(5′S) 5′,8-cyclo-2′-deoxyadenosine Cannot Stop BER. Clustered DNA Lesion Studies
Abstract
1. Introduction
2. Results and Discussion
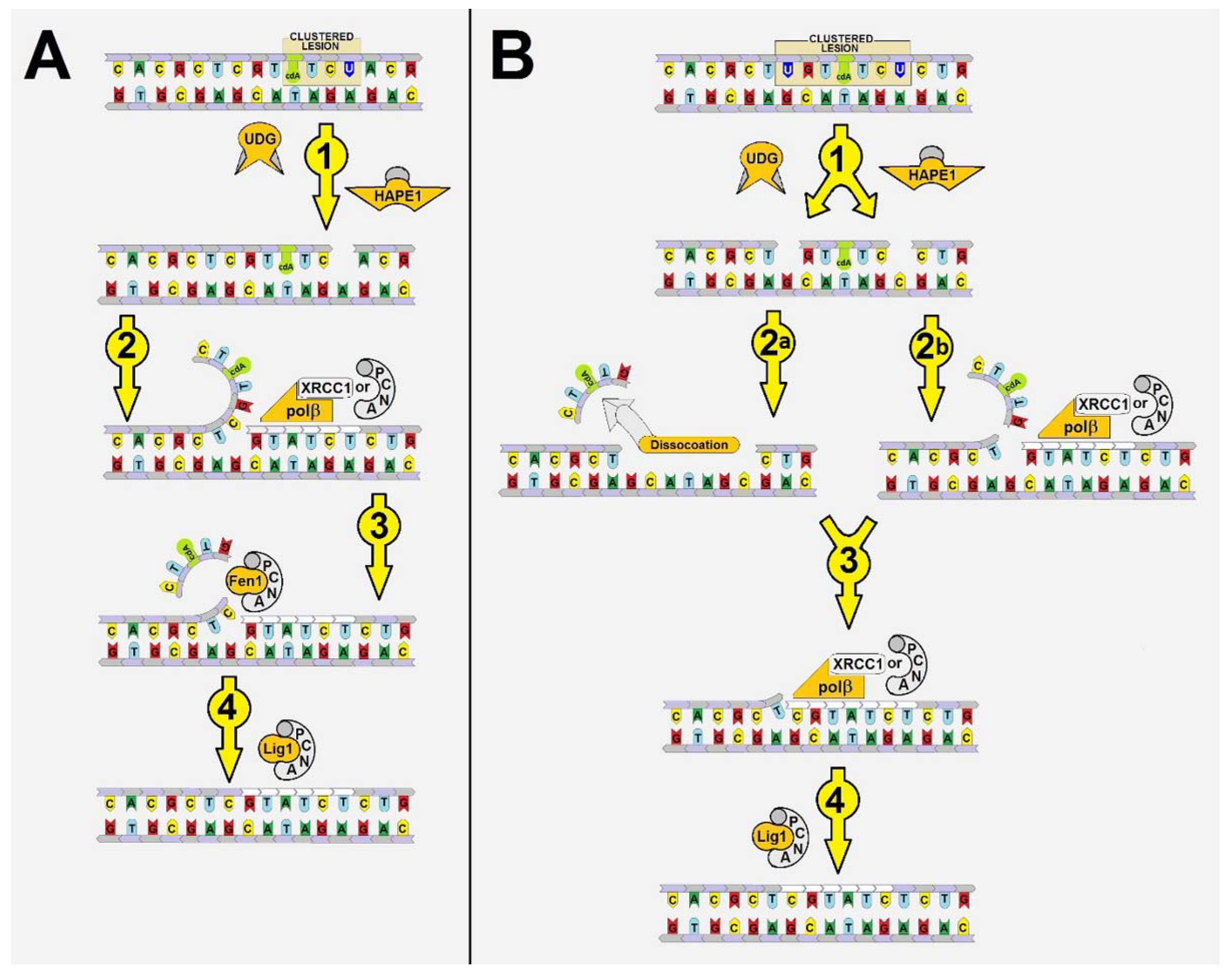
2.1. Hypothesis: How and When BER Proteins Remove (5′S)cdA from the Genome?
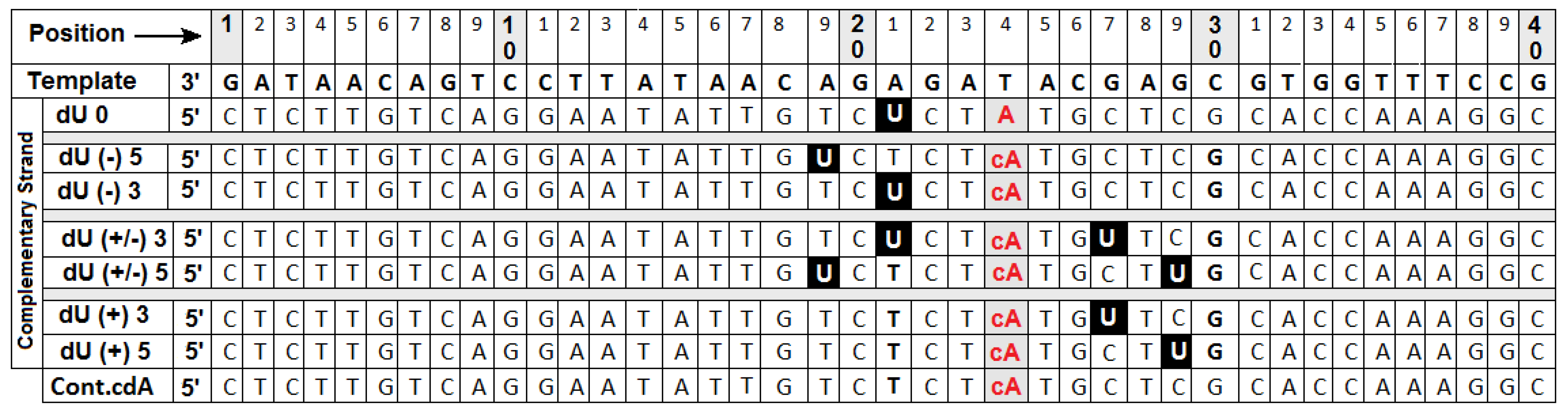
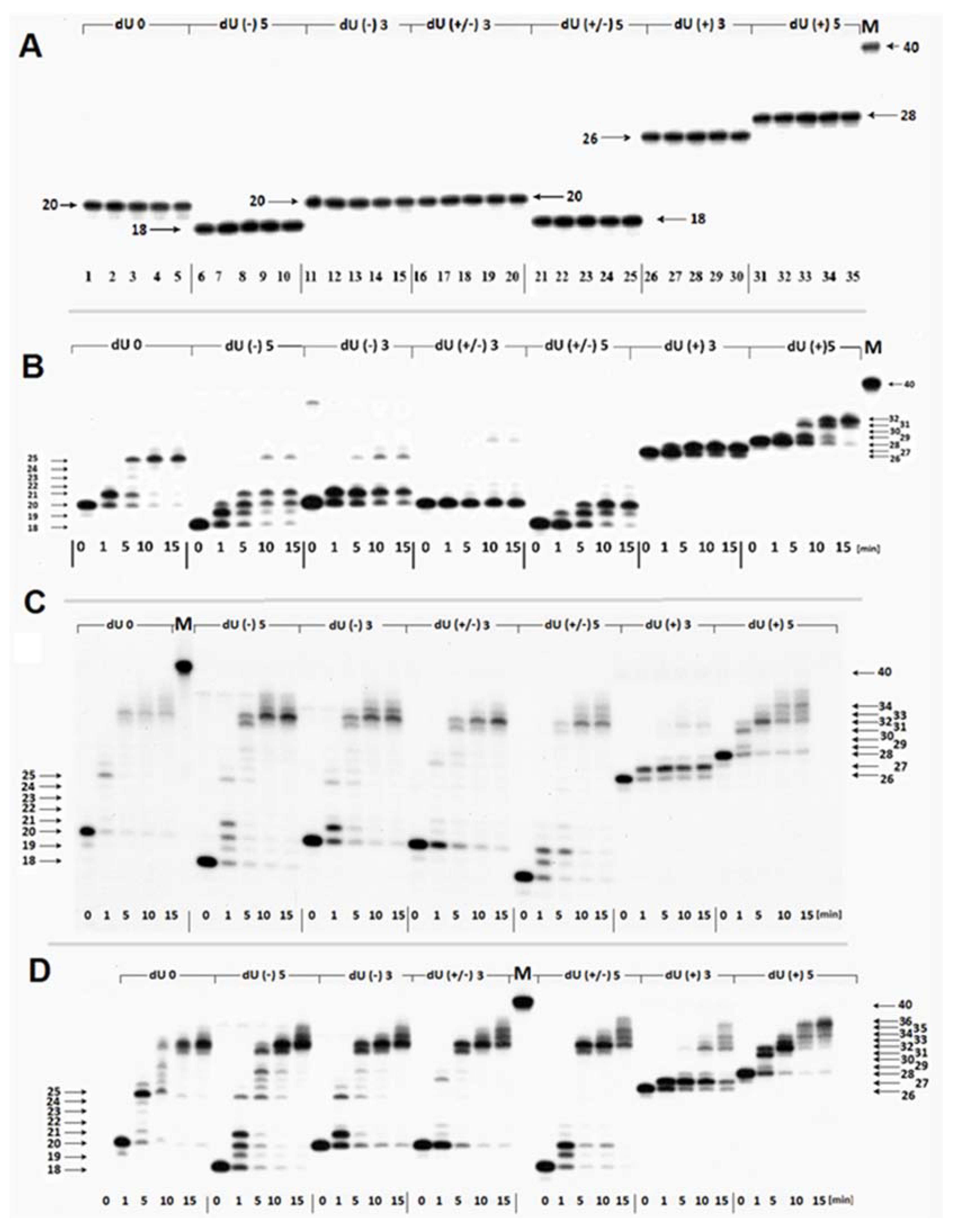
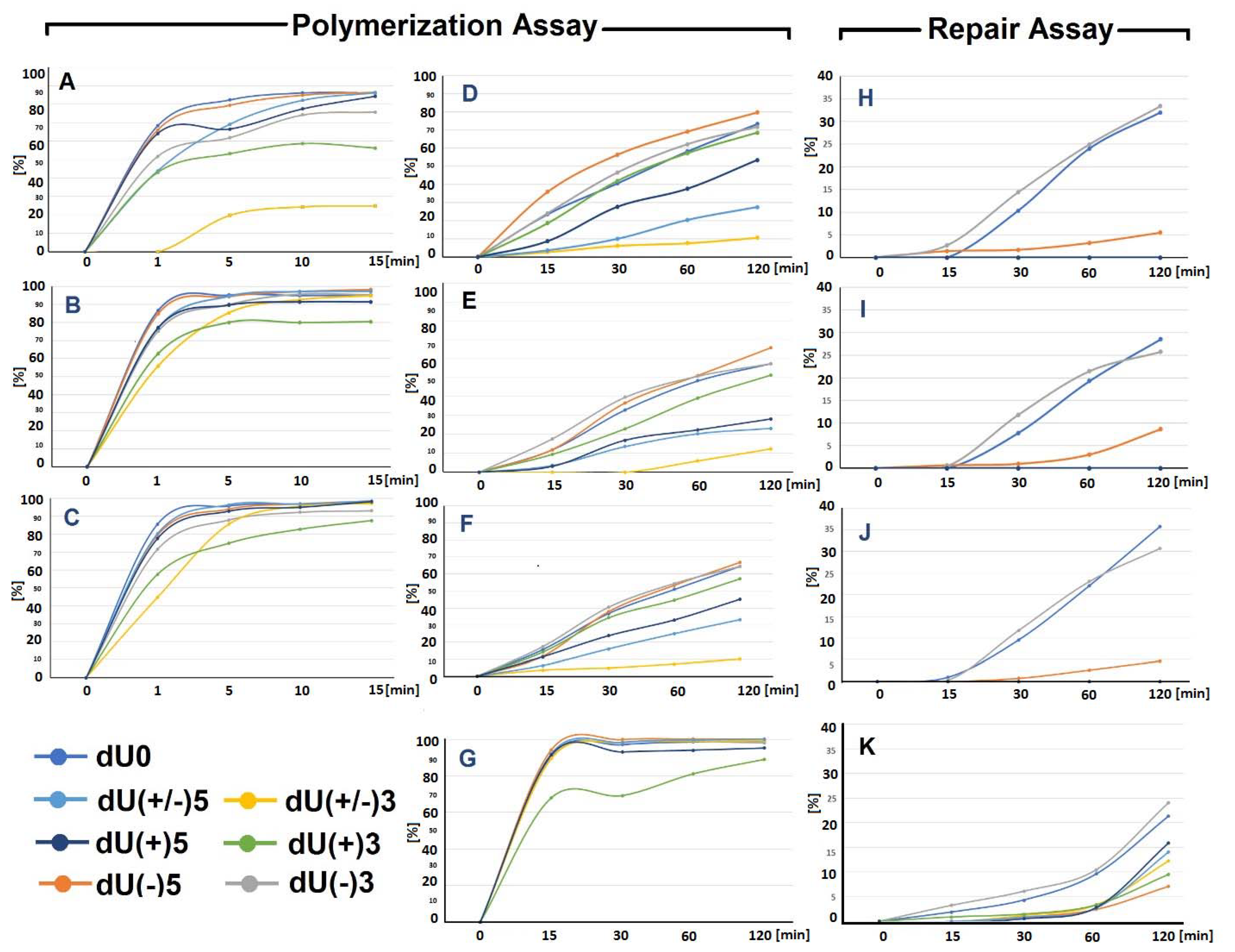
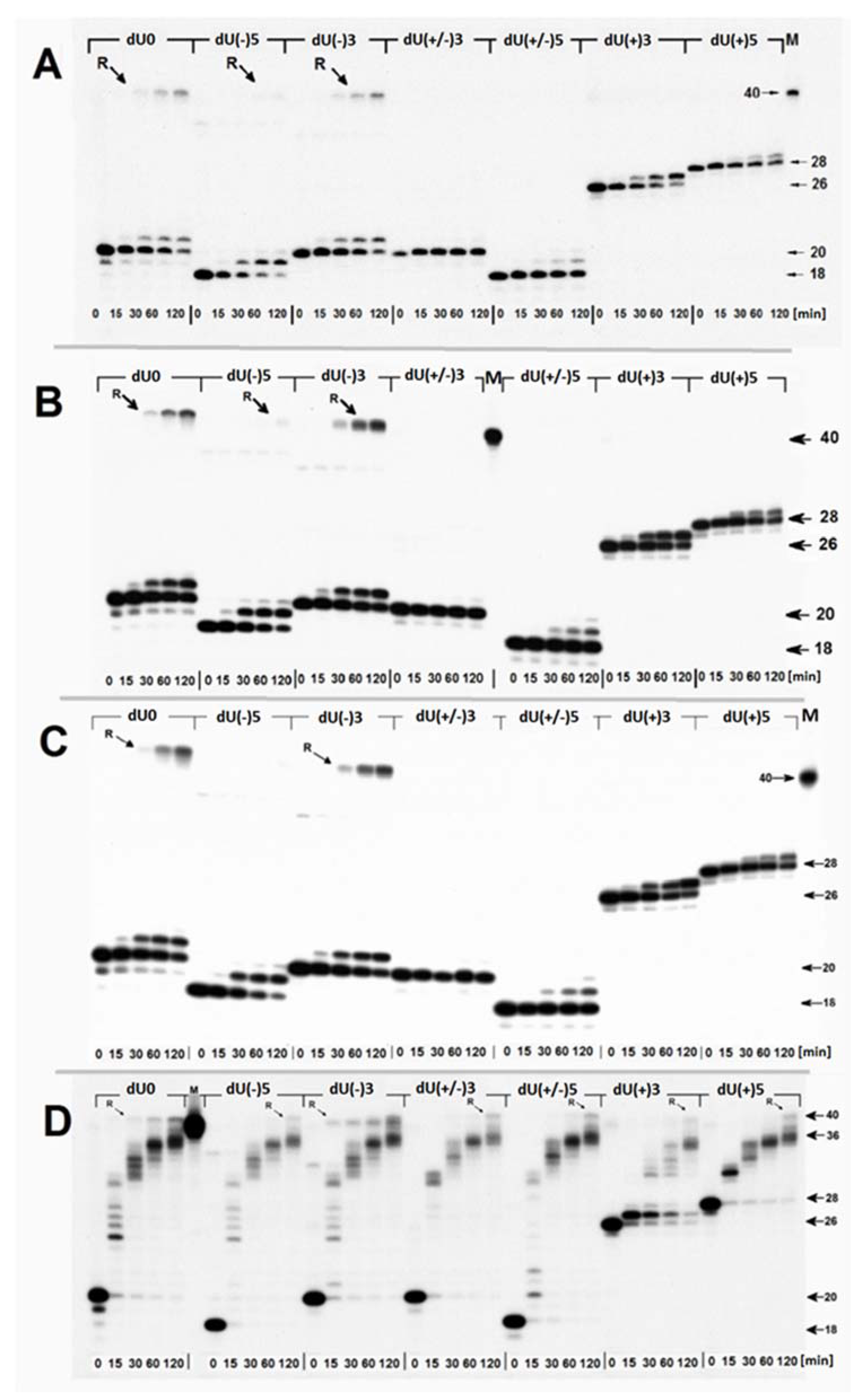
2.2. The Influence of the (5′S)cdA on Polβ Activity
- Firstly, the cooperation between XRCC1 and Polβ leads to an extended elongation of the primer strand. Depending on the relative (5′S)cdA and SSBs positions, up to 12 to 14 nucleotides may be inserted, which is commonly accepted for long patch BER (LP-BER) (in the case of control duplex (dU0) 12 nucleotide units were added). Under the same experimental conditions, Polβ solely added a maximum of seven nucleotides. These observations indicate that the descending strand containing (5′S)cdA was flipped out of the double helix and became a suitable substrate for further FEN1 action (Figure 1A—step 3).
- Secondly, a significant elongation of the primer strand was observed for dU(+/−)3, which was not prone to elongation by Polβ, due to its Okazaki-like fragment nature.
2.3. The Repair of Single-Stranded Clustered DNA Lesions by the xrs5 Nuclear Extract
3. Conclusions
4. Materials and Methods
- The influence of the presence of LMDS in ds-DNA on the strand-displacement activity of Polβ
- The effect of XRCC1 on Polβ activity on the strand-displacement activity of ds-DNA containing LMDS
- The effect of PNCA on Polβ activity on the strand-displacement activity of ds-DNA containing LMDS
- The influence of the presence of LMDS in ds-DNA on strand-displacement and repair activity of xrs5 nuclear extract
- The effect of PCNA (artificially supplemented) on xrs5 nuclear extract activity on the strand-displacement and repair activity of ds-DNA containing LMDS
- The effect of XRCC1 (artificially supplemented) on xrs5 nuclear extract activity on the strand-displacement and repair activity of ds-DNA containing LMDS
- The effect of Polβ (artificially supplemented) on xrs5 nuclear extract activity on the strand-displacement and repair activity of ds-DNA containing LMDS
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef]
- Chakarov, S.; Petkova, R.; Russev, G.; Zhelev, N. DNA Damage and Mutation. Types of DNA Damage. BioDiscovery 2014, 1. [Google Scholar] [CrossRef]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA Damage: Mechanisms, Mutation, and Disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef] [PubMed]
- Imoto, S.; Bransfield, L.A.; Croteau, D.L.; Van, B.H.; Greenberg, M.M. DNA Tandem Lesion Repair by Strand Displacement Synthesis and Nucleotide Excision Repair. Biochemistry 2008, 47, 4306–4316. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sudhir Ambekar, S.; Shrisel Hattur, S.; Budha Bule, P. DNA: Damage and Repair Mechanisms in Humans. Glob. J. Pharm. Pharm. Sci. 2017, 3, 1–8. [Google Scholar] [CrossRef]
- Vilenchik, M.M.; Knudson, A.G. Endogenous DNA Double-Strand Breaks: Production, Fidelity of Repair, and Induc-tion of Cancer. Proc. Natl. Acad. Sci. USA 2003, 100, 12871–12876. [Google Scholar] [CrossRef]
- Ward, J.F.; Evans, J.W.; Limoli, C.L.; Calabro-Jones, P.M. Radiation and Hydrogen Peroxide Induced Free Radical Damage to DNA. Br. J. Cancer 1987, 55, 105–112. [Google Scholar]
- Muniandy, P.A.; Liu, J.; Majumdar, A.; Liu, S.T.; Seidman, M.M. DNA Interstrand Crosslink Repair in Mammalian Cells: Step by Step. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 23–49. [Google Scholar] [CrossRef]
- McHugh, P.J.; Spanswick, V.J.; Hartley, J.A. Repair of DNA Interstrand Crosslinks: Molecular Mechanisms and Clinical Relevance. Lancet Oncol. 2001, 2, 483–490. [Google Scholar] [CrossRef]
- Olive, P.L.; Johnston, P.J. DNA Damage from Oxidants: Influence of Lesion Complexity and Chromatin Organization. Oncol. Res. 1997, 9, 287–294. [Google Scholar]
- Burkart, W.; Jung, T.; Frash, G. Damage Pattern as a Function of Radiation Quality and Other Factors. C. R. de l’Académie des Sci. 1999, 322, 89–101. [Google Scholar] [CrossRef]
- Semenenko, V.A.; Stewart, R.D. Fast Monte Carlo Simulation of DNA Damage Formed by Electrons and Light Ions. Phys. Med. Biol. 2006, 51, 1693–1706. [Google Scholar] [CrossRef] [PubMed]
- Semenenko, V.A.; Stewart, R.D. A Fast Monte Carlo Algorithm to Simulate the Spectrum of DNA Damages Formed by Ionizing Radiation. Radiat. Res. 2004, 161, 451–457. [Google Scholar] [CrossRef]
- Gulston, M.; de Lara, C.; Jenner, T.; Davis, E.; O’Neill, P. Processing of Clustered DNA Damage Generates Additional Double-Strand Breaks in Mammalian Cells Post-Irradiation. Nucleic Acids Res. 2004, 32, 1602–1609. [Google Scholar] [CrossRef]
- Sage, E.; Shikazono, N. Radiation-Induced Clustered DNA Lesions: Repair and Mutagenesis. Free Radic. Biol. Med. 2017, 107, 125–135. [Google Scholar] [CrossRef]
- Sage, E.; Harrison, L. Clustered DNA Lesion Repair in Eukaryotes: Relevance to Mutagenesis and Cell Survival. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2011, 711, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Qing, X.; Shi, D.; Lv, X.; Wang, B.; Chen, S.; Shao, Z. Prognostic Significance of 8-Hydroxy-2′-Deoxyguanosine in Solid Tumors: A Meta-Analysis. BMC Cancer 2019, 19, 997. [Google Scholar] [CrossRef]
- Cadet, J.; Davies, K.J.A.; Medeiros, M.H.G.; di Mascio, P.; Wagner, J.R. Formation and Repair of Oxidatively Generated Damage in Cellular DNA. Free Radic. Biol. Med. 2016, 48, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Pouget, J.-P.; Frelon, S.; Ravanat, J.-L.; Testard, I.; Odin, F. Formation of Modified DNA Bases in Cells Exposed Either to Gamma Radiation or to High-LET Particles. Cadet Source Radiat. Res. 2002, 157, 589–595. [Google Scholar] [CrossRef]
- Krokan, H.E.; Bjoras, M. Base Excision Repair. Cold Spring Harb Perspect Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- Carter, R.J.; Parsons, J.L. Base Excision Repair, a Pathway Regulated by Posttranslational Modifications. Mol. Cell. Biol. 2016, 36, 1426–1437. [Google Scholar] [CrossRef] [PubMed]
- Tsao, D.; Kalogerinis, P.; Tabrizi, I.; Dingfelder, M.; Stewart, R.D.; Georgakilas, A.G. Induction and Processing of Oxidative Clustered DNA Lesions in 56Fe-Ion-Irradiated Human Monocytes. Radiat. Res. 2007, 168. [Google Scholar] [CrossRef] [PubMed]
- Lomax, M.E.; Folkes, L.K.; O’Neill, P. Biological Consequences of Radiation-Induced DNA Damage: Relevance to Radiotherapy. Clin. Oncol. 2013, 25, 578–585. [Google Scholar] [CrossRef]
- Chaudhry, M.A.; Weinfeld, M. Reactivity of Human Apurinic/Apyrimidinic Endonuclease and Escherichia Coli Ex-onuclease III with Bistranded Abasic Sites in DNA. J. Biol. Chem. 1997, 272, 15650–15655. [Google Scholar] [CrossRef]
- Harrison, L.; Hatahet, Z.; Wallace, S.S. In Vitro Repair of Synthetic Ionizing Radiation-Induced Multiply Damaged DNA Sites. J. Mol. Biol. 1999, 290, 667–684. [Google Scholar] [CrossRef]
- Lomax, M.E.; Cunniffe, S.; O’Neill, P. Efficiency of Repair of an Abasic Site within DNA Clustered Damage Sites by Mammalian Cell Nuclear Extracts. Biochemistry 2004, 43, 11017–11026. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I.; Bender, C.; Romieu, A.; Cadet, J.; Wood, R.D.; Lindahl, T. Removal of Oxygen Free-Radical-Induced 5′,8-Purine Cyclodeoxynucleosides from DNA by the Nucleotide Excision-Repair Pathway in Human Cells. Proc. Natl. Acad. Sci. USA 2000, 97, 3832–3837. [Google Scholar] [CrossRef]
- Miaskiewicz, K.; Miller, J.H.; Fuciarellil, A.F. Theoretical Analysis of DNA Intrastrand Cross Linking by Formation of 8,5′-Cyclodeoxyadenosine. Nucleic Acids Res. 1995, 23, 515–521. [Google Scholar] [CrossRef]
- Kraemer, K.H.; Patronas, N.J.; Schiffmann, R.; Brooks, B.P.; Tamura, D.; DiGiovanna, J.J. Xeroderma Pigmentosum, Trichodystrophy and Cockayne Syndrome: A Complex Genotype-Phenotype Relationship. Neuroscience 2007, 145, 1388–1396. [Google Scholar] [CrossRef] [PubMed]
- Krokidis, M.G.; D’Errico, M.; Pascucci, B.; Parlanti, E.; Masi, A.; Ferreri, C.; Chatgilialoglu, C. Oxygen-Dependent Accumulation of Purine DNA Lesions in Cockayne Syndrome Cells. Cells 2020, 9, 1671. [Google Scholar] [CrossRef]
- Karwowski, B.T. The Influence of (5′R)- and (5′S)-5′,8-Cyclo-2′-Deoxyadenosine on UDG and HAPE1 Activity. Tandem Lesions Are the Base Excision Repair System’s Nightmare. Cells 2019, 8, 1303. [Google Scholar] [CrossRef]
- Mazouzi, A.; Vigouroux, A.; Aikeshev, B.; Brooks, P.J.; Saparbaev, M.K.; Morera, S.; Ishchenko, A.A. Insight into Mechanisms of 3′-5′ Exonuclease Activity and Removal of Bulky 8,5′-Cyclopurine Adducts by Apurinic/Apyrimidinic Endonucleases. Proc. Natl. Acad. Sci. USA 2013, 110. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Clauson, C.L.; Robbins, P.D.; Niedernhofer, L.J.; Wang, Y. The Oxidative DNA Lesions 8,5′-Cyclopurines Accumulate with Aging in a Tissue-Specific Manner. Aging Cell 2012, 11, 714–716. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Ferreri, C.; Krokidis, M.G.; Masi, A.; Terzidis, M.A. On the Relevance of Hydroxyl Radical to Purine DNA Damage. Free Radic. Res. 2021, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Marietta, C.; Gulam, H.; Brooks, P.J. A Single 8,5-Cyclo-2-Deoxyadenosine Lesion in a TATA Box Prevents Binding of the TATA Binding Protein and Strongly Reduces Transcription In Vivo. DNA Repair 2002, 1, 967–975. [Google Scholar] [CrossRef]
- Kuraoka, I.; Robins, P.; Masutani, C.; Hanaoka, F.; Gasparutto, D.; Cadet, J.; Wood, R.D.; Lindahl, T. Oxygen Free Radical Damage to DNA: Translesion Synthesis by Human DNA Polymerase η and Resistance to Exonuclease Action at Cyclopurine Deoxynucleoside Residues. J. Biol. Chem. 2001, 276, 49283–49288. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lai, Y.; Jiang, Z.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Liu, Y. A 5′, 8-Cyclo-2′-Deoxypurine Lesion In-duces Trinucleotide Repeat Deletion via a Unique Lesion Bypass by DNA Polymerase β. Nucleic Acids Res. 2014, 42, 13749–13763. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Xu, M.; Lai, Y.; Laverde, E.E.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Liu, Y. Bypass of a 5′,8-Cyclopurine-2′-Deoxynucleoside by DNA Polymerase β during DNA Replication and Base Excision Repair Leads to Nucleotide Misinsertions and DNA Strand Breaks. DNA Repair 2015, 33, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T.; Bellon, S.; O’Neill, P.; Lomax, M.E.; Cadet, J. Effects of (5′S)-5′,8-Cyclo-2′-Deoxyadenosine on the Base Excision Repair of Oxidatively Generated Clustered DNA Damage. A Biochemical and Theoretical Study. Org. Biomol. Chem. 2014, 12, 8671–8682. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M.J.; Kunkel, T.A. Eukaryotic DNA Replication Fork. Annu. Rev. Biochem. 2017, 86, 417–438. [Google Scholar] [CrossRef]
- Zheng, L.; Shen, B. Okazaki Fragment Maturation: Nucleases Take Centre Stage. J. Mol. Cell Biol. 2011, 3, 23–30. [Google Scholar] [CrossRef]
- Cui, J.; Gizzi, A.; Stivers, J.T. Deoxyuridine in DNA Has an Inhibitory and Promutagenic Effect on RNA Transcription by Diverse RNA Polymerases. Nucleic Acids Res. 2019, 47, 4153–4168. [Google Scholar] [CrossRef]
- Vasilenko, N.L.; Nevinsky, G.A. Pathways of Accumulation and Repair of Deoxyuridine Residues in DNA of Higher and Lower Organisms. Biochemistry (Moscow) 2003, 68, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Alsøe, L.; Sarno, A.; Carracedo, S.; Domanska, D.; Dingler, F.; Lirussi, L.; Sengupta, T.; Tekin, N.B.; Jobert, L.; Ale-xandrov, L.B.; et al. Uracil Accumulation and Mutagenesis Dominated by Cytosine Deamination in CpG Dinucleotides in Mice Lacking UNG and SMUG1. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Boguszewska, K.; Szewczuk, M.; Kaźmierczak-Barańska, J.; Karwowski, B.T. How (5′S) and (5′R) 5′,8-Cyclo-2′-Deoxypurines Affect Base Excision Repair of Clustered DNA Damage in Nuclear Extracts of Xrs5 Cells? A Biochemical Study. Cells 2021, 10, 725. [Google Scholar] [CrossRef]
- Canitrot, Y.; Fréchet, M.; Servant, L.; Cazaux, C.; Hoffmann, J. Overexpression of DNA Polymerase β: A Genomic Instability Enhancer Process. FASEB J. 1999, 13, 1107–1111. [Google Scholar] [CrossRef]
- Kibbe, W.A. OligoCalc: An Online Oligonucleotide Properties Calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Prasad, R.; Wilson, S.H.; Kraut, J.; Pelletier, H. Crystal Structures of Human DNA Polymerase Com-plexed with Gapped and Nicked DNA: Evidence for an Induced Fit Mechanism. Biochemistry 1997, 36, 11205–11215. [Google Scholar] [CrossRef]
- Cuneo, M.J.; London, R.E. Oxidation State of the XRCC1 N-Terminal Domain Regulates DNA Polymerase β Binding Affinity. Proc. Natl. Acad. Sci. USA 2010, 107, 6805–6810. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. XRCC1 and DNA Strand Break Repair. DNA Repair 2003, 2, 955–969. [Google Scholar] [CrossRef]
- London, R.E. The Structural Basis of XRCC1-Mediated DNA Repair. DNA Repair 2015, 30, 90–103. [Google Scholar] [CrossRef]
- Horton, J.K.; Watson, M.; Stefanick, D.F.; Shaughnessy, D.T.; Taylor, J.A.; Wilson, S.H. XRCC1 and DNA Polymerase β in Cellular Protection against Cytotoxic DNA Single-Strand Breaks. Cell Res. 2008, 18, 48–63. [Google Scholar] [CrossRef]
- Kubota, Y.; Nash, R.A.; Klungland, A.; Schar, P.; Barnes, D.E.; Lindahl, T. Reconstitution of DNA Base Excision-Repair with Purified Human Proteins: Interaction between DNA Polymerase and the XRCC1 Protein. EMBO J. 1996, 15, 6662–6670. [Google Scholar] [CrossRef]
- Kedar, P.S.; Kim, S.J.; Robertson, A.; Hou, E.; Prasad, R.; Horton, J.K.; Wilson, S.H. Direct Interaction between Mammalian DNA Polymerase β and Proliferating Cell Nuclear Antigen. J. Biol. Chem. 2002, 277, 31115–31123. [Google Scholar] [CrossRef]
- Klungland, A.; Lindahl, T. Second Pathway for Completion of Human DNA Base Excision-Repair: Reconstitution with Purified Proteins and Requirement for DNase IV (FEN1). EMBO J. 1997, 16, 3341–3348. [Google Scholar] [CrossRef]
- Boehm, E.M.; Gildenberg, M.S.; Washington, M.T. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes 2016, 39, 231–254. [Google Scholar] [CrossRef]
- Podust, V.N.; Hubscher, U. Lagging Strand DNA Synthesis by Calf Thymus DNA Polymerases Ae, F3, 6 and e in the Presence of Auxiliary Proteins. Nucleic Acids Res. 1993, 21, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Zaliznyak, T.; Lukin, M.; de Los Santos, C. Structure and Stability of Duplex DNA Containing (5′S)-5′,8-Cyclo-2′-Deoxyadenosine: An Oxidatively Generated Lesion Repaired by NER. Chem. Res. Toxicol. 2012, 25, 2103–2111. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T. The Role of (5′R) and (5′S) 5′,8-Cyclo-2′-Deoxyadenosine in Ds-DNA Structure. A Comparative QM/MM Theoretical Study. Comput. Theor. Chem. 2013, 1010, 38–44. [Google Scholar] [CrossRef]
- Huang, H.; Das, R.S.; Basu, A.K.; Stone, M.P. Structures of (5′S)-8,5′-Cyclo-2′-Deoxyguanosine Mismatched with DA or DT. Chem. Res. Toxicol. 2012, 25, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Calsou, P.; Frit, P.; Salles, B. Double Strand Breaks in DNA Inhibit Nucleotide Excision Repair In Vitro. J. Biol. Chem. 1996, 271, 27601–27607. [Google Scholar] [CrossRef] [PubMed]
- Errami, A.; Smider, V.; Rathmell, W.K.; He, D.M.; Hendrickson, E.A.; Zdzienicka, M.Z.; Chu, G. Ku86 Defines the Genetic Defect and Restores X-Ray Resistance and V(D)J Recombination to Complementation Group 5 Hamster Cell Mutants. Mol. Cell. Biol. 1996, 16, 1519–1526. [Google Scholar] [CrossRef]
- Hashimoto, M.; Donald, C.D.; Yannone, S.M.; Chen, D.J.; Roy, R.; Kow, Y.W. A Possible Role of Ku in Mediating Sequential Repair of Closely Opposed Lesions. J. Biol. Chem. 2001, 276, 12827–12831. [Google Scholar] [CrossRef] [PubMed]
- Sweasy, J.B.; Chen, M.; Loeb, L.A. DNA Polymerase b Can Substitute for DNA Polymerase Iin the Initiation of Plasmid DNA Replication. J. Bacteriol. 1995, 177, 2923–2925. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Romieu, A.; Gasparutto, D.; Cadet, J. Synthesis and Characterization of Oligonucleotides Containing 5′,8-Cyclopurine 2′-Deoxyribonucleosides: (5′R)-5′,8-Cyclo-2′-Deoxyadenosine, (5′S)-5′,8-Cyclo-2′-Deoxyguanosine, and (5′R)-5′,8-Cyclo-2′-Deoxyguanosine. Chem. Res. Toxicol. 1999, 12, 412–421. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karwowski, B.T. (5′S) 5′,8-cyclo-2′-deoxyadenosine Cannot Stop BER. Clustered DNA Lesion Studies. Int. J. Mol. Sci. 2021, 22, 5934. https://doi.org/10.3390/ijms22115934
Karwowski BT. (5′S) 5′,8-cyclo-2′-deoxyadenosine Cannot Stop BER. Clustered DNA Lesion Studies. International Journal of Molecular Sciences. 2021; 22(11):5934. https://doi.org/10.3390/ijms22115934
Chicago/Turabian StyleKarwowski, Boleslaw T. 2021. "(5′S) 5′,8-cyclo-2′-deoxyadenosine Cannot Stop BER. Clustered DNA Lesion Studies" International Journal of Molecular Sciences 22, no. 11: 5934. https://doi.org/10.3390/ijms22115934
APA StyleKarwowski, B. T. (2021). (5′S) 5′,8-cyclo-2′-deoxyadenosine Cannot Stop BER. Clustered DNA Lesion Studies. International Journal of Molecular Sciences, 22(11), 5934. https://doi.org/10.3390/ijms22115934






