A Dual-Color Tyr-FISH Method for Visualizing Genes/Markers on Plant Chromosomes to Create Integrated Genetic and Cytogenetic Maps
Abstract
:1. Introduction
2. Results
2.1. Probe Preparation and Labeling: Reducing Background and Increasing Signal-to-Noise Ratio
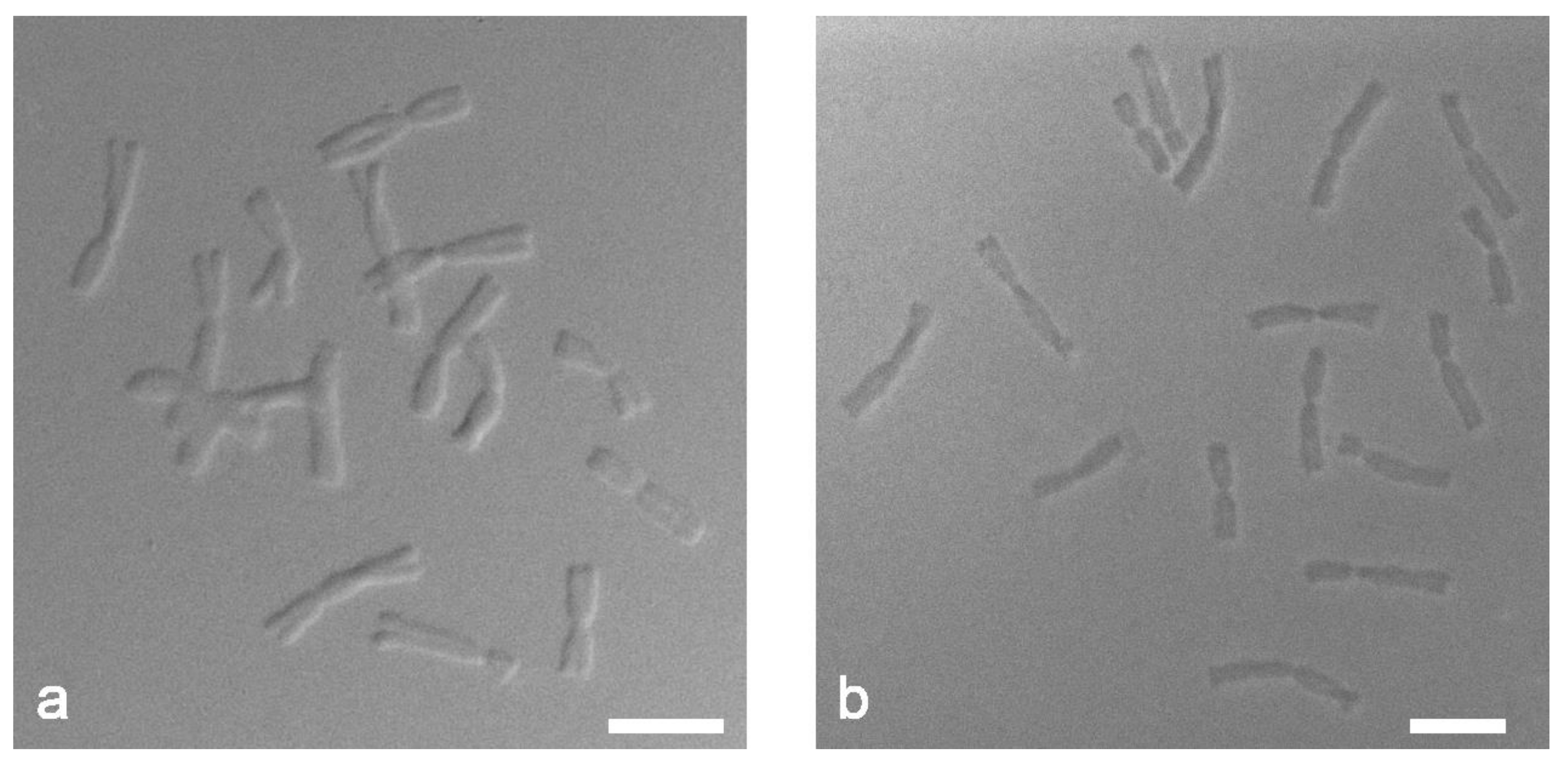
2.2. Chromosome Preparation Is a Key Step in Successful Tyr-FISH
2.3. Quenching Endogenous Peroxidase and Horseradish Peroxidase (HRP)
2.4. A Dual-Color Tyr-FISH Visualization of Markers
2.5. The Integration of Recombination and Cytogenetic Maps
3. Discussion
3.1. Method of Chromosome Preparation Strongly Influences the Signal Detection with Tyramide-Fluorophore
3.2. A Dual-Color Tyr-FISH for Integration of Recombination and Cytogenetic Maps and Genome Assembly
4. Materials and Methods
4.1. Plant Materials
4.2. Chromosome Preparations
4.2.1. “SteamDrop” Method
4.2.2. Squash Method
4.3. Probe Preparation
4.4. Dual-Color Sequential Tyr-FISH
- 1
- Incubate the slides in 4% (w/v) paraformaldehyde in water at RT for 10 min (USE THE FUME HOOD!).
- 2
- Wash slides in 2xSSC at RT three times for 5 min.
- 3
- Dehydrate slides for 2 min each in 70, 90, and 100% ethanol and air dry.
- 4
- Prepare the hybridization mixtures (40 μL per slide) *
- 20 μL formamide
- 8 μL 50% dextransulphate
- 4 μL 20x SSC
- 1 μL 10% SDS
- x μL probe DNA (50 ng/slide)
- z μL probe DNA (50 ng/slide)
- y μL water
- 5
- Denature hybridization mix at 75 °C for 5 min and plunge directly into the ice for 3 min.
- 6
- Add the appropriate hybridization mix to each slide and cover with 24 × 25 coverslip.
- 7
- Denature the slides at 75 °C for 5 min.
- 8
- Place the slides in prewarmed humid chamber and incubate overnight at 37 °C.
- 9
- Wash slides in 2xSSC at 42 °C twice for 5 min (with agitation).
- 10
- Wash slides in 0.1xSSC at 55 °C twice for 7 min (without agitation).
- 11
- Wash slides in 2xSSC at 42 °C for 3 min (with agitation).
- 12
- Take Coplin jar out of water bath and leave to cool to RT for 20–25 min.
- 13
- Wash slides in 4xSSC at RT for 3 min.
- 14
- Wash slides in 2xSSC at RT for 2 min (with agitation).
- 15
- Wash slides in fresh TNT (0.1 M Tris-HCl, 0.15 M NaCl, pH 7.5, 0.05% Tween 20) buffer at RT for 5 min with agitation.
- 16
- Block slides with 100 μL TNB buffer (1% blocking reagent in TN buffer) and place a coverslip (25 × 50 mm) to reduce evaporation. Incubate the slides in a humid chamber at RT for 15 min.
- 17
- Drain off TNB buffer. Add 100 μL of Streptavidin-HRP (PerkinElmer, Waltham, MA 02451 USA) (1:500 diluted in TNB) to each slide and place a coverslip (25 × 50 mm) on top to reduce evaporation. Incubate the slides in a humid chamber at RT for 40 min.
- 18
- Wash slides three times in fresh TNT at RT with agitation for 5 min.
- 19
- Pipet 200 μL of the TSA PLUS Cy3 Reagent (Akoya Biosciences, Menlo Park, CA 94025 USA) (1:50 in 10% dextransulphate in 1x Amplification) onto each slide. Incubate the slides at RT for 3 to 10 min **. (per one slide: 4 μL stock Tyramide-CY3 + 40 μL 50% dextransulphate + 156 μL 1xAmplification Diluent).
- 20
- Wash slides three times in fresh TNT at RT with agitation for 5 min.
- 21
- Deactivate the remaining HRP activity by adding 100 μL 3% H2O2 in 1xTN (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl) for 30 min.
- 22
- Wash the slides three times for 5 min each in TNT at RT with agitation.
- 23
- Block slides with 100 μL 1% TNB buffer and place a coverslip (25 × 50 mm) to reduce evaporation. Incubate the slides in a humid chamber at RT for 15 min.
- 24
- Drain off 1% TNB buffer. Add 100 μL of anti-digoxigenin HRP (Akoya Biosciences) (1:500 diluted in 1% TNB) to each slide and place a coverslip (25 × 50 mm) on top to reduce evaporation. Incubate the slides in a humid chamber at RT for 40 min.
- 25
- Wash slides three times in fresh TNT at RT with agitation for 5 min.
- 26
- Pipet 200 μL of the TSA PLUS Fluorescein Reagent (Akoya Biosciences) (1:50 in 10% dextransulphate in 1x Amplification) onto each slide. Incubate the slides at RT for 3 to 10 min (per one slide: 4 μL stock tyramide-FITC + 40 μL 50% dextransulphate + 156 μL 1xAmplification Diluent).
- 27
- Wash slides three times in fresh TNT at RT with agitation for 5 min.
- 28
- Wash slides in 2xSSC at RT three times for 5 min.
- 29
- Dehydrate slides for 2 min each in 70, 90, and 100% ethanol and air dry in dark place.
- 30
- Prepare fresh 1:20 dilution of DAPI in Vectashield. Add 20 µL per slide. Apply a 24 × 50 mm coverslip.
4.5. Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sturtevant, A.H. The Linear Arrangement of Six Sex-linked Factors in Drosophila, as Shown by their Mode of Association. J. Exp. Zool. 1913, 14, 43–59. [Google Scholar] [CrossRef]
- Wu, H.; Yao, D.; Chen, Y.; Yang, W.; Zhao, W.; Gao, H.; Tong, C. De novo genome assembly of Populus simonii further supports that Populus simonii and Populus trichocarpa belong to different sections. G3 Genes Genomes Genet. 2020, 10, 455–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccaferri, M; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zhang, Y.; Wang, Y.; Zhang, L.; Brinkman, E.K.; Adam, S.A.; Goldman, R.; van Steensel, B.; Ma, J.; Belmont, A.S. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 2018, 217, 4025–4048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, D.H.; Jo, S.H.; Bang, J.W.; Park, H.M.; Lee, S.; Choi, D. Integration of cytogenetic and genetic linkage maps unveils the physical architecture of tomato chromosome 2. Genetics 2008, 179, 1211–1220. [Google Scholar] [CrossRef] [Green Version]
- Szinay, D.; Chang, S.-B.; Khrustaleva, L.; Peters, S.; Schijlen, E.; Bai, Y.; Stiekema, W.J.; van Ham, R.C.H.J.; de Jong, H.; Lankhorst, M.R.K. High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6. Plant J. 2008, 56, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhao, H.; Kou, Q.; Jiang, J.; Guo, S.; Zhang, H.; Hou, W.; Zou, X.; Sun, H.; Gong, G.; et al. A high resolution genetic map anchoring scaffolds of the sequenced watermelon genome. PLoS ONE 2012, 7, e29453. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Koo, D.H.; Li, D.; Zhang, T.; Jiang, J.; Luan, F.; Renner, S.S.; Hénaff, E.; Sanseverino, W.; Garcia-Mas, J. Next-generation sequencing, FISH mapping and synteny-based modeling reveal mechanisms of decreasing dysploidy in Cucumis. Plant J. 2014, 77, 16–30. [Google Scholar] [CrossRef]
- Bielski, W.; Książkiewicz, M.; Šimoníková, D.; Hřibová, E.; Susek, K.; Naganowska, B. The Puzzling Fate of a Lupin Chromosome Revealed by Reciprocal Oligo-FISH and BAC-FISH Mapping. Genes 2020, 11, 1489. [Google Scholar] [CrossRef]
- Danilova, T.V.; Friebe, B.; Gill, B.S. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Appl. Genet. 2014, 127, 715–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khrustaleva, L; Kudryavtseva, N.; Romanov, D.; Ermolaev, A.; Kirov, E. Comparative Tyramide-FISH mapping of the genes controlling flavor and bulb color in Allium species revealed an altered gene order. Sci. Rep. 2019, 9, 12007. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Burton, J.N.; Adey, A.; Patwardhan, R.P.; Qiu, R.; Kitzman, J.O.; Shendure, J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013, 31, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Dudchenko, O.; Batra, S.S.; Omer, A.D.; Nyquist, S.K.; Hoeger, M.; Durand, N.C.; Aiden, E.L. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 2017, 356, 92–95. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Lyons, E.; Town, C.D. Optical mapping in plant comparative genomics. GigaScience 2015, 4, s13742-015-0044-y. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, P.N.; Michael, T.P.; Gilbert, S.; Chu, P.; Motley, S.T.; Appenroth, K.J.; Schubert, I.; Lam, E. Generating a high-confidence reference genome map of the Greater Duckweed by integration of cytogenomic, optical mapping and Oxford Nanopore technologies. Plant J. 2018, 96, 670–684. [Google Scholar] [CrossRef] [Green Version]
- Shearer, L.A.; Anderson, L.K.; de Jong, H.; Smit, S.; Goicoechea, J.L.; Roe, B.A.; Hua, A.; Giovannoni, J.J.; Stack, S.M. Fluorescence in situ hybridization and optical mapping to correct scaffold arrangement in the tomato genome. G3 Genes Genomes Genet. 2014, 4, 1395–1405. [Google Scholar] [CrossRef] [Green Version]
- Febrer, M.; Goicoechea, J.L.; Wright, J.; McKenzie, N.; Song, X.; Lin, J.; Bevan, M.W. An integrated physical, genetic and cytogenetic map of Brachypodium distachyon, a model system for grass research. PLoS ONE 2010, 5, e13461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, P.T.; Fiebig, A.; Novák, P.; Macas, J.; Cao, H.X.; Stepanenko, A.; Schubert, I. Chromosome-scale genome assembly for the duckweed Spirodela intermedia, integrating cytogenetic maps, PacBio and Oxford Nanopore libraries. Sci. Rep. 2020, 10, 19230. [Google Scholar] [CrossRef] [PubMed]
- Raap, A.; Van De Corput, M.; Vervenne, R.; Van Gijlswijk, R.; Tanke, H.; Wiegant, J. Ultra-sensitive FISH using peroxidase-mediated deposition of biotin- or fluorochrome tyramides. Hum. Mol. Genet. 1995, 4, 529–534. [Google Scholar] [CrossRef]
- Khrustaleva, L.I.; Kik, C. Localization of single copy T-DNA insertion in transgenic shallots (Allium cepa) by using ultra-sensitive FISH with tyramide signal amplification. Plant J. 2001, 25, 699–707. [Google Scholar] [CrossRef]
- Stephens, J.L.; Brown, S.E.; Lapitan, N.L.; Knudson, D.L. Physical mapping of barley genes using an ultrasensitive fluorescence in situ hybridization technique. Genome 2004, 47, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Pérez, R.; de Bustos, A.; Jouve, N.; Cuadrado, A. Localization of Rad50, a single-copy gene, on group 5 chromosomes of wheat, using a FISH protocol employing tyramide for signal amplification (Tyr-FISH). Cytogenet. Genome Res. 2009, 125, 321–328. [Google Scholar] [CrossRef]
- Sanz, M.J.; Loarce, Y.; Ferrer, E.; Fominaya, A. Use of Tyramide-Fluorescence in situ Hybridization and Chromosome Microdissection for Ascertaining Homology Relationships and Chromosome Linkage Group Associations in Oats. Cytogenet. Genome Res. 2012, 136, 145–156. [Google Scholar] [CrossRef]
- Kirov, I.; Van Laere, K.; De Riek, J.; De Keyser, E.; Van Roy, N.; Khrustaleva, L. Anchoring linkage groups of the Rosa genetic map to physical chromosomes with Tyramide-FISH and EST-SNP markers. PLoS ONE 2014, 9, e95793. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Gill, B.S. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 2006, 49, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Fujito, S.; Akyol, T.Y.; Mukae, T.; Wako, T.; Yamashita, K.; Tsukazaki, H.; Hirakawa, H.; Tanaka, K.; Mine, Y.; Sato, S.; et al. Construction of an ultrahigh-density linkage map and graphical representation of the arrangement of transcriptome-based unigene markers on the chromosomes of Allium cepa L. BMC Genom. (under review). [CrossRef]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. Bmc Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, L.E.; Ramakrishnan, R.; Ruffalo, T.M.; Wilber, K.A. Labeling fluorescence in situ hybridization probes for genomic targets. Methods Mol. Biol. 2002, 204, 21–40. [Google Scholar] [CrossRef]
- Kirov, I.; Divashuk, M.; Van Laere, K.; Soloviev, A.; Khrustaleva, L. An easy “SteamDrop” method for high quality plant chromosome preparation. Mol. Cytogenet. 2014, 7, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, D. Differential interference contrast (DIC) microscopy. In Fundamentals of Light Microscopy and Digital Imaging, 2nd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2001; pp. 153–168. [Google Scholar]
- Bobrow, M.N.; Harris, T.D.; Shanghnessy, K.J.; Litt, G.J. Catalazed reporter deposition, a novel method of signal amplification. Application to immunoassays. J. Immunol. Methods 1989, 125, 279–285. [Google Scholar] [CrossRef]
- Gross, R.A.; Kumar, A.; Kalra, B. Polymer synthesis by in vitro enzyme catalysis. Chem. Rev. 2001, 101, 2097–2124. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.L.; Indiani, C.; Henriksen, A.; Feis, A.; Becucci, M.; Gajhede, M.; Smulevich, G.; Welinder, K.G. Differential activity and structure of highly similar peroxidases. Spectroscopic, crystallographic, and enzymatic analyses of lignifying Arabidopsis thaliana peroxidase A2 and horseradish peroxidase A2. Biochemistry 2001, 40, 11013–11021. [Google Scholar] [CrossRef]
- Pandey, V.P.; Awasthi, M.; Singh, S.; Tiwari, S.; Dwivedi, U.N. A comprehensive review on function and application of plant peroxidases. Biochem. Anal. Biochem. 2007, 6, 1009–2161. [Google Scholar] [CrossRef]
- Kirov, I.; Khrustaleva, L.; Van Laere, K.; Soloviev, A.; Meeus, S.; Romanov, D.; Fesenko, I. DRAWID: User-friendly java software for chromosome measurements and idiogram drawing. Comp. Cytogenet. 2017, 11, 747–775. [Google Scholar] [CrossRef]
- Wang, C.J.R.; Harper, L.; Cande, W.Z. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 2006, 18, 529–544. [Google Scholar] [CrossRef] [Green Version]
- Mohanta, D.; Santra, S.; Jana, M. Conformational disorder and solvation properties of the key-residues of a protein in water–ethanol mixed solutions. Phys. Chem. Chem. Phys. 2017, 19, 32636–32646. [Google Scholar] [CrossRef]
- Halder, R; Jana, B. Exploring the role of hydrophilic amino acids in unfolding of protein in aqueous ethanol solution. Proteins 2021, 89, 116–125. [Google Scholar] [CrossRef]
- Solovei, I.; Cavallo, A.; Schermelleh, L.; Jaunin, F.; Scasselati, C.; Cmarko, D.; Cremer, C.; Fakan, S.; Cremer, T. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp. Cell Res. 2002, 276, 10–23. [Google Scholar] [CrossRef] [Green Version]
- De Jong, J.H.; Fransz, P.; Zabel, P. High resolution FISH in plants–techniques and applications. Trends Plant Sci. 1999, 4, 258–263. [Google Scholar] [CrossRef]
- Jiang, J.; Gill, B.S.; Wang, G.L.; Ronald, P.C.; Wang, D.C. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 1995, 92, 4487–4491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raap, A.K. Advances in fluorescence in situ hybridization. Mutat. Res. 1998, 400, 287–298. [Google Scholar] [CrossRef]
- Schriml, L.M.; Padilla-Nash, H.M.; Coleman, A.; Moen, P.; Nash, W.G.; Menninger, J.; Jones, G.; Ried, T.; Dean, M. Tyramide signal amplification (TSA)-FISH applied to mapping PCR-labeled probes less than 1 Kb in size. Biotechniques 1999, 27, 608–613. [Google Scholar] [CrossRef]
- Yamato, K.T.; Ishizaki, K.; Fujisawa, M.; Okada, S.; Nakayama, S.; Fujishita, M.; Bando, H.; Yodoya, K.; Hayashi, K.; Bando, T.; et al. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc. Natl. Acad. Sci. USA 2007, 104, 6472–6477. [Google Scholar] [CrossRef] [Green Version]
- Seifertova, E.; Zimmerman, L.B.; Gilchrist, M.J.; Macha, J.; Kubickova, S.; Cernohorska, H.; Zarsky, V.; Owens, N.D.L.; Sesay, A.K.; Tlapakova, T.; et al. Efficient high-throughput sequencing of a laser microdissected chromosome arm. BMC Genom. 2013, 14, 357. [Google Scholar] [CrossRef] [Green Version]
- Kint, S.; Van Criekinge, W.; Vandekerckhove, L.; De Vos, W.H.; Bomsztyk, K.; Krause, D.S.; Denisenko, O. Single cell epigenetic visualization assay. Nucleic Acids Res. 2021, 49, e43. [Google Scholar] [CrossRef]
- Kirov, I.V.; Van Laere, K.; Khrustaleva, L.I. High resolution physical mapping of single gene fragments on pachytene chromosome 4 and 7 of Rosa. BMC Genet. 2015, 16, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khrustaleva, L.; Jiang, J.; Havey, M.J. High-resolution tyramide-FISH mapping of markers tightly linked to the male fertility restoration (Ms) locus of onion. Theor. Appl. Genet. 2016, 129, 535–545. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov/genome/browse#!/eukaryotes/plants (accessed on 16 April 2021).
- Jackson, S.; Rounsley, S.; Purugganan, M. Comparative Sequencing of Plant Genomes: Choices to Make. Plant Cell 2006, 18, 1100–1104. [Google Scholar] [CrossRef]
- YouTube. Available online: https://www.youtube.com/watch?v=UiXIPJDtN2E (accessed on 16 April 2021).
- Wallace, D.M. Precipitation of nucleic acids. Methods Enzymol. 1987, 152, 41–48. [Google Scholar] [CrossRef]
- Kalkman, E.R. Analysis of the C-banded karyotype of Allium cepa L. standard system of nomenclature and polymorphism. Genetics 1984, 65, 141–148. [Google Scholar] [CrossRef]
- De Vries, J.H. Onion chromosome nomenclature and homoeology relationships—Workshop report. Euphytica 1990, 49, 1–3. [Google Scholar] [CrossRef]








| Probe Name | Direction | Primer Sequence (5′ to 3′) | Size |
|---|---|---|---|
| Unigene572 | F | AGTGGTGCAGTTCTTCAGCA | 3.2 Kb |
| R | AACCGATTGGCAGGGAAGTT | ||
| Unigene5305 | F | TGTTTCACAAACGCTTCGTCC | 1.6 Kb |
| R | TGTCGCCACCACTCATTCAA |
| Method | Probe | Number of Analyzed Metaphases | Number of Metaphases with Signal | Signal Detection Frequency, % | ||||
|---|---|---|---|---|---|---|---|---|
| Total | On One Homolog | On Both Homologs | Total MetaPhases with Signal | On One Homolog | On Both Homologs | |||
| Squashed | Unigene572 (3.2 Kb) | 34 | 12 | 11 | 1 | 35.3 | 32.4 | 2.9 |
| Unigene5305 (1.6 Kb) | 5 | 5 | 0 | 14.7 | 14.7 | 0.0 | ||
| SteamDrop | Unigene572 (3.2 Kb) | 62 | 48 | 22 | 26 | 77.4 | 35.5 | 41.9 |
| Unigene5305 (1.6 Kb) | 32 | 24 | 8 | 51.6 | 38.7 | 12.9 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudryavtseva, N.; Ermolaev, A.; Karlov, G.; Kirov, I.; Shigyo, M.; Sato, S.; Khrustaleva, L. A Dual-Color Tyr-FISH Method for Visualizing Genes/Markers on Plant Chromosomes to Create Integrated Genetic and Cytogenetic Maps. Int. J. Mol. Sci. 2021, 22, 5860. https://doi.org/10.3390/ijms22115860
Kudryavtseva N, Ermolaev A, Karlov G, Kirov I, Shigyo M, Sato S, Khrustaleva L. A Dual-Color Tyr-FISH Method for Visualizing Genes/Markers on Plant Chromosomes to Create Integrated Genetic and Cytogenetic Maps. International Journal of Molecular Sciences. 2021; 22(11):5860. https://doi.org/10.3390/ijms22115860
Chicago/Turabian StyleKudryavtseva, Natalya, Aleksey Ermolaev, Gennady Karlov, Ilya Kirov, Masayoshi Shigyo, Shusei Sato, and Ludmila Khrustaleva. 2021. "A Dual-Color Tyr-FISH Method for Visualizing Genes/Markers on Plant Chromosomes to Create Integrated Genetic and Cytogenetic Maps" International Journal of Molecular Sciences 22, no. 11: 5860. https://doi.org/10.3390/ijms22115860
APA StyleKudryavtseva, N., Ermolaev, A., Karlov, G., Kirov, I., Shigyo, M., Sato, S., & Khrustaleva, L. (2021). A Dual-Color Tyr-FISH Method for Visualizing Genes/Markers on Plant Chromosomes to Create Integrated Genetic and Cytogenetic Maps. International Journal of Molecular Sciences, 22(11), 5860. https://doi.org/10.3390/ijms22115860








