 213–321. [Google Scholar] [CrossRef]










| CYP3A7 | CPR-Supported | CuOOH-Supported | ||||
|---|---|---|---|---|---|---|
| Vmax (min−1) | Km (μM) | Vmax/Km (min−1 μM−1) | Vmax (min−1) | Km (μM) | Vmax/Km (min−1 μM−1) | |
| WT | 0.0036 ± 0.0006 | 17 ± 3 | 2.1 × 10−4 | 0.035 ± 0.004 | 10 ± 2 | 3.5 × 10−3 |
| Mutant | 0.0011 ± 0.0003 | 9 ± 2 | 1.2 × 10−4 | 0.034 ± 0.005 | 11 ± 1 | 3.1 × 10−3 |
| Data statistics | ||
| Space group | C222 | |
| Unit cell parameters | a = 72 Å, b = 205 Å, c = 157 Å; α, β, γ = 90° | |
| Molecules per asymmetric unit | 2 | |
| Resolution range (Å) | 85.79–2.15 (2.27–2.15) a | |
| Total reflections | 529,291 (79,567) | |
| Unique reflections | 63,247 (9183) | |
| Redundancy | 8.4 (8.7) | |
| Completeness | 99.5 (99.6) | |
| Average I/σI | 10.7 (1.4) | |
| Rmerge | 0.119 (1.628) | |
| Rpim | 0.044 (0.579) | |
| CC ½ | 0.999 (0.635) | |
| Refinement statistics | ||
| Rwork/Rfree b | 20.3/25.3 | |
| Number of atoms: | ||
| Protein: | molecule A | 3727 |
| molecule B | 3794 | |
| Solvent | 230 | |
| R.m.s. deviations: | ||
| Bond lengths, Å | 0.007 | |
| Bond angles, ° | 0.923 | |
| Wilson B-factor, Å2 | 43 | |
| Average B-factor, Å2: | ||
| Molecule A | 59 | |
| Molecule B | 65 | |
| Ligand A | 86 | |
| Ligand B | 100 | |
| Solvent | 57 | |
| Ramachandran plot c (residues; %) | ||
| Preferred | 891 (95.8%) | |
| Allowed | 37 (4.0%) | |
| Outliers | 2 (0.2%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sevrioukova, I.F. Structural Basis for the Diminished Ligand Binding and Catalytic Ability of Human Fetal-Specific CYP3A7. Int. J. Mol. Sci. 2021, 22, 5831. https://doi.org/10.3390/ijms22115831
Sevrioukova IF. Structural Basis for the Diminished Ligand Binding and Catalytic Ability of Human Fetal-Specific CYP3A7. International Journal of Molecular Sciences. 2021; 22(11):5831. https://doi.org/10.3390/ijms22115831
Chicago/Turabian StyleSevrioukova, Irina F. 2021. "Structural Basis for the Diminished Ligand Binding and Catalytic Ability of Human Fetal-Specific CYP3A7" International Journal of Molecular Sciences 22, no. 11: 5831. https://doi.org/10.3390/ijms22115831
APA StyleSevrioukova, I. F. (2021). Structural Basis for the Diminished Ligand Binding and Catalytic Ability of Human Fetal-Specific CYP3A7. International Journal of Molecular Sciences, 22(11), 5831. https://doi.org/10.3390/ijms22115831






